Abstract
Background
Increased levels of hepcidin, the master regulator of iron homeostasis, contribute to the diversion of iron underlying the anemia of chronic disease. Yet hepcidin levels are low in anemia of chronic disease with concomitant true iron deficiency. Here we clarify the different underlying pathways regulating hepcidin expression under these conditions in vivo.
Design and Methods
We used rat models of iron deficiency anemia, anemia of chronic disease and anemia of chronic disease with concomitant true iron deficiency and investigated upstream signaling pathways controlling hepcidin transcription in the liver. Protein and mRNA levels of iron metabolism genes and genes involved in SMAD1/5/8 and STAT3 signaling were determined by RT-PCR, Western blotting and immunohistochemistry.
Results
SMAD1/5/8 phosphorylation and in parallel hepcidin mRNA expression were increased in anemia of chronic disease but significantly down-regulated in anemia of chronic disease with concomitant iron deficiency, either on the basis of phlebotomy or dietary iron restriction. Iron deficiency resulted in reduced bone morphogenetic protein-6 expression and impaired SMAD1/5/8 phosphorylation and trafficking, two key events for hepcidin transcription. Reduced SMAD1/5/8 activity in association with phlebotomy was paralleled by increased expression of the inhibitory factor, SMAD7, dietary iron restriction appeared to impair hepcidin transactivating SMAD pathways via reduction of membrane bound hemojuvelin expression.
Conclusions
This study evaluated hepcidin signaling pathways in anemia of chronic disease with/without concomitant iron deficiency in vivo. While iron deficiency in general decreased bone morphogenetic protein-6 expression, phlebotomy or dietary iron restriction inhibited inflammation driven SMAD1/5/8 mediated hepcidin formation by different pathways, indicating alternate hierarchic signaling networks as a function of the mode and kinetics of iron deficiency. Nonetheless, iron deficiency inducible regulatory pathways can reverse inflammation mediated stimulation of hepcidin expression.
Keywords: anemia of chronic disease, iron deficiency, hepcidin, inflammation, bone morphogenetic proteins, SMAD
Introduction
Anemia of chronic disease (ACD, or anemia of inflammation) is the most prevalent anemia in hospitalized patients 1–3 and develops in subjects with diseases involving acute or chronic immune activation, such as infections, malignancies or autoimmune disorders.1 At least three major immunity driven mechanisms contribute to ACD among which the retention of iron within the mononuclear phagocytic system with subsequent development of hypoferremia, along with a limited availability of iron for erythroid progenitor cells, are of pivotal importance.1,4 This diversion of iron traffic is induced via regulatory effects of cytokines on iron uptake and release by macrophages5,6 and by the activity of the iron and cytokine inducible liver derived peptide hepcidin.7 Hepcidin affects cellular iron homeostasis after binding to the only known iron export protein ferroportin, resulting in its degradation and blockage of iron transfer from monocytes/macrophages to the circulation.8–10 This effect is further aggravated in inflammatory macrophages/monocytes by the autocrine formation of hepcidin.11,12
The role of hepcidin in the pathogenesis of human ACD is supported by the finding that hepcidin levels: i) are significantly increased in patients with ACD7,13 and in subjects injected with LPS;14 ii) are correlated to iron retention in monocytes/macrophages in vivo;15 and iii) by the observation that administration of anti-hepcidin antibodies ameliorates the therapy of anemia in mice suffering from brucellosis.16 However, a significant number of patients with ACD suffer from concomitant true iron deficiency anemia (IDA) as a consequence of chronic blood loss (ACD/IDA).1,17–18
The differentiation between ACD and ACD/IDA is of clinical importance because ACD and ACD/IDA patients may need contrasting therapies in terms of iron substitution.1 When investigating patients suffering from ACD and ACD/IDA, we found that ACD/IDA patients had significantly lower serum hepcidin levels than subjects with ACD, although the degree of inflammation was comparable.13 This observation was confirmed in a rat model of ACD and ACD/IDA13 and in a mouse model of critical illness associated anemia,19 showing significantly decreased Hamp mRNA expression in livers of animals with ACD/IDA compared to animals with ACD alone. This indicates contrasting signaling networks for hepcidin expression in ACD and ACD/IDA.
Several pathways are involved in the regulation of hepcidin expression in the liver. The inflammatory cytokine IL-6 induces hepcidin expression via induction of STAT3 phosphorylation.20–22 Iron mediated induction of hepcidin expression is affected by the membrane bound peptide hemojuvelin (mHJV)23 which has been characterized as a bone morphogenetic protein (BMP) co-receptor.24 Recently, the membrane bound serine protease TMPRSS6 (Matriptase 2) has been shown to degrade membrane anchored HJV,25–27 thereby reducing hepcidin expression.
In addition, BMPs were characterized as potent inducers of hepcidin formation,28,29 and genetic ablation of BMP6 resulted in tissue iron overload as a consequence of reduced hepcidin expression.30,31
The interactions of BMPs with BMP receptors result in phosphorylation of a subset of SMAD proteins (SMAD1, SMAD5 and SMAD8) and subsequent formation of a heteromeric complex with SMAD4 which translocates to the nucleus and induces the transcription of target genes32, 33 The activation of BMP signaling through the SMAD1/5/8 pathway is negatively affected by the inhibitory SMADs, SMAD6 and SMAD7, and proteins such as TOB1 and TOB2.34
In addition to iron and inflammation, the expression of hepcidin is controlled by hypoxia, endoplasmatic reticulum stress, erythropoietic activity and erythropoiesis driven signals.25,35–40 Interestingly, STAT3 mediated transcriptional hepcidin activation is decreased in LPS treated mice exposed to erythropoietin41 pointing to a hierarchy of different signals for hepcidin induction. However, so far no data are available on the regulatory networks controlling hepcidin expression in inflammatory anemia and how they are affected by concomitant iron deficiency in vivo.
Design and Methods
Animals
Female Lewis rats (Charles River Laboratories, Sulzfeld, Germany) were kept on a standard rodent diet (180 mg Fe/kg, C1000 from Altromin, Lage, Germany) until they reached an age of 8–10 weeks. The animal experiments were approved by the Medical University of Innsbruck and the Austrian Federal Ministry of Science and Research (BMWF-66.011/0146-11/10b/2008 and BMWF-66.011/0074-11/10b/2008).
In a first set of experiments, rats were inoculated on day 0 with an i.p. injection of PG-APS (group A streptococcal peptidoglycan-polysaccharide) (Lee Laboratories, Grayson, GA, USA) suspended in 0.85% saline with a total dose of 15 μg rhamnose/g body weight which has been shown to induce chronic arthritis and anemia in rats.13 Carrier-immunized control rats received i.p. injections of sterile 0.85% saline.
