Abstract
The runt-related transcription factor 1, RUNX1, is crucial in the development of myeloid and lymphoid cell lineages and has been reported to be mutated in myeloid malignancies in approximately 30% of cases. In this study, the mutational status of RUNX1 was investigated in 128 acute lymphoblastic leukemia patients. We detected a mutation rate of 18.3% (13 of 71) in patients with T-cell acute lymphoblastic leukemia, 3.8% (2 of 52) in patients with B-cell acute lymphoblastic leukemia and no mutation (0 of 5) in patients with natural killer cell leukemia, respectively. In T-cell acute lymphoblastic leukemia patients, RUNX1 mutations were significantly associated with higher age (P=0.017) and lower white blood cell count (P=0.038). Moreover, an inferior outcome was observed in the subgroup of early T-cell acute lymphoblastic leukemia patients carrying RUNX1 mutations for overall survival (P=0.043). In conclusion, RUNX1 mutations are an important novel biomarker for a comprehensive characterization of T-cell acute lymphoblastic leukemia with poor prognostic impact and have implications for use also in monitoring disease.
Keywords: T-cell ALL, prognosis, RUNX1 mutations
Introduction
Acute lymphoblastic leukemia (ALL) is characterized by an excessive accumulation of lymphoblasts and their progenitor cells.1 ALL is the most frequent childhood cancer and accounts for approximately 25% of adult acute leukemias.1 In approximately 80% of cases, ALL arises from B-cell lineage progenitor cells, whereas 20% are derived from T-cell lineage precursors.2
The diagnosis of ALL is based on immunophenotyping which allows lineage assignment as well as the identification of prognostically important disease subtypes.3,4 Furthermore, chromosomal aberrations have been shown to provide information of great prognostic relevance and are used to stratify patients according to different treatment regimens.1 Today, virtually all patients with ALL can be classified according to specific genetic abnormalities.5 In addition, molecular analyses have shown that ALL subtypes harbor specific gene expression signatures, e.g. depending on the cell lineage or cytogenetic abnormalities,6 carry specific DNA copy number alterations,7 or molecular alterations such as mutations in single genes, e.g. IKZF1.5,8
T-ALL, in particular, has been demonstrated to represent a heterogeneous disease with genetic subgroups that correspond to specific T-cell development stages, genomic alterations leading to deregulated expression of genes, e.g. BAALC, ERG, TAL1 or LMO2, as well as harboring deletions or mutations of genes such as PTEN, NOTCH1 or FBXW7.8
The runt-related transcription factor 1, RUNX1, located on chromosome 21q22, is essential in the development of all hematopoietic cell lineages, including lymphoid cells,9 and contains an 128-amino acid Runt domain (RUNT) and an 80-amino acid transactivation domain (TAD).10 Through the RUNT domain, RUNX1 is able to bind to a common DNA motif of its targets and this domain also mediates the heterodimerization with CBFβ, which increases DNA binding activity to RUNX proteins.10 Furthermore, RUNX1 both increases and inhibits transcriptional activity of target genes, depending on the cellular background and pathway.10 RUNX1 has been reported to be mutated in AML (32%),11 MDS (23%),12 and CMML (37%),13 and is associated with a shorter overall and event-free survival in AML.11,14 Moreover, the gene is involved in a multitude of chromosomal translocations, e.g. the translocation t(12;21)(p13;q22) (ETV6-RUNX1) in B-cell ALL cases, in which the TAD domain but not the RUNT domain of RUNX1 is retained in the fusion gene. In contrast, in the majority of other translocations involving RUNX1, e.g. t(8;21)(q22;q22) (RUNX1-RUNX1T1), the RUNT domain is retained in the fusion genes.15 Truncated RUNX1 proteins act as dominant negative inhibitors of the wild-type protein by binding of DNA through the RUNT domain. Therefore, the wild-type protein loses its ability to regulate target genes.16
In a recent study investigating 39 patients with chronic myeloid leukemia in blast crisis (BC-CML) with either myeloid or lymphoid features, 3 out of 10 patients with a lymphoid BC-CML harbored a RUNX1 mutation indicating a potential role of RUNX1 alterations in lymphatic malignancies which has not yet been discussed.17 Here, we analyzed the RUNX1 mutation status in a cohort of 128 adult patients harboring T-ALL, B-ALL, or natural killer (NK) cell leukemia to further study the impact of RUNX1 alterations in de novo acute lymphoblastic leukemias.
