Abstract
Background
Mutations in the PHF6 gene were recently described in patients with T-cell acute lymphoblastic leukemia and in those with acute myeloid leukemia. The present study was designed to determine the prevalence of PHF6 gene alterations in T-cell acute lymphoblastic leukemia.
Design and Methods
We analyzed the incidence and prognostic value of PHF6 mutations in 96 Chinese patients with T-cell acute lymphoblastic leukemia. PHF6 deletions were screened by real-time quantitative polymerase chain reaction and array-based comparative genomic hybridization. Patients were also investigated for NOTCH1, FBXW7, WT1, and JAK1 mutations together with CALM-AF10, SET-NUP214, and SIL-TAL1 gene rearrangements.
Results
PHF6 mutations were identified in 11/59 (18.6%) adult and 2/37 (5.4%) pediatric cases of T-cell acute lymphoblastic leukemia, these incidences being significantly lower than those recently reported. Although PHF6 is X-linked and mutations have been reported to occur almost exclusively in male patients, we found no sex difference in the incidences of PHF6 mutations in Chinese patients with T-cell acute lymphoblastic leukemia. PHF6 deletions were detected in 2/79 (2.5%) patients analyzed. NOTCH1 mutations, FBXW7 mutations, WT1 mutations, JAK1 mutations, SIL-TAL1 fusions, SET-NUP214 fusions and CALM-AF10 fusions were present in 44/96 (45.8%), 9/96 (9.4%), 4/96 (4.1%), 3/49 (6.1%), 9/48 (18.8%), 3/48 (6.3%) and 0/48 (0%) of patients, respectively. The molecular genetic markers most frequently associated with PHF6 mutations were NOTCH1 mutations (P=0.003), SET-NUP214 rearrangements (P=0.002), and JAK1 mutations (P=0.005). No differences in disease-free survival and overall survival between T-cell acute lymphoblastic leukemia patients with and without PHF6 mutations were observed in a short-term follow-up.
Conclusions
Overall, these results indicate that, in T-cell acute lymphoblastic leukemia, PHF6 mutations are a recurrent genetic abnormality associated with mutations of NOTCH1, JAK1 and rearrangement of SET-NUP214.
Keywords: T-cell acute lymphoblastic leukemia, mutation, PHF6, NOTCH1, SET-NUP214
Introduction
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive malignancy of thymocytes with multiple genetic abnormalities, accounting for 15% and 25% of newly diagnosed cases of ALL in children and adults, respectively. T-ALL often presents with more unfavorable clinical features than precursor B-ALL, such as a high white blood cell count, bulky adenopathy, and central nervous system infiltration.1 Although the outcome has improved with current intensive therapy and hematopoietic stem cell transplantation, about 10% of children with T-ALL2–5 and 37% of adult T-ALL patients relapse and ultimately die.6 Further improvements in survival rates will require new therapies and targets. In the wake of the Human Genome Project, genes deregulated in specific disease entities may now be readily identified by a fast evolving variety of molecular genetic methods, relevant to disease stratification.
In recent years, genome-wide profiling of DNA copy-number variations and candidate gene sequencing have revealed abundant recurrent gene abnormalities affecting RAS,7 FLT3,8 LMO2,9 NOTCH1,10 MYB,11 FBXW7,12 JAK1,13 WT1,14 PTPN2,15 and LEF116.
Inactivating mutations of the plant homeodomain (PHD)-like finger 6 (PHF6) gene were originally linked to a form of syndromic X-linked mental retardation, Börjeson-Forssman-Lehmann syndrome (BFLS), originally observed in males.17 Recently, PHF6 was identified as a new key tumor suppressor gene in T-ALL. PHF6 mutations were detected in 38% (16/42) of adult and 16% (14/89) of pediatric T-ALL patients and rarely also in adults with acute myeloid leukemia.18,19 PHF6 is X-linked, and tumorigenic mutations have been almost exclusively detected in male patients with either T-ALL or acute myeloid leukemia. This gender-bias has been proposed as contributing to the 3-fold higher incidence of T-ALL in males over females.18
In this study, we investigated the incidence and prognostic value of PHF6 mutations by polymerase chain reaction (PCR) and direct sequencing in a cohort of 96 T-ALL patients. Patients were also assessed for the presence of NOTCH1, FBXW7, WT1, and JAK1 mutations and CALM-AF10, SET-NUP214 and SIL-TAL1 rearrangements.