One group of PG-APS treated and one group of carrier-immunized rats was phlebotomized, starting one week before sacrifice; 1.8 mL of blood were taken daily for five consecutive days. Each group consisted of 6 rats. Three weeks after PG-APS administration, rats were euthanized and tissue was harvested for RNA and protein extraction. In Lewis rats, total body iron is 4–5 mg/100 g body weight. We used rats with 200–250 mg body weight. During the experiment 9 mL of whole blood were taken. This corresponds to a loss of 4.5 mg iron. Thus phlebotomized rats lose approximately 30–50% of total body iron during the phlebotomy period.
In a second set of experiments, one group of PG-APS treated rats was put on an iron deficient diet (5.2 mg Fe/kg; C1038 from Altromin) one week before PG-APS administration and kept on the iron deficiency diet during the entire observation period. Each group consisted of 6 rats. Again, three weeks after PG-APS administration, rats were euthanized and tissue was harvested for RNA-and protein extraction.
RNA preparation from tissue, reverse transcription and quantitative real-time PCR
Total RNA preparation from nitrogen-frozen rat tissue, reverse transcription of 4 μg RNA and TaqMan or SYBR Green PCR were performed as previously described.13 TaqMan and SYBR Green PCR primer and probes:
Hamp: 5′- TGAGCAGCGGTGCCTATCT -3′, 5′- CCATGC-CAAGGCTGCAG -3′,
FAM-CGGCAACAGACGAGACAGACTACGGC -BHQ1);
Gusb(beta-glucoronidase): 5′-ATTACTCGAACAATCGGTTG-CA-3′, 5′- GACCGGCATGTCCAAGGTT-3′,
FAM-CGTAGCGGCTGCCGGTACCACT-BHQ1;
BMP2: 5′-ATCACGAAGAAGCCATCGAGGAAC-3′, 5′-GGACAGAACTTAAATTGAAGAAGAAGCGTC-3′;
BMP4: 5′-GTTTGTTCAAGATTGGCTCCCAA-3′, 5′-GCATTCGGTTACCAGGAATCATG-3′;
BMP6: 5′-TGGTCATGAGCTTTGTGAACCTGG-3′, 5′-CTGCCTCACCCTCGGGAATCT-3′;
BMP7: 5′-CAGTGTGGCAGAAAACAGCAGCA-3′, 5′-TGC-GATGATCCAGTCCTGCCAG-3′;
BMP9: 5′-CGGAGCCACCCCAGTACATGAT-3′, 5′-GCT-GTCGATATAGCATCTTCCACGC-3′
HJV: 5′-CCACCATCCGGAAGATCACTATC -3′, 5′-TTCAAAGGCTGCAGGAAGATTG-3′
SMAD7: 5′-AAATCCATCGGGTATCTGGAG -3′, 5′-TGCTGTGAATCTTACGGGAAG-3′
Western blotting
Cytosolic protein extracts were prepared from nitrogen frozen tissue as previously described.12 Nuclear extracts were prepared from freshly isolated tissue using a commercially available Kit (NE-PER, Thermo Scientific, Rockford, USA). Membrane protein extracts were prepared from nitrogen frozen tissue as previously described.42 Western blotting of cellular extracts was performed as described.12
STAT3-antibody (final concentration 0.1 μg/mL), phospho-STAT3 (Ser727)-antibody (0.1 μg/mL), phospho-SMAD1/SMAD5/SMAD8-antibody (0.1 μg/mL), SMAD5-antibody (0.1 μg/mL), SMAD4-antibody (0.1 μg/mL, all five from Cell Signaling Technology, Inc., Danvers, USA), SMAD6-antibody (0.5 μg/mL), SMAD7-antibody (0.5 μg/mL, both from Acris Antibodies, Herford, Germany), hemojuvelin-antibody (1:100),43 TOB1-antibody (0.5 μg/mL) and TOB2-antibody (0.5 μg/mL, both from Santa Cruz Biotechnology, Heidelberg, Germany), rat/HRP-antibody (0.1 μg/mL, Dako, Glostrup, Denmark) or β-actin-antibody (2 μg/mL, Sigma, Munich, Germany) were used as described.13 Protein levels were quantified by densitometry using Quantity One Basic software (Bio-Rad, CA, USA). Online Supplementary Figures S1 and S2 show the detailed blots used for densitometric quantification.
Information from the manufacturer confirms that all antibodies are specific for the respective rat antigen. As an additional control, we also performed blocking experiments with the antibodies used in our study to ensure their specificity.
Immunohistochemistry
Analyses were performed as described.44 In brief, formalin fixed sections of paraffin-embedded tissues were mounted on glass slides. Antigen retrieval was performed by incubating tissue sections with trypsin (1 mg/mL) for 8 min at 37°C. Endogenous per-oxidase activity was quenched by incubating specimens with Dako REAL Peroxidase Blocking Solution (Dako, Glostrup, Denmark). To inactivate unspecific avidin/biotin binding the slides were blocked with Biotin/Avidin Blocking System (Dako, Glostrup, Denmark). Tissue sections were then blocked with normal horse blocking serum (Vector Laboratories, Burlingame, CA, USA) for 1 h at room temperature and incubated overnight at 4°C with the primary anti-BMP6 (N-19) antibody (1:100; Santa Cruz Biotechnology, Santa Cruz, CA, USA) diluted in PBS/1% BSA/1% FCS. A biotin anti-goat antibody (1:300; Vector Laboratories, Burlingame, CA, USA) diluted in PBS/1% BSA was used as a secondary antibody and incubated for 30 min at room temperature. Immunohistochemical staining was performed using a Streptavidin/HRP antibody (1:300; Vector Laboratories, Burlingame, CA, USA) diluted in PBS/1% BSA for 30 min at room temperature, Vectastain ABC Kit (Vector Laboratories, Burlingame, CA, USA) and peroxidase substrate Kit AEC (Vector Laboratories, Burlingame, CA, USA) according to the manufacturer’s instructions. Sections were counterstained with hematoxylin (Vector Laboratories, Burlingame, CA, USA). A Zeiss Axioscope 40 microscope with a 40x lens and an AxioCam MRc5 was used for evaluation. Representative fields were photodocumented using a pixellink® system.
Immunofluorescence staining
Fresh tissue was embedded in OCT compound (TISSUE-TEK®, Sakura Finetek), snap-frozen in liquid nitrogen and then sliced into 5 μm sections. Slides were incubated using a hemojuvelin antibody (1:100) which had been previously shown to cross react with rat hemojuvelin43 for 30 min at 37°C. For fluorescence microscopy, slides were incubated with the appropriate goat anti-rabbit antibody (Alexa 488; 1:200; Invitrogen) diluted in PBS-/1% BSA for 30 min at 37°C.43 A Zeiss Axioscope 40 microscope with a 40x lens and an AxioCam MRc5 was used for evaluation. Representative fields were photodocumented using a pixellink® system.
Data analysis
All parameters were tested for normality by the Kolmogorov-Smirnov test. Calculations for statistical differences between the various groups were carried out by ANOVA with Bonferroni’s correction for multiple tests. Spearman’s-rho test was used for correlation analyses. A value of P<0.05 was considered statistically significant. All statistical analyses were carried out using the Statistical Package for the Social Sciences (SPSS) software package version 15.0 (SPSS Inc., Chicago, IL, USA).