Design and Methods
Peripheral blood or bone marrow mononuclear cells were collected between October 2005 and December 2010 from the purified fraction of mononuclear cells after Ficoll density centrifugation from 128 thoroughly characterized patients with T-ALL (n=71), BALL (n=52), or natural killer (NK) cell leukemia (n=5). T-ALL cases were differentiated by immunophenotyping into early T-ALL (n=30), cortical T-ALL (n=30), and mature T-ALL (n=3). A distinction according to pre-and pro-subtypes is given in the Online Supplementary Spreadsheet 1 (data not available for 8 cases). The expression intensity of T-cell markers, clinical, pathological and cytogenetic data for these patients are also available (Online Supplementary Spreadsheet 1). Study design adhered to the tenets of the Declaration of Helsinki and was approved before initiation by the institutional review board.
Multiparameter flow cytometry (MFC) was performed by applying five-color stainings and isotype controls with monoclonal antibodies conjugated with the fluorochromes FITC, PE, ECD, PC-5, and PC-7. Samples were processed as previously described.18 MFC analyses were performed using FC500 and Navios flow cytometers (Beckman Coulter, Miami, FL, USA). List mode files were analyzed using CXP Software version 2.0 and Kaluza version 1.0 (Beckman Coulter). Diagnoses were assigned according to EGIL and WHO classifications.1,3
For molecular analyses, a sensitive next-generation amplicon deep-sequencing assay (NGS) was applied, using the small volume Titanium amplicon chemistry (454 Life Sciences, Branford, CT, USA). A sequencing library was prepared interrogating the complete coding region of RUNX1, split into 7 distinct PCR amplicons (transcript identifier: ENST00000344691). Data on primer sequences, molecular barcodes used and the PCR amplification protocol are available in the Online Supplementary Tables S1, S2, and S3. NGS data were analyzed using the GS Variant Analyzer Software 2.5.3 (454 Life Sciences) and Sequence Pilot version 3.4.2 (JSI Medical Systems, Kippenheim, Germany). All variants reported were independently validated using Sanger sequencing or a separate setup of NGS PCR, emPCR and sequencing run. Statistical analyses were performed using SPSS version 14.0.1 (SPSS Inc., Chicago, IL, USA). Reported P values are two-sided and not corrected for multiple testing.
Results and Discussion
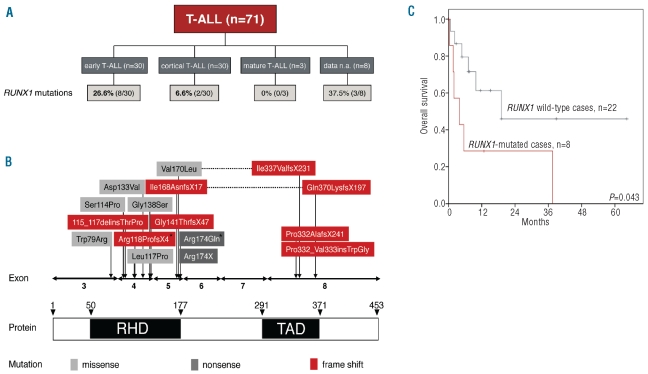
RUNX1 was successfully analyzed in all cases, i.e. in total 896 PCR amplicons were generated for the subsequent characterization by next-generation deep-sequencing. A median of 776 reads per amplicon and patient (range 217–1,654) were obtained thus yielding sufficient coverage for mutation detection with high sensitivity (<5%). Overall, 17 mutations were detected in 15 patients. In the cohort of B-cell ALL, 2 of 52 cases were found to be RUNX1 mutated, both of them exclusively detected in the subgroup of patients harboring a BCR-ABL1 translocation (n=2 of 22 BCR-ABL mutated B-cell ALL). In T-ALLs, 15 distinct mutations were observed in 13 of 71 cases (18.3%). Interestingly, 8 cases were harboring an early T-ALL (8 of 30, 26.6%) and only 2 cases a cortical TALL (2 of 30, 6.6%); subgroup data of 3 RUNX1 mutated cases were not available (Figure 1A).