Design and Methods
Patients’ samples and cell lines
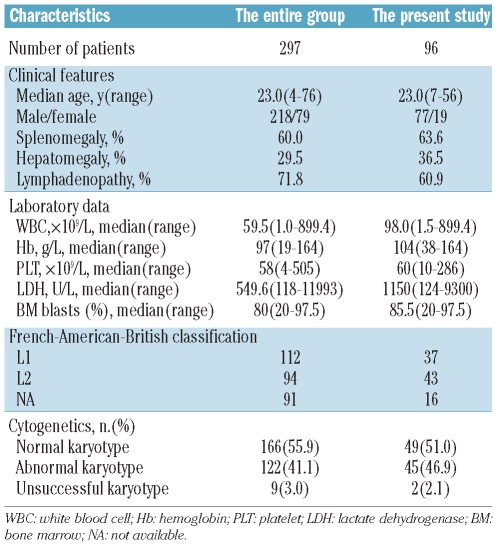
Between June 1998 and August 2010, 297 T-ALL patients were identified following admission to the Jiangsu Institute of Hematology. Overall, genomic DNA from 96 patients was available for use in the present study. The median age of the case series was 23 years (range, 7–56 years), the majority were male (80.2%) and all were from the same ethnic group: the Han Chinese. The main characteristics of the patients in this study and the entire group are summarized in Table 1. The clinical characteristics of these 96 T-ALL patients were representative of the entire group. T-cell phenotype was defined by positivity for CD3, either on the cell surface or in the cytoplasm, with at least two of the B-cell antigens (CD19, CD20, CD22, CD79a) and three of the myeloid markers (CD11b, CD13, CD33, myeloperoxidase) testing negative.
Table 1.
Clinical characteristics of 96 patients with T-ALL.
Sample preservation and genetic analysis required informed consent and approval by the Ethics Committee of the First Affiliated Hospital of Soochow University in accordance with the Declaration of Helsinki. Genomic DNA was extracted from frozen bone marrow mononuclear cells of 68 T-ALL cases after Ficoll gradient centrifugation using standard procedures. In another 28 patients, genomic DNA was extracted from methanol/acetic acid-fixed cells that had been routinely prepared for cytogenetic analysis.
The CCRF-CEM, MOLT-4, and Jurkat T-ALL cell lines (detailed in www.dsmz.de) were cultured in RPMI-1640 medium (Gibco BRL, Grand Island, NY, USA) supplemented with 20% heat-inactivated fetal calf serum (Gibco BRL) at 37°C in a humidified incubator with 5% CO2.
Cytogenetic analysis
Cytogenetic studies were carried out on bone marrow cells at diagnosis. Samples were processed using standard 24-h unstimulated cultures. Conventional R-banding assay was used for karyotypic analysis. When possible, at least 20 metaphases per sample were analyzed. Clonal karyotypic abnormalities are described using the International System for Human Cytogenetic Nomenclature (ISCN, 2005).
Molecular characterization of T-cell acute lymphoblastic leukemia samples
PHF6 mutations were analyzed by PCR amplification of the entire coding region of PHF6 exons 2–10 followed by direct bidirectional DNA sequencing as previously described.18
NOTCH1 mutations were detected by PCR amplification and direct DNA sequencing of exons 26, 27, and 34 of the gene.10 WT1 mutations were screened by PCR amplification and direct DNA sequencing of exons 7 and 9.14 JAK1 mutations were screened for by PCR amplification and direct DNA sequencing of the entire JAK1 coding region.13 FBXW7 mutation analysis was performed by PCR amplification and direct DNA sequencing of exons 9 and 10.20
Expression of SIL-TAL1, SET-NUP214, and CALM-AF10 was assessed by reverse transcription-PCR (RT-PCR) using previously published primers and PCR conditions.17–19 Briefly, total RNA was extracted from bone marrow mononuclear cells of 48 T-ALL cases prepared with Ficoll gradient centrifugation. RNA concentrations were measured by fluorometry. Afterwards cDNA was synthesized using the SuperScript II reverse transcriptase kit (Invitrogen, Carlsbad, CA, USA) with random hexamers. A single PCR was performed in a volume of 25 μL using 1 μL of cDNA. PCR products were separated on 2% agarose and visualized with GelRed. The quality of RNA was checked by amplification of GAPDH gene transcripts. Leukemic cells from two T-ALL patients known to contain the SIL-TAL1 or SET-NUP214 fusion transcripts and the U937 cell line with CALM-AF10 fusion transcript were utilized as positive controls. Primer sequences and PCR conditions are shown in Online Supplementary Table S1.