Results
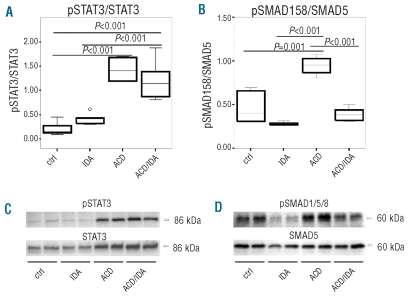
Based on our previous observation of increased hepcidin expression in humans and rats suffering from ACD, and of reduced hepcidin levels in ACD individuals with concomitant true iron deficiency (ACD/IDA),13 we aimed to clarify the pathways underlying contrasting hepcidin expression under these conditions. We used rats injected with PG-APS which resulted in the development of a chronic arthritis associated with a chronic persisting inflammatory anemia, bearing the typical features of ACD.13 Additional iron deficiency was induced by phlebotomy (ACD/IDA) (see Design and Methods section). To study differences in hepcidin upstream signaling pathways between ACD and ACD/IDA animals, we first investigated the inflammation inducible STAT3 pathway in rat liver (Figure 1A and C). We found that STAT3 phosphorylation (pSTAT3) was significantly increased in the livers of ACD (P<0.001) and ACD/IDA (P<0.001) rats when compared to control and IDA rats. In contrast, we observed no significant difference in STAT3 activation between ACD and ACD/IDA animals (Figure 1A and C).
Figure 1.
Changes in liver STAT3 and SMAD1/5/8 phosphorylation in different anemia groups. ACD was induced by i.p. injection of PG-APS (n=6) as detailed in the Design and Methods section; controls received solvent injections (n=6). One group of PG-APS treated (n=6) and one of solvent treated (n=6) rats was phlebotomized, starting one week before sacrifice, to create a combination of ACD and iron deficiency anemia (ACD/IDA) or IDA alone, respectively. Liver nuclear cell extracts were immunoblotted with antibodies against pSTAT3 (A and C) and pSMAD1/5/8 (B and D). Blots were stripped and reprobed with antibodies directed against STAT3 and SMAD5. Protein levels were quantified by densitometry and results are expressed as ratios of phospho-protein/total protein (A and B). Data are depicted as lower quartile, median and upper quartile (boxes) and minimum/maximum ranges (whiskers). Statistic outliers are displayed as circles. Calculations for statistical differences between the various groups were carried out by ANOVA technique and Bonferroni’s correction for multiple tests.
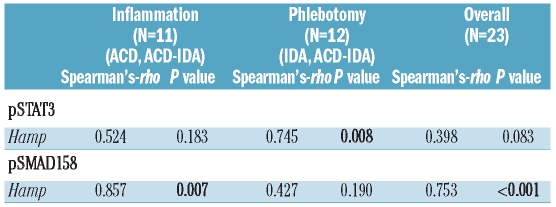
We then analyzed the signaling activity via the SMAD1/5/8 pathway (Figure 1B and D). We found that SMAD1/5/8 phosphorylation (pSMAD1/5/8) was lower in IDA than in control animals. In contrast, we observed significantly increased SMAD1/5/8 phosphorylation in rats with ACD compared to control (P=0.001) and ACD/IDA rats (P<0.001) (Figure 1B and D). The differences in pSMAD1/5/8 mirrored the relative changes in Hamp mRNA expression between the various anemia groups.13 Accordingly, we found a significant correlation of SMAD1/5/8 activity with liver Hamp mRNA expression (r=0.753, P<0.001), as determined by means of Spearman’s rank correlation coefficient (Table 1). This correlation was not found for STAT3 phosphorylation (Table 1). However, when analyzing the phlebotomized subgroup, including IDA and ACD/IDA rats, and the PG-APS treated inflammation subgroups, including ACD and ACD/IDA, separately, we found that in the phlebotomized group changes in STAT3 phosphorylation showed a very good correlation with alterations of Hamp mRNA expression (r=0.745, P=0.008) while no association between SMAD1/5/8 phosphorylation and Hamp mRNA expression was found (Table 1). In contrast, when analyzing rats with ACD and ACD/IDA, individual Hamp mRNA expression levels did not correlate to STAT3 phosphorylation (r=0.524, P=0.183) but were strongly associated with SMAD1/5/8 phosphorylation (r=0.857, P=0.007) (Table 1). These results suggest that under inflammatory conditions Hamp mRNA expression is influenced rather by alterations in SMAD1/5/8 phosphorylation than by STAT3 phosphorylation.
Table 1.
Correlation of pSTAT3 and pSMAD1/5/8 levels with Hamp mRNA liver expression during inflammation, phlebotomy and under both conditions. Correlation analyses were carried out using Spearman’s-rho test. Correlation coefficients and two-sided P values are reported.
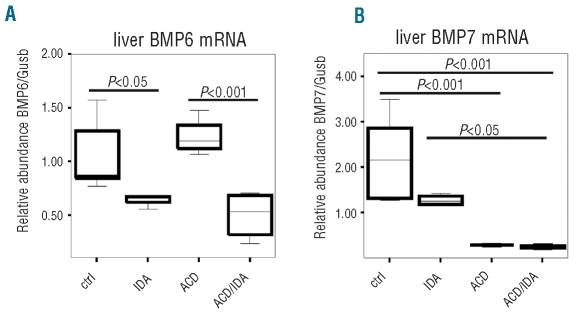
To further investigate the mechanisms causing differences in hepatic SMAD1/5/8 phosphorylation between ACD and ACD/IDA rats, we analyzed known pathways affecting SMAD1/5/8 phosphorylation and its nuclear trafficking. While there was no significant difference in BMP2, 4 and 9 mRNA levels between the various groups (data not shown), BMP6 mRNA concentrations were significantly lower in IDA (P<0.05) and ACD/IDA (P<0.001) rats compared to control and ACD animals (Figure 2A). BMP7 mRNA levels were found to be significantly decreased in association with inflammation in ACD (P<0.001) and ACD/IDA (P<0.001) rats when compared with control rats, but no difference was observed between ACD and ACD/IDA rats (Figure 2B).
Figure 2.
Liver BMP mRNA expression in different anemia groups. ACD was induced by i.p. injection of PG-APS (n=6) as detailed in the Design and Methods section; controls received solvent injections (n=6). One group of PG-APS treated (n=6) and one of solvent treated (n=6) rats were phlebotomized, starting one week before sacrifice, to create a combination of ACD and iron deficiency anemia (ACD/IDA) or IDA alone, respectively. BMP 6 and 7 mRNA (A and B) expression was determined by quantitative RT-PCR and normalized to the mRNA expression level of the housekeeping gene β-glucoronidase (Gusb) (A and B). Data are depicted as lower quartile, median and upper quartile (boxes) and minimum/maximum ranges (whiskers). Calculations for statistical differences between the various groups were carried out by ANOVA technique and Bonferroni’s correction for multiple tests.