Figure 1.
(A) Distribution of RUNX1 mutations between the three subgroups of T-ALL. (B) Distribution of RUNX1 mutations in ALL. Location of the 17 mutations in RUNX1 according to the functional domains as detected in 15 patients. Vertical arrows indicate the location of the mutations; corresponding concurrent mutation pairs of the same patient are connected by horizontal lines. Two mutations marked with an asterisk indicate the 2 B-ALL cases. (C) Kaplan–Meier overall survival estimates including 30 early T-ALL cases. Data are shown for overall survival of T-ALL patients separated into two groups of RUNX1-mutated (n=8) and RUNX1 wild-type patients (n=22; alive at two years 28.6% vs. 46.0%).
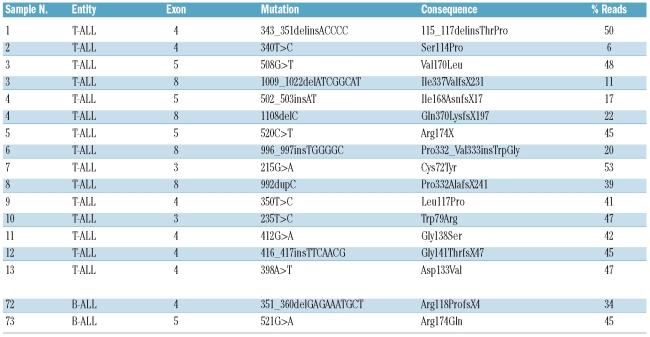
In more detail, 17 different RUNX1 mutations were observed in 15 patients (Figure 1B): 8 missense alterations, one nonsense mutation, 7 frame shift alterations, and one in-frame insertion. Two of the 15 affected patients concomitantly harbored two distinct RUNX1 mutations. In both cases, these were located on two separate amplicons thus not allowing the discrimination between a mono-or biallelic state. As shown in Figure 1B, the RUNX1 mutations were generally distributed across several exons, but exclusively clustered in the RUNT (amino acid 50–177, 13 of 17 mutations) and TAD domain (amino acid 291–371, 4 of 17 mutations). The double-mutated cases were harboring a mutation affecting each of the two domains (Figure 1B).
Table 1.
RUNX1 mutations and functional consequences.
As assessed by the percentage of single sequencing reads carrying the alteration, the median burden of RUNX1 mutations was 42% (range 6–53%) per patient. Information on blast count and clonality as detected by FISH or cytogenetics are given in the Online Supplementary Spreadsheet 1. Moreover, for one patient carrying the Ser114Pro mutation (case n. 2), a paired DNA sample from the remission and relapse state was available. At remission, the Ser114Pro mutation was undetectable, thus confirming the somatic nature as disease-specific alteration at diagnosis. In the relapse sample the mutation had emerged again with a clonality of 10%.
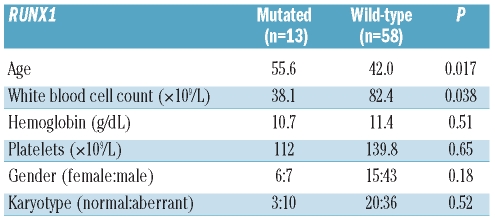
Here, we not only demonstrated for the first time the very high incidence of RUNX1 mutations in T-ALL, but also addressed the clinical impact of RUNX1 mutations detected in great detail. We observed that RUNX1 mutations in TALL were associated with a higher age (mean±SD 42±18 vs. 56±18 years; P=0.017), and lower white blood cell count (mean±SD 82±96 109/L vs. 38±45×109/L cell count; P=0.038), but not with platelet count, hemoglobin level, gender or karyotype (Table 2). Furthermore, a trend for a higher incidence of RUNX1 mutations in the subgroup of early T-ALL was detected (P=0.038).