Real-time quantification of DNA copy number
Real-time quantitative DNA PCR analysis was performed to screen for genomic PHF6 deletions in 79 T-ALL patients for whom genomic DNA was available, using TaqMan Universal PCR Mastermix (Applied Biosystems) and the 7500 real-time PCR system (Applied Biosystems) as previously reported.18 Data were analyzed using the comparative ΔCt method (Applied Biosystems).
Array-based comparative genomic hybridization analysis
Array-based comparative genomic hybridization (array-CGH) analysis was performed in 19 T-ALL patients for whom at least 1.5 μg genomic DNA was available. Agilent human CGH microarrays (Agilent Technologies, Santa Clara, CA, USA) containing 244 000 probes (244k) with an average spatial resolution of 6.4 Kb were used for the array-CGH experiments according to protocols provided by the manufacturer. Image analysis, normalization and annotation were based on Feature Extraction 9.1 (Agilent), while data were visualized with Agilent Genomic Workbench Lite Edition 6.5 software.
Statistical analysis
The patients’ characteristics were analyzed by the chi-squared (χ2) or Fisher’s exact tests for univariate analysis. Overall survival was calculated from the date of diagnosis until the date of death or last follow-up through October 31, 2010. Relapse-free survival was measured from the date of complete remission to the date of first relapse. Kaplan-Meier analysis was used to evaluate patients’ survival. The log-rank test was used to compare differences in survival. P values less than 0.05 were deemed statistically significant. All calculations were performed using the SPSS software package (version 13.0).
Results
PHF6 mutations and deletions in T-cell acute lymphoblastic leukemia
PHF6 mutations were identified in 11/59 (18.6%) adult and 2/37 (5.4%) pediatric T-ALL patients analyzed. While just falling short of statistical significance (χ2 P=0.065), our data indicate an adult preference for PHF6 mutations in TALL. However, we were unable to confirm the 13-fold male excess reported recently in T-ALL.18 Although 11/13 (84.6%) cases with PHF6 mutations were male and only 2 of 13 (15.4%) female, this skewing reflected that of our population of patients, which comprised 77 males and 19 females (P>0.5), in turn attributable to the approximately 3-fold excess of T-ALL commonly seen in males.
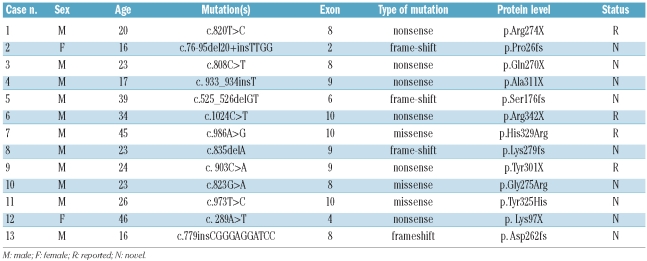
Overall, 13 PHF6 mutations were documented in this study (Table 2), involving exons 2, 4 and 6 (one case each), and exons 8, 9 and 10 (4, 3 and 3 cases, respectively). PHF6 mutations were not detected in CCRF-CEM, MOLT-4, or Jurkat cell lines.
Table 2.
Characteristics of 13 T-ALL patients with a PHF6 mutation.
The PHF6 mutations detected in the present study are listed in Table 2 and graphically depicted in Figure 1. Among these, nonsense, frameshift, and missense mutations accounted for, respectively, 46.2% (6/13), 30.8% (4/13) and 23.0% (3/13) of all PHF6 mutations. The mutation R342X in exon 10 was previously described in the germline of a child with BFLS who developed T-ALL.24 Two nonsense mutations and one missense mutation have already been described.18 The remaining nine mutations have not been reported previously. The patients’ characteristics are listed in Online Supplementary Table S2. In all four of 13 T-ALL cases with PHF6 mutations tested, analysis of matched newly-diagnosed and remission genomic DNA confirmed the somatic origin of PHF6 mutations (nonsense mutations: Arg274X, Tyr301X, Ala311X; missense mutations: Gly275Arg).