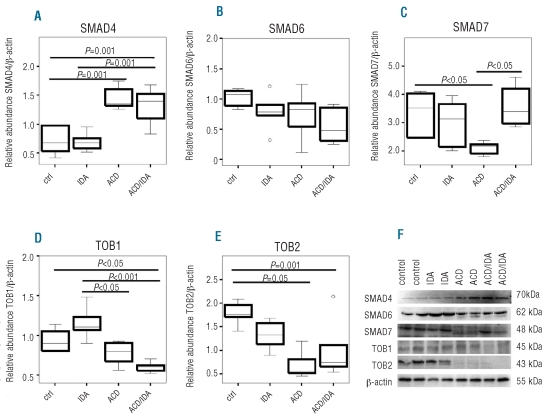
Because the co-SMAD SMAD4 forms a complex with pSMAD1/5/8 before translocating to the nucleus, we determined SMAD4 levels in the different anemia groups. SMAD4 protein expression was induced in ACD (P<0.001) and ACD/IDA (P=0.001) as compared to controls, while no differences were observed between ACD and ACD/IDA (Figure 3A and F).
Figure 3.
Hepatic levels of SMAD proteins and inhibitors of SMAD expression. ACD was induced by i.p. injection of PG-APS (n=6) as detailed in the Design and Methods section, controls received solvent injections (n=6). One group of PG-APS treated (n=6) and one of solvent treated (n=6) rats were phlebotomized, starting one week before sacrifice, to create a combination of ACD and iron deficiency anemia (ACD/IDA) or IDA alone, respectively. Liver tissue samples were subjected to immunoblot analysis using antibodies against (A) SMAD4, (B) SMAD6, (C) SMAD7, (D) TOB1 and (E) TOB2. Protein levels were quantified by densitometry. Expression levels were normalized to the housekeeping gene β-actin. Data are depicted as lower quartile, median and upper quartile (boxes) and minimum/maximum ranges (whiskers). Statistic outliers are displayed as circles. Calculations for statistical differences between the various groups were carried out by ANOVA technique and Bonferroni’s correction for multiple tests. Representative blots for each protein are shown in panel (F). For blots used for quantification by densitometry please see Online Supplementary Figure S1.
Next, we analyzed the expression of SMAD6 and SMAD7; two inhibitory proteins in the BMP/SMAD signal transduction pathway. While no statistically significant differences in SMAD6 expression were found between the different groups (Figure 3B and F), SMAD7 protein expression was significantly lower in ACD than in control rats (P<0.05). In contrast, ACD/IDA animals presented with significantly higher SMAD7 levels than ACD rats (P<0.05) (Figure 3C and F).
The expression of TOB1 and TOB2, two additional negative regulators of the BMP signaling pathway, was lower in ACD and ACD/IDA when compared with control or IDA rats, respectively. However, no significant differences were found when comparing ACD with ACD/IDA animals (Figure 3D-F) pointing to a modulation of TOB1 and TOB2 expression by inflammation.
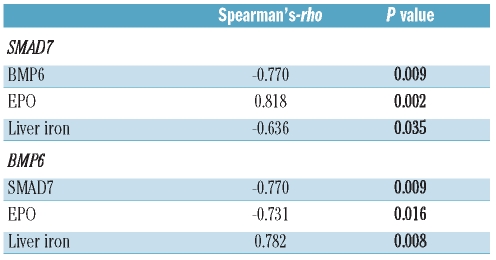
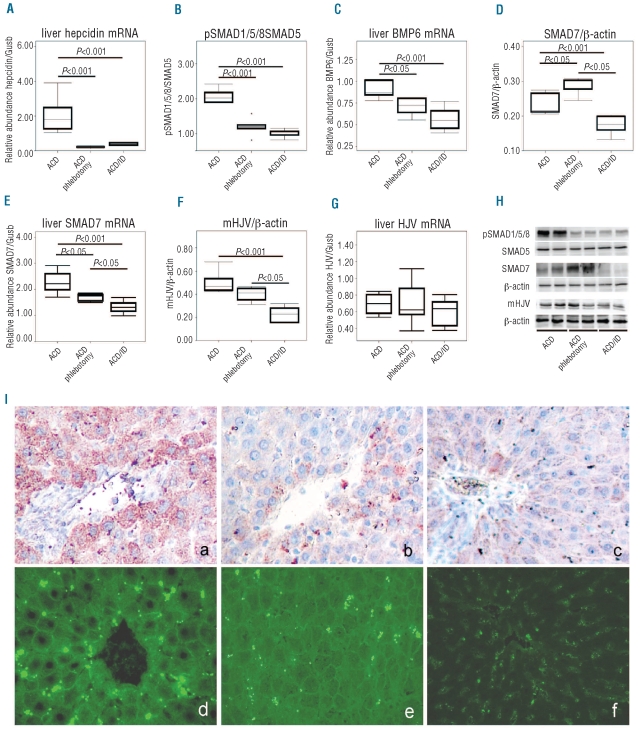
There was a significant difference in both BMP6 mRNA expression and in SMAD7 protein levels between ACD and ACD/IDA rats. These parameters were significantly correlated to Hamp mRNA levels in the liver (r=−0.738, P=0.037 for SMAD7; r=0.857, P=0.014 for BMP6). However, using the phlebotomy model to induce ACD/IDA we could not establish whether the alterations in BMP6 and SMAD7 expression are caused by changes in liver iron concentrations or erythropoietic activity, although there was no difference in hemoglobin levels between ACD and ACD/IDA animals.13 As a surrogate for erythropoietic activity, we determined serum erythropoietin levels which were of interest because erythropoietin has previously been demonstrated to inhibit hepcidin formation.10,37,40 As shown in Table 2, SMAD7 levels correlate positively (r=0.818, P=0.002) and BMP6 expression inversely with serum EPO levels (r=−0.731, P=0.016), while the opposite was true for correlations between liver iron concentrations with BMP6 (r=0.782, P=0.008) or SMAD7 (r=−0.636, P=0.035) expression, mirroring the differences in serum iron concentrations.13 We could not estimate to what extent alterations in SMAD7 and BMP6 expression can be related to either changes in erythropoietic activity or in hepatic iron availability. Therefore, we then compared rats with inflammatory anemia and two different forms of concomitant iron deficiency, i.e. ACD rats undergoing phlebotomies versus ACD rats with dietary iron restriction. When inflammatory anemia was induced in animals kept on an iron deficient diet, the relative changes in Hamp mRNA expression and the corresponding changes in SMAD1/5/8 activity observed three weeks after PG-APS injection were exactly the same as in the ACD group undergoing phlebotomy (Figure 4 A, B and H). BMP6 mRNA and protein levels were reduced in both groups of true iron deficient ACD animals compared to ACD rats (Figure 4C and I a-c). In contrast, SMAD7 protein levels were differently affected upon phlebotomy or iron deficient diet. While regular phlebotomy significantly increased SMAD7 protein levels (P<0.05), dietary iron restriction decreased SMAD7 expression in ACD rats (P<0.001) (Figure 4D and H). Interestingly, we found SMAD7 mRNA levels to be significantly decreased in rats treated with phlebotomy (P<0.05) and on an iron restricted diet (P<0.001) (Figure 4E). This indicates that the reported differences in SMAD7 protein levels as a function of alternative iron replacing strategies, and phlebotomy versus dietary iron restriction, respectively, are likely due to a post-transcriptional regulation of SMAD7 in an iron independent manner, since there was no difference in liver iron levels between the two iron deficient groups (data not shown). However, we found significantly elevated serum EPO levels in the phlebotomized group compared to the group receiving an iron deficient diet (P<0.001; Online Supplementary Table S1).
