Acute lymphoblastic and myeloid leukemia (ALL, AML) are neoplasms characterized by a clonal, abnormal and self-maintaining proliferation of hematopoietic cells. These acute leukemias can be categorized into several subtypes, based principally on phenotype and according to specific genetic abnormalities. Nowadays, the outcome of most pediatric patients with acute leukemia is favorable; 5-year event-free survival is 76–86% for ALL and 49–63% for AML1.1 However, new therapies are needed to cure all leukemia patients and to reduce the burden of treatment-related complications. To this end, the development of new drugs, as well as a better understanding of the biology of these malignancies, is warranted.
The comprehension of leukemogenesis passes through the identification of the genetic alterations causing abnormal proliferation of cancer cells. Molecular characterization of pediatric leukemias revealed a number of recurrent genetic lesions, and the advent of the large-scale DNA re-sequencing has allowed the identification of new somatically mutated genes.2 Recently, improvements in sequencing and bioinformatic technologies have provided the means of performing analyses of whole cancer genomes from AML adult patients and somatic mutations have been found at the DNA methyltransferase 3a (DNMT3A) gene. Twenty-two percent of adult AML showed DNMT3A mutations; in particular, the substitution at the R882 codon represents 15% of all mutations identified.3 DNMT3A recurrent somatic mutations have been described also in a significant proportion of patients with myelodysplastic syndromes4 and acute monocytic leukemia.5
Two studies reported the frequency of DNMT3A mutations in childhood AML. A report from Thol et al.6 found a total of 2 patients with a mutation in codon R882 in a cohort of 195 pediatric AML patients, and the Children’s Oncology Group identified no DNMT3A mutation in a cohort of 180 children with AML, showing that DNMT3A mutations are rare in childhood AML.7
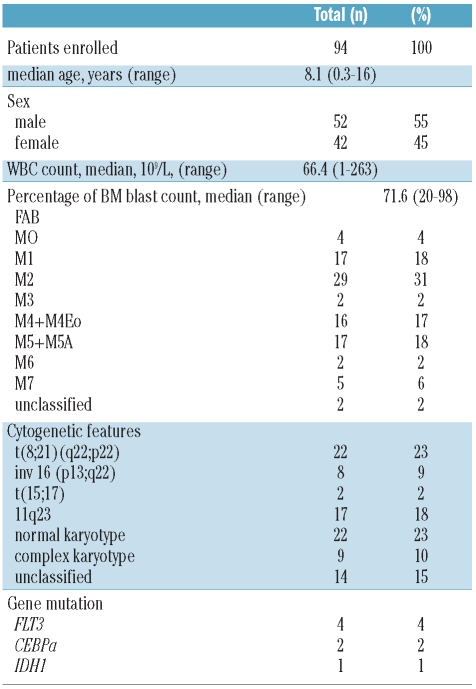
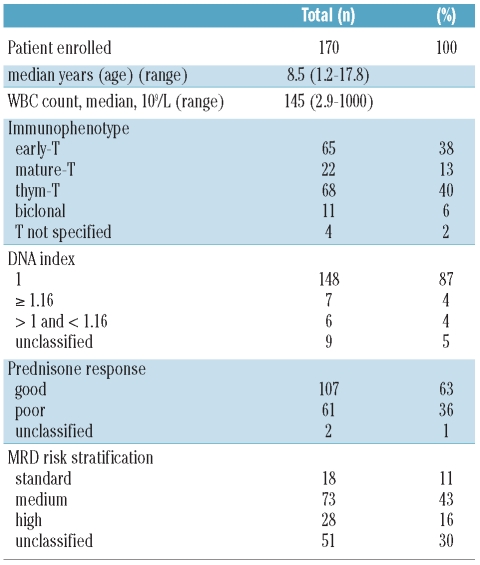
Here, we investigated the mutational status of DNMT3A exon 23 in a pediatric cohort of 264 Italian patients. For this analysis, 94 pediatric patients with AML and 170 pediatric patients with T-ALL (Tables 1 and 2) were selected on the basis of availability of DNA samples. Pediatric patients with diagnosis of de novo AML or T-ALL were enrolled in the AIEOP (Italian Association of Pediatric Hematology and Oncology) treatment protocols.8 Bone marrow (BM) samples of children with AML and T-ALL were collected and stored in the national reference laboratories from 2000 to 2008 and from 2000 to 2006, respectively. Morphological evaluation, immunophenotyping and immunoglobulin and T-cell receptor (TCR) rearrangement analysis were performed at the laboratory of Pediatric Hematology of the University-Hospital in Padova and at the San Gerardo Hospital in Monza; these are both centralized national reference laboratories. In accordance with the principles of the Declaration of Helsinki, consent was obtained from parents or legal guardians of all study participants. Both cohorts, AML and T-ALL, were chosen because recent publications showed that these leukemias shared biological features, such as several translocations and somatically acquired mutations in crucial genes, e.g. RAS, WT1, FLT3, NF1, and PHF6.9 Moreover, aberrant methylation events have recently been identified as being highly relevant also in T-ALL since they are associated with clinicopathological features, as is already known for AML.10 We performed a comprehensive analysis of mutations occurring in the “hot-spot” locus of DNMT3A, by processing and sequencing the target region of exon 23 using the Roche 454-Junior platform with the Universal Tailed Amplicon Sequencing. Its experimental design allows the same amplicon to be rapidly sequenced in a large number of samples. Since the technology of 454 amplicon sequencing is still relatively new, we also analyzed 20 adult AML patients at