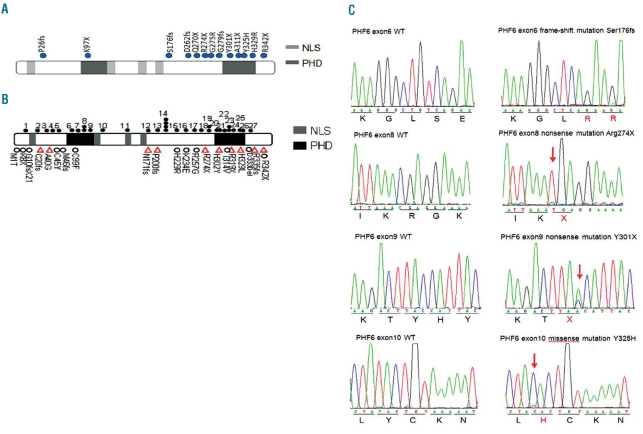
Figure 1.
PHF6 mutations in T-ALL. (A) Structure of the PHF6 protein, including four nuclear localization signals (NLS) and two imperfect PHD zinc-finger domains. Filled circles indicate PHF6 mutations. (B) PHF6 mutations previously reported in T-ALL, AML and BFLS. Filled circles represent mutations in T-ALL. Triangles represent mutations in AML. Open circles represent mutations in BFLS. 1, G10fs. 2, C28fs. 3, A41fs. 4, H43fs. 5, H44fs. 6, T98fs. 7, Y105fs. 8, R116X. 9, G122X. 10, H135fs. 11, S158fs. 12, F172fs. 13, S191fs. 14, C215F and C215Y. 15, R225X. 16, K235X. 17, R257X. 18, G263fs. 19, R274X. 20, C280Y. 21, C283R. 22, T300A. 23, Y303fs and Y303X. 24, A311P. 25, S320X. 26, H329R. 27, D333fs. (C) Representative DNA sequencing chromatograms of T-ALL genomic DNA samples showing mutations in exons 6, 8, 9 and 10 of PHF6.
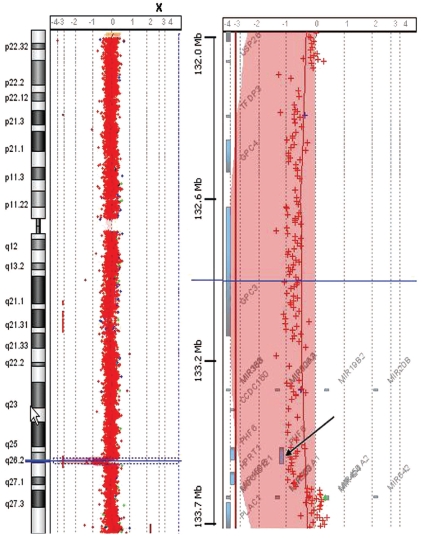
In addition, PHF6 gene deletions were identified in 2/79 (2.5%) T-ALL patients (one male and one female) by quantitative PCR analysis and a 1.4 Mb deletion at Xq26 including PHF6 confirmed in the female patient by array-CGH (Figure 2).
Figure 2.
Whole-genome array CGH. Image shows array CGH (244K) analysis in one female T-ALL patient which revealed a 1.4 Mb deletion at Xq26 including the PHF6 gene (black arrow).
NOTCH1, FBXW7, WT1, and JAK1 mutations in T-cell acute lymphoblastic leukemia
NOTCH1 mutations were detected in 44 (45.8%) of the 96 T-ALL patients in the following domains and with the following frequencies: HD, 59.1%; PEST, 25.0%; HD+PEST 11.4%; and TAD, 4.5%. FBXW7 mutations were identified in 9/96 (9.4%) T-ALL cases; alone in three, and associated with NOTCH1 HD mutations in the other six cases. WT1 mutations were found in 4/96 (4.2%) cases; one patient harbored a missense mutation at codon 462 (R462W) located in exon 9, while the others all had frameshift mutations located in exon 7. JAK1 mutations were found in 3/49 (6.1%) cases; the mutation was missense in two cases, and a frameshift in another. In all eight available T-ALL cases with NOTCH1 or FBXW7 mutations, analysis of matched newly-diagnosed and remission genomic DNA confirmed the somatic origin of mutations (NOTCH1 mutations: R1597P, L1599P, K1606fs, I1615V, delF@1605+insLGEN@1605/1606, del9@1605-1613+insLGGK, delF@1605+insLGVTH@1605/1606; FBXW7 mutations: R465Y).