Table 2.
Correlation of SMAD7 protein and BMP6 mRNA expression in the liver with hepatic iron concentrations and serum EPO levels in ACD and ACD/IDA. Correlation analyses were carried out using Spearman’s-rho test. Correlation coefficients and two-sided P values are reported (n=11).
Figure 4.
Differential regulation of Hepcidin transcription in various ACD groups. ACD was induced in Lewis rats by i.p. injection of PG-APS as detailed in the Design and Methods section and animals were divided into three groups. One group of PG-APS treated rats was phlebotomized (ACD/phlebotomy; n=6), starting one week before sacrifice, whereas another group of rats was put on an iron deficient diet one week before PG-APS administration (ACD/ID; n=6). Liver (A) Hamp, (C) BMP6, (E) SMAD7 and (G) HJV mRNA expression were determined by quantitative RT-PCR and normalized to the expression of the housekeeping gene β-glucoronidase (Gusb) (A, C, E, and G). Liver nuclear cell extracts were immunoblotted with an antibody against pSMAD1/5/8 (B and H). Liver cytoplasmatic extracts were subjected to immunoblot analysis using antibodies against (D and H) SMAD7 and (F and H) mHJV. Protein levels were quantified by densitometry (B, D and F). A representative blot is shown in panel (H). The blots used for quantification by densitometry are shown in the Online Supplementary Figure S2. Data are depicted as lower quartile, median and upper quartile (boxes) and minimum/maximum ranges (whiskers). Statistic outliers are displayed as circles. Calculations for statistical differences between the various groups were carried out by ANOVA technique and Bonferroni’s correction for multiple tests. Immunohistochemical determination of liver BMP6 expression in ACD rats (I a), in phlebotomized ACD rats (I b) and in ACD rats on an iron deficient diet (I c) using affinity purified BMP6 antibody.43 Tissue distribution of mHJV in liver determined by immunoflourescence technique in (I d) ACD rats, (I e) phlebotomized ACD rats, and (I f) ACD rats on an iron deficient diet, using affinity purified mHJV antibody.43 A Zeiss Axioscope 40 microscope with a 40x lens and an AxioCam MRc5 was used for evaluation. Representative fields were photodocumented by a pixellink® system. The fluorescent pictures were taken under the same conditions.
As TMPRSS6 protein expression has been reported to be rapidly induced by oral iron deprivation,45 we analyzed TMPRSS6 in ACD rats with true iron deficiency due to phlebotomy or an iron deficient diet. In accordance with previous data,45 we found no changes in hepatic TMPRSS6 mRNA expression between the different anemia groups (data not shown). Unfortunately, no commercially available TMPRSS6 antibody works with rat samples, therefore, we were not able to analyze TMPRSS6 protein expression. However, as a surrogate for TMPRSS6 activity we investigated mHJV expression using liver membrane fractions. In phlebotomized rats, only a trend toward lower hepatic mHJV expression was found (Figure 4F, H and I), while a significantly reduced mHJV expression was observed in ACD rats on an iron deficient diet (Figure 4F, H and I d-e) as compared to ACD rats. In contrast, neither phlebotomy nor an iron deficient diet significantly changed HJV mRNA expression (Figure 4G).
Discussion
We and others reported increased hepcidin expression in patients suffering from ACD and in mammalian models mimicking ACD.7,13,14,19
In contrast, serum hepcidin levels and/or liver Hamp mRNA expression were significantly lower in patients and mammals suffering from ACD with true iron deficiency (ACD/IDA) when compared with ACD alone,13,19 indicating different signaling pathways and hierarchies between inflammatory anemia (ACD) and inflammatory anemia with associated true iron deficiency (ACD/IDA).
In agreement with a previous observation made in LPS challenged mice,41 we observed increased STAT3 phosphorylation in ACD rats which was not altered by concomitant true iron deficiency after phlebotomy. When analyzing the BMP/SMAD pathway, we found increased SMAD1/5/8 phosphorylation in inflammatory anemia (ACD) compared to controls while, most interestingly, SMAD1/5/8 activation significantly decreased with concomitant iron deficiency (ACD/IDA). This is in accordance with data showing that STAT3 inducible hepcidin expression is influenced by BMP dependent SMAD activation but not vice versa46 and with data indicating that SMAD1/5/8 phosphorylation is affected by iron status.47 In addition, transcriptional activation of hepcidin is not only abrogated in SMAD4-deficient hepatocytes in response to iron overload and BMPs, but also in response to IL-6,48 indicating that the BMP/SMAD pathway is able to modulate the IL-6 inducible STAT3 pathway. In ACD, STAT3 and SMAD1/5/8 phosphorylation as well as SMAD4 expression were increased. However, while STAT3 phosphorylation and SMAD4 expression were also high in animals with ACD/IDA, SMAD1/5/8 phosphorylation was reduced in the latter. This suggests that during inflammatory anemia a concomitant true iron deficiency (ACD/IDA) reduces pSMAD1/5/8 mediated transcriptional activation leading to lower hepcidin levels despite massive STAT3 activation.
Consecutively, we studied the mechanisms underlying contrasting SMAD1/5/8 phosphorylation in ACD and ACD/IDA. We found that the hepatic BMP6 expression was significantly lower in ACD/IDA than in ACD rats. These changes showed a significant correlation to SMAD1/5/8 phosphorylation and Hamp mRNA levels but were inversely mirrored by changes in SMAD7 expression. Interestingly, BMP7, which has been reported to be the strongest inducer of hepcidin transcription in vitro,29 was decreased in ACD and ACD/IDA suggesting that this BMP plays no essential role in hepcidin formation in ACD in vivo.