Table 2.
Clinical associations.
Moreover, the most interesting observation from a clinical point of view was the negative impact of RUNX1 mutations on the overall survival in T-ALL. First, when taking the complete cohort into account, we observed a generally poor survival for RUNX1 mutated cases (alive at two years: 44.4% mutated vs. 66.6% wild-type patients; P=0.011; Online Supplementary Figure S1). Secondly, we confirmed that the cortical immunophenotype had a positive effect on the overall survival of T-ALL patients (including the 63 cases with the known subtype; alive at two years 82.1% vs. 42.8%, P=0.002; Online Supplementary Figure S2).19 A multivariable analysis further demonstrated the cortical T-ALL subtype to be an independent prognostic parameter (P=0.016) and in RUNX1 mutations indicated a strong trend for an independent prognostic impact (P=0.062). Since in the cortical T-ALL patients only 6.6% (2 of 30) were carrying RUNX1 mutations and no mutation was observed in the 3 cases with the mature subtype, we then restricted the analysis of the prognostic impact of RUNX1 mutations to early T-ALL. In this subtype, 26.6% (8 of 30) of patients carried a mutation in RUNX1. Again, mutated RUNX1 conferred a poor prognosis (alive at two years 28.6% mutated vs. 46.0% wild-type patients, P=0.043; Figure 1C).
RUNX1 mutations have so far been described in myeloid malignancies such as AML and MDS.11–13 In our T-ALL cohort, the location of mutations was observed to be similar in comparison to AML and MDS, as well as in families carrying RUNX1 germline mutations predisposing to platelet disorders and other myeloid malignancies.20 Generally, with respect to its involvement in hematopoietic processes, RUNX1 has been shown to play a fundamental part in differentiation and that its role would not be limited to myeloid differentiation only.21 As reported by Wong et al., RUNX1 has been demonstrated to be involved in lymphoid differentiation, which is particularly true for T-cell differentiation.9 Moreover, expression of RUNX1 is observed in naïve CD4-positive T cells and RUNX1 proteins repress the expression of GATA3 and IL4, thereby influencing their further development into Th2 T-helper cells. Our data reporting on 18.3% RUNX1 mutations in T-ALL leading to a disrupted RUNX1 function clearly supports a rationale for a pathogenetic role of RUNX1 alterations in T-ALL. Finally, in B-cell ALL, RUNX1 mutations were exclusively detected in BCR-ABL1 positive cases, which are known to frequently co-express myeloid markers such as CD13 and CD33.20 This may further point to a potential correlation of RUNX1 mutations that can be found not only in myeloid neoplasms but in lymphatic leukemias also demonstrating some myeloid features.
Moreover, it has been proposed that mutated RUNX1 represent a clinically useful biomarker to indicate disease progression from MDS to s-AML,22 as well as a characteristic to monitor minimal residual disease (MRD) during AML treatment.23 While clonal rearrangements of T-cell receptor (TCR) genes also enable MRD assessment; these analyses are time-consuming and expensive.24 As such, in addition to the significant relevance of RUNX1 mutations with respect to survival, monitoring of MRD in T-ALL can also be accomplished by investigating RUNX1 alterations in combination with next-generation deep-sequencing and generating thousands of reads for a patient-specific mutation.
In conclusion, here we have demonstrated for the first time the fundamental importance of RUNX1 mutation assessment in hematologic diseases, not only in myeloid but also in lymphoid malignancies. We detected RUNX1 mutations in 13 of 71 (18.3%) patients with T-ALL and thereby provide a better molecular understanding of underlying disease mechanisms. The high incidence of RUNX1 mutations in early T-ALL may indicate an important role in the early steps of hematopoietic development. However, testing in larger independent patient cohorts is required. Moreover, RUNX1 mutations were significantly associated with higher age and lower white blood cell count and characterized a novel as yet undefined poor prognosis risk group, i.e. in the subtype of early T-ALL. Therefore, RUNX1 mutation screening is proposed for future clinical studies of T-ALL and may thus allow for an even more individualized diagnosis and treatment strategy to be adopted.