SET-NUP214, SIL-TAL1, and CALM-AF10 fusions in T-cell acute lymphoblastic leukemia
The expression of SIL-TAL1, SET-NUP214, and CALM-AF10 fusion transcripts was determined by RT-PCR in 48 patients. SIL-TAL1 was positive in 9/48 (18.8%) patients. The nine cases with SIL-TAL1 fusion were younger than the 39 cases without SIL-TAL1 fusion (P=0.018). SET-NUP214 fusion transcript was detected in 3/48 (6.3%) patients. All three patients with a SET-NUP214 fusion harbored PHF6 mutations. No CALM-AF10 fusion was detected in 48 T-ALL patients tested.
Correlation of PHF6 mutations with clinical features and prognosis
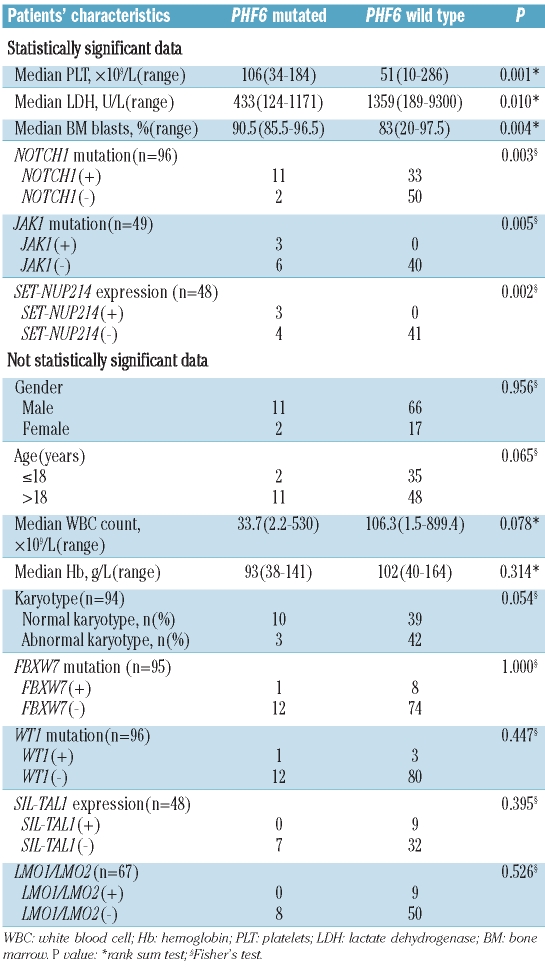
The clinical features of PHF6 mutant and PHF6 wild-type patients are compared in Table 3. PHF6 mutations were significantly associated with lower lactate dehydrogenase serum levels (P=0.01). Cases with PHF6 mutations had a higher platelet count (P=0.001) and higher bone marrow blast count (P=0.004), than cases without PHF6 mutations. No significant differences between the cases with and without PHF6 mutations were found for median white blood cell count (P=0.078) or for median hemoglobin concentration (P=0.314).
Table 3.
Patients’ characteristics and PHF6 mutations.
Of the 96 T-ALL patients undergoing conventional induction chemotherapy, 46/66 patients (69.7%) achieved complete remission. The complete remission rates of cases with and without PHF6 mutations were not significantly different (80% versus 67.9%, respectively; P=0.692). Similarly, no significant difference in 1-year overall survival was found between patients with and without PHF6 mutations (30.0% versus 51.1%, respectively; P=0.059) with a short-term follow-up (median follow-up of 12.5 months; range, 1.0–62 months).
Association of PHF6 mutation with other cytogenetic and molecular abnormalities
Among 96 T-ALL patients undergoing cytogenetic analysis, karyotypic data were available for 94 cases; karyotyping failed in two cases because of lack of metaphases. Among the 94 patients with cytogenetic data, abnormal karyotypes were detected in 45 cases (47.9%). These included numerical changes (6 cases, 13.3%), structural changes (30 cases, 66.7%) and both types of changes (9 cases, 20%). Although PHF6 mutations were more frequent in T-ALL patients with normal karyotypes (10/49, 20.4%) than in those with abnormal ones (3/45, 6.7%), the difference just failed to reach statistical significance (P=0.054).
Patients with PHF6 mutations were more likely than PHF6 wild-type patients to harbor the following: NOTCH1 mutations (84.6% versus 39.8%, P=0.003), JAK1 mutations (33.3% versus 0%, P=0.005) and SET-NUP214 fusions (42.9% versus 0%, P=0.002). (Table 3) There were no differences in the incidence of FBXW7 mutated cases (P=1.000), WT1 mutated cases (P=0.447), and SIL-TAL1 fusion (P=0.395), between patients with and without PHF6 mutations. Primer sequences and PCR conditions are shown in Online Supplementary Table S1.