SMAD7 has recently been found to inhibit Hamp mRNA expression by triggering the dephosphorylation and degradation of BMP receptors and by blocking phosphorylation of SMAD1/5/8.34 Also, SMAD7 binds to the hepcidin promoter thus preventing the attachment of the SMAD4 containing activator complexes.34 While we found SMAD7 expression to be up-regulated by phlebotomy, the expression levels of other negative regulators of SMAD1/5/8 phosphorylation, such as TOB1 and TOB2, remained unchanged. However, as dietary iron restriction has previously been reported to decrease SMAD7 expression,47 we concluded that other factors induced by phlebotomy may stimulate SMAD7 expression in addition to changes in iron status. We, therefore, analyzed the effects of two forms of true iron deficiency on Hamp transcription and SMAD signaling in ACD rats which underwent either phlebotomy or were exposed to dietary iron restriction. There was no difference in BMP6 mRNA levels between both truly iron deficient ACD groups. As there was no difference in liver iron concentrations between either group but there was a difference in serum EPO levels, BMP6 expression appears to be affected by tissue iron levels. This agrees with the observed iron mediated induction of BMP6.47,49,50 In contrast, SMAD7 expression was differentially regulated between the two groups. This could not be traced back to differences in liver iron content but phlebotomized rats showed significantly higher EPO levels as an indicator of stimulated erythropoiesis compared to ACD rats on an iron deficient diet. These data suggest that SMAD7 expression is influenced by both changes in iron status and erythropoiesis activity, although in opposite directions.43
The expression of the inhibitory SMAD7 is decreased in rats on an iron deficient diet yet SMAD1/5/8 phosphorylation is low and comparable to that of phlebotomized rats. We, therefore, explored alternative pathways for SMAD transactivation. We found less mHJV in liver cell membranes of low-iron fed ACD rats than in ACD rats on a normal diet. This agrees with data indicating increased TMPRSS6 activity in association with dietary iron restriction.45,50 In addition, we found no differences in HJV mRNA expression in association with iron deficiency in accordance with recent data from Bondi et al.51 However, definitive cause effect relationships and the sequence of events in this network are hard to demonstrate in a chronic disease model like the one used here because of the numerous cross-regulatory feedback loops involved in the regulation of iron homeostasis.33,52 Nevertheless, the associations of distinct signaling pathways with hepcidin expression, iron availability, anemia and inflammation found here, together with evidence in literature, led to the prediction of the following model for hepcidin regulation in ACD and ACD/IDA.
STAT3 activation is associated with inflammatory anemia, but the fine tuning of hepcidin expression in inflammation as a function of iron availability is exerted via modulation of SMAD1/5/8 phosphorylation and formation of the SMAD activation complex. While its activity is high in ACD, SMAD1/5/8 phosphorylation is reduced by true iron deficiency in the setting of inflammatory anemia (ACD/IDA) independent of the cause of iron deficiency. This is on the one hand due to reduced expression of BMP6 as a consequence of low iron availability. On the other hand, phlebotomy but not dietary iron deficiency induces SMAD7 protein expression, an inhibitor of the SMAD1/5/8 transactivating pathway. This appears, therefore, not to be the consequence of low iron availability, because SMAD7 levels were reduced in ACD rats receiving a low iron diet, but rather of the induction of erythropoietic activity following phlebotomy, as evidenced by increased circulating erythropoietin levels or hypoxia as a consequence of blood loss. As iron deficiency induced by phlebotomy as well as by a low iron diet inhibits SMAD7 transcription, the differences in SMAD7 protein expression were unexpected. However, SMAD7 has been reported to be regulated post-transcriptionally53 and post-translationally.54 The coactivator p300 acetylates and stabilizes SMAD7, thus preventing its ubiquitination and degradation in the proteasome.53 P300 interacts with central mediators of erythropoiesis,55–56 suggesting a possible interaction between erythropoiesis and post-transcriptional regulation of SMAD7 expression.
Strikingly, dietary iron restriction reduced the expression of the BMP-R co-factor mHJV, thus impairing SMAD1/5/8 activity. This demonstrates that iron deficiency, erythropoietic activity, hypoxia and inflammation induce different regulatory pathways which control hepcidin expression. Importantly, these regulatory pathways appear to underlie a specific hierarchy because inflammation mediated induction of Hamp transcription can be partly reversed in vivo by the regulatory cascades induced by true iron deficiency.46,48
This work shows the regulatory mechanisms affecting hepcidin expression under inflammatory conditions. At the moment, different hepcidin lowering therapy regimes are under evaluation to treat anemia of chronic disease. The data provided in this paper and in those of others highlight BMP6, mHJV and SMAD7 as putative targets of such therapeutic strategies.
Footnotes
Funding: this study was supported by grants from the Austrian Research Funds FWF (P-19964, TRP-188) (G.W.), the “Verein zur Förderung von Forschung in Infektiologie und Immunologie, Innsbruck “, a research fund from the Medical University of Innsbruck MFI (2007-416) (I.T.) and a research fund from the OENB (14182) (I.T.)
The online version of this article has a Supplementary Appendix.
Authorship and Disclosures
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.Weiss G, Goodnough LT. Anemia of chronic disease. N Engl J Med. 2005;352(10):1011–23. doi: 10.1056/NEJMra041809. [DOI] [PubMed] [Google Scholar]
- 2.Matzner Y, Levy S, Grossowicz N, Izak G, Hershko C. Prevalence and causes of anemia in elderly hospitalized patients. Gerontology. 1979;25(2):113–9. doi: 10.1159/000212328. [DOI] [PubMed] [Google Scholar]
- 3.Means RT., Jr Recent developments in the anemia of chronic disease. Curr Hematol Rep. 2003;2(2):116–21. [PubMed] [Google Scholar]
- 4.Spivak JL. Iron and the anemia of chronic disease. Oncology (Williston Park) 2002;16(9 Suppl 10):25–33. [PubMed] [Google Scholar]