Footnotes
The online version of this article has a Supplementary Appendix.
Funding: WK, SS, CH, and TH have equity ownership of MLL Munich Leukemia Laboratory. VG, SH, TA, SJ and AK are employed by MLL Munich Leukemia Laboratory.
Authorship and Disclosures
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. WHO classification of tumours of haematopoietic and lympoid tissues. Lyon, France: IARC; 2008. [Google Scholar]
- 2.Zhao WL. Targeted therapy in T-cell malignancies: dysregulation of the cellular signaling pathways. Leukemia. 2010;24(1):13–21. doi: 10.1038/leu.2009.223. [DOI] [PubMed] [Google Scholar]
- 3.Bene MC, Castoldi G, Knapp W, Ludwig WD, Matutes E, Orfao A, et al. Proposals for the immunological classification of acute leukemias. European Group for the Immunological Characterization of Leukemias (EGIL) Leukemia. 1995;9(10):1783–6. [PubMed] [Google Scholar]
- 4.Gokbuget N, Hoelzer D, Arnold R, Bohme A, Bartram CR, Freund M, et al. Treatment of Adult ALL according to protocols of the German Multicenter Study Group for Adult ALL (GMALL) Hematol Oncol Clin North Am. 2000;14(6):1307–25. ix. doi: 10.1016/s0889-8588(05)70188-x. [DOI] [PubMed] [Google Scholar]
- 5.Pui CH, Carroll WL, Meshinchi S, Arceci RJ. Biology, risk stratification, and therapy of pediatric acute leukemias: an update. J Clin Oncol. 2011;29(5):551–65. doi: 10.1200/JCO.2010.30.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yeoh EJ, Ross ME, Shurtleff SA, Williams WK, Patel D, Mahfouz R, et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1(2):133–43. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- 7.Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446(7137):758–64. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 8.Baldus CD, Martus P, Burmeister T, Schwartz S, Gokbuget N, Bloomfield CD, et al. Low ERG and BAALC expression identifies a new subgroup of adult acute T-lymphoblastic leukemia with a highly favorable outcome. J Clin Oncol. 2007;25(24):3739–45. doi: 10.1200/JCO.2007.11.5253. [DOI] [PubMed] [Google Scholar]
- 9.Wong WF, Kohu K, Chiba T, Sato T, Satake M. Interplay of transcription factors in T-cell differentiation and function: the role of Runx. Immunology. 2011;132(2):157–64. doi: 10.1111/j.1365-2567.2010.03381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen MM., Jr Perspectives on RUNX genes: an update. Am J Med Genet A. 2009;149A(12):2629–46. doi: 10.1002/ajmg.a.33021. [DOI] [PubMed] [Google Scholar]
- 11.Schnittger S, Dicker F, Kern W, Wendland N, Sundermann J, Alpermann T, et al. RUNX1 mutations are frequent in de novo AML with noncomplex karyotype and confer an unfavorable prognosis. Blood. 2011;117(8):2348–57. doi: 10.1182/blood-2009-11-255976. [DOI] [PubMed] [Google Scholar]
- 12.Harada H, Harada Y, Niimi H, Kyo T, Kimura A, Inaba T. High incidence of somatic mutations in the AML1/RUNX1 gene in myelodysplastic syndrome and low blast percentage myeloid leukemia with myelodysplasia. Blood. 2004;103(6):2316–24. doi: 10.1182/blood-2003-09-3074. [DOI] [PubMed] [Google Scholar]