Discussion
The PHF6 gene is located on Xq26 - a chromosome region undergoing random X-inactivation in female embryos. It has been confirmed that PHF6 does not (like a few other X-linked genes) escape inactivation.18 Germline PHF6 mutations cause BFLS, an X-linked recessively inherited mental retardation syndrome.17 Though X-linked and recessive, BFLS may also occur in women. Remarkably, a 13-fold male excess of PHF6 mutated cases in T-ALL and a 7-fold male excess in acute myeloid leukemia have been reported recently,18,19 implying skewed inactivation of PHF6 mutations, X-homologs in females, or some other unknown mechanism for maintaining PHF6 expression. However, unlike Van Vlierberghe,18 we observed no male excess of PHF6 mutations in T-ALL patients, and although we were able to confirm the reported excess of adult (18.6%) over pediatric (5.4%) cases of PHF6 mutations in T-ALL, both our estimates were significantly lower than the corresponding estimates of 38.1% and 15.7% in previous reports. Further series are needed to clarify the issue, and in particular whether ethnic factors might contribute to the discrepancies in gender risks between the respective series.
PHF6 contains four nuclear localization sequences and two imperfect plant homeodomain (PHD) zinc finger domains suggesting a role for PHF6 in transcriptional regulation and/or chromatin remodeling.17,25 The PHF6 mutations identified in our series were clustered in exons 8, 9, and 10 (Figures 1A, B) while those reported recently in TALL were more evenly distributed. Overall, 77% of the PHF6 mutations were loss of function alleles comprising four frameshift and six nonsense mutations. Missense mutations accounted for the remaining 23% of PHF6 mutations. Most PHF6 mutations (9/13) are described here for the first time: notably, two nonsense mutations (Ala311X and Tyr301X), and two missense mutations (His329Arg and Tyr325His) involving the second (C-terminal) PHD-like domain of PHF6. Two nonsense mutations (Arg274X and Gln270X) and three frameshift mutations (Ser176fs, Lys279fs, and Asp262fs) occurred upstream of the second PHD-like domain causing its loss from the predicted protein (Figure 1A). Clustering of PHF6 mutations in the second PHD-like domain or immediately upstream highlights a possible role for this region in tumor suppression by PHF6.
T-ALL is an aggressive malignancy requiring multiple genetic abnormalities, which are now being identified in increasing numbers thanks to new sequencing technologies. The associations displayed by these pathological alterations already aid disease stratification and should improve intelligent therapeutic targeting. Of note, our study demonstrated that PHF6 mutations were particularly prevalent in T-ALL patients harboring mutations of NOTCH1 and JAK1 and those with a SET-NUP214 rearrangement. NOTCH1 mutation is the most common genetic alteration in T-ALL leading to constitutive activation of NOTCH signaling. JAK1 is mutated in about 20% of adult patients with T-ALL and connotes an unfavorable prognosis. The SET-NUP214 fusion is associated with elevated expression of HOXA genes in T-ALL. We also found that PHF6 mutations were significantly associated with lower serum lactate dehydrogenase levels, higher platelet counts, and higher bone marrow blast counts. Our studies suggest that PHF6 mutations may cooperate with these genetic abnormalities during T-cell leukemogenesis.
Collectively, this study demonstrated PHF6 mutations in a significant proportion of adult T-ALL patients and in a small percentage of pediatric T-ALL patients, occurring less frequently than hitherto reported. PHF6 mutations were clustered inside or immediately before DNA sequences encoding the second (C-terminal) PHD domain highlighting the importance of this region in tumor suppression. We found no difference in the rates of PHF6 mutated cases between male and female patients from China, in contrast to a previous study18 which examined an ethnically undefined variety of patients. Again in contrast to previous findings, we found positive associations between both PHF6 and NOTCH1 mutation, and also JAK1 mutation and rearrangement of SET-NUP214 implying leukemic cooperation. Taken together with previous findings, our data call for closer examination of PHF6 mutations in T-ALL patients of both sexes in ethnically defined populations.
Footnotes
Funding: this work was supported in part by grants from National Key Scientific Projects of China (2011CB933501), the National Natural Science Foundation of China (81070416), the Priority Academic Program Development of Jiangsu Higher Education Institutions, Jiangsu Province Natural Science Foundation (BK2010204), and the Foundation of Jiangsu Province Health Department (H200915).