- 5.Ludwiczek S, Aigner E, Theurl I, Weiss G. Cytokine-mediated regulation of iron transport in human monocytic cells. Blood. 2003;101(10):4148–54. doi: 10.1182/blood-2002-08-2459. [DOI] [PubMed] [Google Scholar]
- 6.Yang F, Liu XB, Quinones M, Melby PC, Ghio A, Haile DJ. Regulation of reticuloendothelial iron transporter MTP1 (Slc11a3) by inflammation. J Biol Chem. 2002;277(42):39786–91. doi: 10.1074/jbc.M201485200. [DOI] [PubMed] [Google Scholar]
- 7.Ganz T. Hepcidin--a regulator of intestinal iron absorption and iron recycling by macrophages. Best Pract Res Clin Haematol. 2005;18(2):171–82. doi: 10.1016/j.beha.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 8.Fleming RE. Iron and inflammation: cross-talk between pathways regulating hepcidin. J Mol Med. 2008;86(5):491–4. doi: 10.1007/s00109-008-0349-8. [DOI] [PubMed] [Google Scholar]
- 9.Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, et al. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306(5704):2090–3. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 10.Nicolas G, Chauvet C, Viatte L, Danan JL, Bigard X, Devaux I, et al. The gene encoding the iron regulatory peptide hepcidin is regulated by anemia, hypoxia, and inflammation. J Clin Invest. 2002;110(7):1037–44. doi: 10.1172/JCI15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peyssonnaux C, Zinkernagel AS, Datta V, Lauth X, Johnson RS, Nizet V. TLR4-dependent hepcidin expression by myeloid cells in response to bacterial pathogens. Blood. 2006;107(9):3727–32. doi: 10.1182/blood-2005-06-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Theurl I, Theurl M, Seifert M, Mair S, Nairz M, Rumpold H, et al. Autocrine formation of hepcidin induces iron retention in human monocytes. Blood. 2008;111(4):2392–9. doi: 10.1182/blood-2007-05-090019. [DOI] [PubMed] [Google Scholar]
- 13.Theurl I, Aigner E, Theurl M, Nairz M, Seifert M, Schroll A, et al. Regulation of iron homeostasis in anemia of chronic disease and iron deficiency anemia: diagnostic and therapeutic implications. Blood. 2009;113(21):5277–86. doi: 10.1182/blood-2008-12-195651. [DOI] [PubMed] [Google Scholar]
- 14.Kemna E, Pickkers P, Nemeth E, van der Hoeven H, Swinkels D. Time-course analysis of hepcidin, serum iron, and plasma cytokine levels in humans injected with LPS. Blood. 2005;106(5):1864–6. doi: 10.1182/blood-2005-03-1159. [DOI] [PubMed] [Google Scholar]
- 15.Theurl I, Mattle V, Seifert M, Mariani M, Marth C, Weiss G. Dysregulated monocyte iron homeostasis and erythropoeitin formation in patients with anemia of chronic disease. Blood. 2006;107(10):4142–8. doi: 10.1182/blood-2005-08-3364. [DOI] [PubMed] [Google Scholar]
- 16.Sasu BJ, Cooke KS, Arvedson TL, Plewa C, Ellison AR, Sheng J, et al. Anti-hepcidin anti-body treatment modulates iron metabolism and is effective in a mouse model of inflammation-induced anemia. Blood. 2010;115(17):3616–24. doi: 10.1182/blood-2009-09-245977. [DOI] [PubMed] [Google Scholar]
- 17.Brugnara C. Iron deficiency and erythropoiesis: new diagnostic approaches. Clin Chem. 2003;49(10):1573–8. doi: 10.1373/49.10.1573. [DOI] [PubMed] [Google Scholar]
- 18.Goodnough LT, Nemeth E, Ganz T. Detection, evaluation, and management of iron-restricted erythropoiesis. Blood. 2010;116(23):4754–61. doi: 10.1182/blood-2010-05-286260. [DOI] [PubMed] [Google Scholar]
- 19.Lasocki S, Millot S, Andrieu V, Letteron P, Pilard N, Muzeau F, et al. Phlebotomies or erythropoietin injections allow mobilization of iron stores in a mouse model mimicking intensive care anemia. Crit Care Med. 2008;36(8):2388–94. doi: 10.1097/CCM.0b013e31818103b9. [DOI] [PubMed] [Google Scholar]
- 20.Wrighting DM, Andrews NC. Interleukin-6 induces hepcidin expression through STAT3. Blood. 2006;108(9):3204–9. doi: 10.1182/blood-2006-06-027631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verga Falzacappa MV, Vujic Spasic M, Kessler R, Stolte J, Hentze MW, Muckenthaler MU. STAT3 mediates hepatic hepcidin expression and its inflammatory stimulation. Blood. 2007;109(1):353–8. doi: 10.1182/blood-2006-07-033969. [DOI] [PubMed] [Google Scholar]
- 22.Pietrangelo A, Dierssen U, Valli L, Garuti C, Rump A, Corradini E, et al. STAT3 is required for IL-6-gp130-dependent activation of hepcidin in vivo. Gastroenterology. 2007;132(1):294–300. doi: 10.1053/j.gastro.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 23.Papanikolaou G, Samuels ME, Ludwig EH, MacDonald ML, Franchini PL, Dube MP, et al. Mutations in HFE2 cause iron overload in chromosome 1q-linked juvenile hemochromatosis. Nat Genet. 2004;36(1):77–82. doi: 10.1038/ng1274. [DOI] [PubMed] [Google Scholar]
- 24.Babitt JL, Huang FW, Wrighting DM, Xia Y, Sidis Y, Samad TA, et al. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet. 2006;38(5):531–9. doi: 10.1038/ng1777. [DOI] [PubMed] [Google Scholar]
- 25.Du X, She E, Gelbart T, Truksa J, Lee P, Xia Y, et al. The serine protease TMPRSS6 is required to sense iron deficiency. Science. 2008;320(5879):1088–92. doi: 10.1126/science.1157121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silvestri L, Pagani A, Nai A, De Domenico I, Kaplan J, Camaschella C. The serine pro-tease matriptase-2 (TMPRSS6) inhibits hepcidin activation by cleaving membrane hemojuvelin. Cell Metab. 2008;8(6):502–11. doi: 10.1016/j.cmet.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finberg KE, Whittlesey RL, Fleming MD, Andrews NC. Down-regulation of Bmp/Smad signaling by Tmprss6 is required for maintenance of systemic iron homeostasis. Blood. 2010;115(18):3817–26. doi: 10.1182/blood-2009-05-224808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Truksa J, Peng H, Lee P, Beutler E. Bone morphogenetic proteins 2, 4, and 9 stimulate murine hepcidin 1 expression independently of Hfe, transferrin receptor 2 (Tfr2), and IL- 6. Proc Natl Acad Sci USA. 2006;103(27):10289–93. doi: 10.1073/pnas.0603124103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Babitt JL, Huang FW, Xia Y, Sidis Y, Andrews NC, Lin HY. Modulation of bone morphogenetic protein signaling in vivo regulates systemic iron balance. J Clin Invest. 2007;117(7):1933–9. doi: 10.1172/JCI31342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andriopoulos B, Jr, Corradini E, Xia Y, Faasse SA, Chen S, Grgurevic L, et al. BMP6 is a key endogenous regulator of hepcidin expression and iron metabolism. Nat Genet. 2009;41(4):482–7. doi: 10.1038/ng.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meynard D, Kautz L, Darnaud V, Canonne-Hergaux F, Coppin H, Roth MP. Lack of the bone morphogenetic protein BMP6 induces massive iron overload. Nat Genet. 2009;41(4):478–81. doi: 10.1038/ng.320. [DOI] [PubMed] [Google Scholar]