- 13.Kuo MC, Liang DC, Huang CF, Shih YS, Wu JH, Lin TL, et al. RUNX1 mutations are frequent in chronic myelomonocytic leukemia and mutations at the C-terminal region might predict acute myeloid leukemia transformation. Leukemia. 2009;23(8):1426–31. doi: 10.1038/leu.2009.48. [DOI] [PubMed] [Google Scholar]
- 14.Gaidzik VI, Bullinger L, Schlenk RF, Zimmermann AS, Rock J, Paschka P, et al. RUNX1 Mutations in Acute Myeloid Leukemia: Results From a Comprehensive Genetic and Clinical Analysis From the AML Study Group. J Clin Oncol. 2011;29(10):1364–72. doi: 10.1200/JCO.2010.30.7926. [DOI] [PubMed] [Google Scholar]
- 15.Pui CH, Carroll WL, Meshinchi S, Arceci RJ. Biology, risk stratification, and therapy of pediatric acute leukemias: An Update. J Clin Oncol. 2011;29(5):551–65. doi: 10.1200/JCO.2010.30.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Braekeleer E, Douet-Guilbert N, Morel F, Le Bris MJ, Férec C, De Braekeleer M. RUNX1 translocations and fusion genes in malignant hemopathies. Future Oncol. 2011;7(1):77–91. doi: 10.2217/fon.10.158. [DOI] [PubMed] [Google Scholar]
- 17.Grossmann V, Kohlmann A, Zenger M, Schindela S, Eder C, Weissmann S, et al. A deep-sequencing study of chronic myeloid leukemia patients in blast crisis (BC-CML) detects mutations in 76.9% of cases. Leukemia. 2011;25(3):557–60. doi: 10.1038/leu.2010.298. [DOI] [PubMed] [Google Scholar]
- 18.Kern W, Voskova D, Schoch C, Hiddemann W, Schnittger S, Haferlach T. Determination of relapse risk based on assessment of minimal residual disease during complete remission by multiparameter flow cytometry in unselected patients with acute myeloid leukemia. Blood. 2004;104(10):3078–85. doi: 10.1182/blood-2004-03-1036. [DOI] [PubMed] [Google Scholar]
- 19.Chiaretti S, Foa R. T-cell acute lymphoblastic leukemia. Haematologica. 2009;94(2):160–2. doi: 10.3324/haematol.2008.004150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Appelmann I, Linden T, Rudat A, Mueller-Tidow C, Berdel WE, Mesters RM. Hereditary thrombocytopenia and acute myeloid leukemia: a common link due to a germline mutation in the AML1 gene. Ann Hematol. 2009;88(10):1037–8. doi: 10.1007/s00277-009-0722-x. [DOI] [PubMed] [Google Scholar]
- 21.Ito Y. Oncogenic potential of the RUNX gene family: 'overview'. Oncogene. 2004;23(24):4198–208. doi: 10.1038/sj.onc.1207755. [DOI] [PubMed] [Google Scholar]
- 22.Dicker F, Haferlach C, Sundermann J, Wendland N, Weiss T, Kern W, et al. Mutation analysis for RUNX1, MLL-PTD, FLT3-ITD, NPM1 and NRAS in 269 patients with MDS or secondary AML. Leukemia. 2010;24(8):1528–32. doi: 10.1038/leu.2010.124. [DOI] [PubMed] [Google Scholar]
- 23.Kohlmann A, Grossmann V, Schindela S, Kern W, Haferlach C, Haferlach T, et al. Ultra-Deep Next-Generation Sequencing Detects RUNX1 Mutations with Unprecedented Sensitivity and Allows to Monitor Minimal Residual Disease In 116 Samples From MDS and AML Patients Clinically Relevant Abstract. Blood. 2010;116(21):1691a. [Google Scholar]
- 24.Droese J, Langerak AW, Groenen PJ, Bruggemann M, Neumann P, Wolvers-Tettero IL, et al. Validation of BIOMED-2 multiplex PCR tubes for detection of TCRB gene rearrangements in T-cell malignancies. Leukemia. 2004;18(9):1531–8. doi: 10.1038/sj.leu.2403428. [DOI] [PubMed] [Google Scholar]