The online version of this article has a Supplementary Appendix.
Authorship and Disclosures
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.Uckun FM, Gaynon PS, Sensel MG, Nachman J, Trigg ME, Steinherz PG, et al. Clinical features and treatment outcome of childhood T-lineage acute lymphoblastic leukemia according to the apparent maturational stage of T-lineage leukemic blasts: a Children’s Cancer Group study. J Clin Oncol. 1997;15(6):2214–21. doi: 10.1200/JCO.1997.15.6.2214. [DOI] [PubMed] [Google Scholar]
- 2.Kox C, Zimmermann M, Stanulla M, Leible S, Schrappe M, Ludwig WD, et al. The favorable effect of activating NOTCH1 receptor mutations on long-term outcome in T-ALL patients treated on the ALL-BFM 2000 protocol can be separated from FBXW7 loss of function. Leukemia. 2010;24(12):2005–13. doi: 10.1038/leu.2010.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breit S, Stanulla M, Flohr T, Schrappe M, Ludwig WD, Tolle G, et al. Activating NOTCH1 mutations predict favorable early treatment response and long-term outcome in childhood precursor T-cell lymphoblastic leukemia. Blood. 2006;108(4):1151–7. doi: 10.1182/blood-2005-12-4956. [DOI] [PubMed] [Google Scholar]
- 4.Clappier E, Collette S, Grardel N, Girard S, Suarez L, Brunie G, et al. NOTCH1 and FBXW7 mutations have a favorable impact on early response to treatment, but not on outcome, in children with T-cell acute lymphoblastic leukemia (T-ALL) treated on EORTC trials 58881 and 58951. Leukemia. 2010;24(12):2023–31. doi: 10.1038/leu.2010.205. [DOI] [PubMed] [Google Scholar]
- 5.Zuurbier L, Homminga I, Calvert V, te Winkel ML, Buijs-Gladdines JG, Kooi C, et al. NOTCH1 and/or FBXW7 mutations predict for initial good prednisone response but not for improved outcome in pediatric T-cell acute lymphoblastic leukemia patients treated on DCOG or COALL protocols. Leukemia. 2010;24(12):2014–22. doi: 10.1038/leu.2010.204. [DOI] [PubMed] [Google Scholar]
- 6.Marks DI, Paietta EM, Moorman AV, Richards SM, Buck G, DeWald G, et al. T-cell acute lymphoblastic leukemia in adults: clinical features, immunophenotype, cytogenetics, and outcome from the large randomized prospective trial (UKALL XII/ECOG 2993) Blood. 2009;114(25):5136–45. doi: 10.1182/blood-2009-08-231217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawamura M, Ohnishi H, Guo SX, Sheng XM, Minegishi M, Hanada R, et al. Alterations of the p53, p21, p16, p15 and RAS genes in childhood T-cell acute lymphoblastic leukemia. Leuk Res. 1999;23(2):115–26. doi: 10.1016/s0145-2126(98)00146-5. [DOI] [PubMed] [Google Scholar]
- 8.Paietta E, Ferrando AA, Neuberg D, Bennett JM, Racevskis J, Lazarus H, et al. Activating FLT3 mutations in CD117/KIT(+) T-cell acute lymphoblastic leukemias. Blood. 2004;104(2):558–60. doi: 10.1182/blood-2004-01-0168. [DOI] [PubMed] [Google Scholar]
- 9.Van Vlierberghe P, van Grotel M, Beverloo HB, Lee C, Helgason T, Buijs-Gladdines J, et al. The cryptic chromosomal deletion del(11)(p12p13) as a new activation mechanism of LMO2 in pediatric T-cell acute lymphoblastic leukemia. Blood. 2006;108(10):3520–9. doi: 10.1182/blood-2006-04-019927. [DOI] [PubMed] [Google Scholar]
- 10.Weng AP, Ferrando AA, Lee W, Morris JP, 4th, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269–71. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 11.Clappier E, Cuccuini W, Kalota A, Crinquette A, Cayuela JM, Dik WA, et al. The C-MYB locus is involved in chromosomal translocation and genomic duplications in human T-cell acute leukemia (T-ALL), the translocation defining a new T-ALL subtype in very young children. Blood. 2007;110(4):1251–61. doi: 10.1182/blood-2006-12-064683. [DOI] [PubMed] [Google Scholar]