- 32.Anderson GJ, Frazer DM. Iron metabolism meets signal transduction. Nat Genet. 2006;38(5):503–4. doi: 10.1038/ng0506-503. [DOI] [PubMed] [Google Scholar]
- 33.Hentze MW, Muckenthaler MU, Galy B, Camaschella C. Two to tango: regulation of Mammalian iron metabolism. Cell. 2010;142(1):24–38. doi: 10.1016/j.cell.2010.06.028. [DOI] [PubMed] [Google Scholar]
- 34.Mleczko-Sanecka K, Casanovas G, Ragab A, Breitkopf K, Muller A, Boutros M, et al. SMAD7 controls iron metabolism as a potent inhibitor of hepcidin expression. Blood. 2009;115(13):2657–65. doi: 10.1182/blood-2009-09-238105. [DOI] [PubMed] [Google Scholar]
- 35.Peyssonnaux C, Zinkernagel AS, Schuepbach RA, Rankin E, Vaulont S, Haase VH, et al. Regulation of iron homeostasis by the hypoxia-inducible transcription factors (HIFs) J Clin Invest. 2007;117(7):1926–32. doi: 10.1172/JCI31370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanno T, Bhanu NV, Oneal PA, Goh SH, Staker P, Lee YT, et al. High levels of GDF15 in thalassemia suppress expression of the iron regulatory protein hepcidin. Nat Med. 2007;13(9):1096–101. doi: 10.1038/nm1629. [DOI] [PubMed] [Google Scholar]
- 37.Pinto JP, Ribeiro S, Pontes H, Thowfeequ S, Tosh D, Carvalho F, et al. Erythropoietin mediates hepcidin expression in hepatocytes through EPOR signaling and regulation of C/EBPalpha. Blood. 2008;111(12):5727–33. doi: 10.1182/blood-2007-08-106195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vecchi C, Montosi G, Zhang K, Lamberti I, Duncan SA, Kaufman RJ, et al. ER stress controls iron metabolism through induction of hepcidin. Science. 2009;325(5942):877–80. doi: 10.1126/science.1176639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, Pantopoulos K. Regulation of cellular iron metabolism. Biochem J. 2011;434(3):365–81. doi: 10.1042/BJ20101825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nicolas G, Viatte L, Bennoun M, Beaumont C, Kahn A, Vaulont S. Hepcidin, a new iron regulatory peptide. Blood Cells Mol Dis. 2002;29(3):327–35. doi: 10.1006/bcmd.2002.0573. [DOI] [PubMed] [Google Scholar]
- 41.Huang H, Constante M, Layoun A, Santos MM. Contribution of STAT3 and SMAD4 pathways to the regulation of hepcidin by opposing stimuli. Blood. 2009;113(15):3593–9. doi: 10.1182/blood-2008-08-173641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ludwiczek S, Theurl I, Artner-Dworzak E, Chorney M, Weiss G. Duodenal HFE expression and hepcidin levels determine body iron homeostasis: modulation by genetic diversity and dietary iron availability. J Mol Med. 2004;82(6):373–82. doi: 10.1007/s00109-004-0542-3. [DOI] [PubMed] [Google Scholar]
- 43.Merle U, Theilig F, Fein E, Gehrke S, Kallinowski B, Riedel HD, et al. Localization of the iron-regulatory proteins hemojuvelin and transferrin receptor 2 to the basolateral membrane domain of hepatocytes. Histochem Cell Biol. 2007;127(2):221–6. doi: 10.1007/s00418-006-0229-7. [DOI] [PubMed] [Google Scholar]
- 44.Kautz L, Meynard D, Besson-Fournier C, Darnaud V, Al Saati T, Coppin H, et al. BMP/Smad signaling is not enhanced in Hfe-deficient mice despite increased Bmp6 expression. Blood. 2009;114(12):2515–20. doi: 10.1182/blood-2009-02-206771. [DOI] [PubMed] [Google Scholar]
- 45.Zhang AS, Anderson SA, Wang J, Yang F, DeMaster K, Ahmed R, et al. Suppression of hepatic hepcidin expression in response to acute iron deprivation is associated with an increase of matriptase-2 protein. Blood. 2011;117(5):1687–99. doi: 10.1182/blood-2010-06-287292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Casanovas G, Mleczko-Sanecka K, Altamura S, Hentze MW, Muckenthaler MU. Bone morphogenetic protein (BMP)-responsive elements located in the proximal and distal hepcidin promoter are critical for its response to HJV/BMP/SMAD. J Mol Med. 2009;87(5):471–80. doi: 10.1007/s00109-009-0447-2. [DOI] [PubMed] [Google Scholar]
- 47.Kautz L, Meynard D, Monnier A, Darnaud V, Bouvet R, Wang RH, et al. Iron regulates phosphorylation of Smad1/5/8 and gene expression of Bmp6, Smad7, Id1, and Atoh8 in the mouse liver. Blood. 2008;112(4):1503–9. doi: 10.1182/blood-2008-03-143354. [DOI] [PubMed] [Google Scholar]
- 48.Wang RH, Li C, Xu X, Zheng Y, Xiao C, Zerfas P, et al. A role of SMAD4 in iron metabolism through the positive regulation of hepcidin expression. Cell Metab. 2005;2(6):399–409. doi: 10.1016/j.cmet.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 49.Corradini E, Garuti C, Montosi G, Ventura P, Andriopoulos B, Jr, Lin HY, et al. Bone morphogenetic protein signaling is impaired in an HFE knockout mouse model of hemochromatosis. Gastroenterology. 2009;137(4):1489–97. doi: 10.1053/j.gastro.2009.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arndt S, Maegdefrau U, Dorn C, Schardt K, Hellerbrand C, Bosserhoff AK. Iron-induced expression of bone morphogenic protein 6 in intestinal cells is the main regulator of hepatic hepcidin expression in vivo. Gastroenterology. 2010;138(1):372–82. doi: 10.1053/j.gastro.2009.09.048. [DOI] [PubMed] [Google Scholar]
- 51.Bondi A, Valentino P, Daraio F, Porporato P, Gramaglia E, Carturan S, et al. Hepatic expression of hemochromatosis genes in two mouse strains after phlebotomy and iron overload. Haematologica. 2005;90(9):1161–7. [PubMed] [Google Scholar]
- 52.Ramey G, Deschemin JC, Vaulont S. Crosstalk between the mitogen activated protein kinase and bone morphogenetic protein/hemojuvelin pathways is required for the induction of hepcidin by holotransferrin in primary mouse hepatocytes. Haematologica. 2009;94(6):765–72. doi: 10.3324/haematol.2008.003541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Monteleone G, Del Vecchio Blanco G, Monteleone I, Fina D, Caruso R, Gioia V, et al. Post-transcriptional regulation of Smad7 in the gut of patients with inflammatory bowel disease. Gastroenterology. 2005;129(5):1420–9. doi: 10.1053/j.gastro.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 54.Yan X, Chen YG. Smad7: not only a regulator, but also a cross-talk mediator of TGF-beta signalling. Biochem J. 2011;434(1):1–10. doi: 10.1042/BJ20101827. [DOI] [PubMed] [Google Scholar]
- 55.Cantor AB, Orkin SH. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 2002;21(21):3368–76. doi: 10.1038/sj.onc.1205326. [DOI] [PubMed] [Google Scholar]
- 56.Han L, Lu J, Pan L, Wang X, Shao Y, Han S, et al. Histone acetyltransferase p300 regulates the transcription of human erythroid-specific 5-aminolevulinate synthase gene. Biochem Biophys Res Commun. 2006;348(3):799–806. doi: 10.1016/j.bbrc.2006.07.147. [DOI] [PubMed] [Google Scholar]