- 12.Malyukova A, Dohda T, von der Lehr N, Akhoondi S, Corcoran M, Heyman M, et al. The tumor suppressor gene hCDC4 is frequently mutated in human T-cell acute lymphoblastic leukemia with functional consequences for Notch signaling. Cancer Res. 2007;67(12):5611–6. doi: 10.1158/0008-5472.CAN-06-4381. [DOI] [PubMed] [Google Scholar]
- 13.Flex E, Petrangeli V, Stella L, Chiaretti S, Hornakova T, Knoops L, et al. Somatically acquired JAK1 mutations in adult acute lymphoblastic leukemia. J Exp Med. 2008;205(4):751–8. doi: 10.1084/jem.20072182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tosello V, Mansour MR, Barnes K, Paganin M, Sulis ML, Jenkinson S, et al. WT1 mutations in T-ALL. Blood. 2009;114(5):1038–45. doi: 10.1182/blood-2008-12-192039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kleppe M, Lahortiga I, El Chaar T, De Keersmaecker K, Mentens N, Graux C, et al. Deletion of the protein tyrosine phosphatase gene PTPN2 in T-cell acute lymphoblastic leukemia. Nat Genet. 2010;42(6):530–5. doi: 10.1038/ng.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutierrez A, Sanda T, Ma W, Zhang J, Grebliunaite R, Dahlberg S, et al. Inactivation of LEF1 in T-cell acute lymphoblastic leukemia. Blood. 2010;115(14):2845–51. doi: 10.1182/blood-2009-07-234377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lower KM, Turner G, Kerr BA, Mathews KD, Shaw MA, Gedeon AK, et al. Mutations in PHF6 are associated with Börjeson-Forssman-Lehmann syndrome. Nat Genet. 2002;32(4):661–5. doi: 10.1038/ng1040. [DOI] [PubMed] [Google Scholar]
- 18.Van Vlierberghe P, Palomero T, Khiabanian H, Van der Meulen J, Castillo M, Van Roy N, et al. PHF6 mutations in T-cell acute lymphoblastic leukemia. Nat Genet. 2010;42(4):338–42. doi: 10.1038/ng.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Vlierberghe P, Patel J, Abdel-Wahab O, Lobry C, Hedvat CV, Balbin M, et al. PHF6 mutations in adult acute myeloid leukemia. Leukemia. 2011;25(1):130–4. doi: 10.1038/leu.2010.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson BJ, Buonamici S, Sulis ML, Palomero T, Vilimas T, Basso G, et al. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med. 2007;204(8):1825–35. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu YM, Zhao WL, Fu JF, Shi JY, Pan Q, Hu J, et al. NOTCH1 mutations in T-cell acute lymphoblastic leukemia: prognostic significance and implication in multifactorial leukemogenesis. Clin Cancer Res. 2006;12(10):3043–9. doi: 10.1158/1078-0432.CCR-05-2832. [DOI] [PubMed] [Google Scholar]
- 22.Van Vlierberghe P, van Grotel M, Tchinda J, Lee C, Beverloo HB, van der Spek PJ, et al. The recurrent SET-NUP214 fusion as a new HOXA activation mechanism in pediatric T-cell acute lymphoblastic leukemia. Blood. 2008;111(9):4668–80. doi: 10.1182/blood-2007-09-111872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Asnafi V, Radford-Weiss I, Dastugue N, Bayle C, Leboeuf D, Charrin C, et al. CALM-AF10 is a common fusion transcript in T-ALL and is specific to the TCRgammadelta lineage. Blood. 2003;102(3):1000–6. doi: 10.1182/blood-2002-09-2913. [DOI] [PubMed] [Google Scholar]
- 24.Chao MM, Todd MA, Kontny U, Neas K, Sullivan MJ, Hunter AG, et al. T-cell acute lymphoblastic leukemia in association with Börjeson-Forssman-Lehmann syndrome due to a mutation in PHF6. Pediatr Blood Cancer. 2010;55(4):722–4. doi: 10.1002/pbc.22574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voss AK, Gamble R, Collin C, Shoubridge C, Corbett M, Gecz J, et al. Protein and gene expression analysis of Phf6, the gene mutated in the Borjeson-Forssman-Lehmann syndrome of intellectual disability and obesity. Gene Expr Patterns. 2007;7(8):858–71. doi: 10.1016/j.modgep.2007.06.007. [DOI] [PubMed] [Google Scholar]