diagnosis as putative positive controls. We obtained 271,253 sequences aligned with DNMT3A exon 23. The average number of sequences per patient was 965±446 for 281 out of the 284 (98.9%) patients studied. None of the pediatric AML or TALL patients analyzed showed mutations at the “hot-spot” DNMT3A region. By contrast, 3 out of the 20 (15%) AML adult patients carried a mutation in exon 23 of DNMT3A, confirming a mutational rate similar to that previously described.3 All three mutations occurred at amino acid R882: 2 patients had the R882H mutation, while one patient carried the R882P mutation. These mutated sequences were present with mean frequencies of 46.16±2.9%, indicating the heterozygous status of the mutation.
Table 1.
Clinical features of AML pediatric patients.
Table 2.
Clinical features of T-ALL pediatric patients.
Our results confirmed that also in our Italian pediatric AML patients, DNMT3A mutations at the R882 locus are rare; moreover, we highlighted the fact that DNMT3A is also not mutated in pediatric T-ALL. We enhanced mutational analysis of methyltransferases by sequencing DNMT3B, also a member of the DNMT3 family of genes.5,11 DNMT3B is considered functionally complementary to DNMT3A with common and differential target specificities, and may be considered to play a role in the aberrant methylation observed in acute leukemia.12 The mutational screening was performed in some of our pediatric cohort. In particular, 60 T-ALL patients and 90 AML patients were sequenced at the homolog amino acid R823 at exon 23 of DNMT3B and no mutations were found, suggesting that neither methylase provides information about pediatric leukemia at the focused genomic locus.
Specifically, the difference between genetic alterations observed in childhood leukemia with respect to adults confirmed that only in a few cases were genes found to be mutated from adult whole-genome sequencing relevant for childhood AML. It is to be expected that other genes play a key role in the pathogenesis of childhood leukemia, and most likely in other pediatric solid tumors. These will be identified in extended somatic whole-genome sequencing of pediatric specimens of diseased cells. This study also highlights how recent improvements in sequencing and bioinformatic technologies provide new, rapid and extremely reliable methods to provide information on gene mutation status in large cohorts of patients. This opportunity to genetically characterize patients opens a new era of study in which the discovery of specific lesions might improve our knowledge on leukemogenesis and allow specific gene lesions to be therapeutically targeted.
Acknowledgments
We thank Emanuela Giarin, Sabrina Gelain, Alessandra Beghin, Samuela Francescato, Barbara Buldini, Anna Leszl, Elena Seganfreddo, Barbara Michielotto, Maria Grazia Giacometti, Silvia Disarò and Katia Polato for their collaboration.
Footnotes
Funding: this study was supported by grants from the Fondazione Città della Speranza-Padova, University of Padova, European Union (EU FP7-228971), Fondazione Veneto Banca and AIL.
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.Pui CH, Carroll WL, Meshinchi S, Arceci RJ. Biology, risk stratification, and therapy of pediatric acute leukemias: an update. J Clin Oncol. 2011;29(5):551–65. doi: 10.1200/JCO.2010.30.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong SA, Look AT. Molecular genetics of acute lymphoblastic leukemia. J Clin Oncol. 2005;23(26):6306–15. doi: 10.1200/JCO.2005.05.047. [DOI] [PubMed] [Google Scholar]
- 3.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363(25):2424–33. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walter MJ, Ding L, Shen D, Shao J, Grillot M, McLellan M, et al. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia. 2011;25(7):1153–8. doi: 10.1038/leu.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan XJ, Xu J, Gu ZH, Pan CM, Lu G, Shen Y, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet. 2011;43(4):309–15. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- 6.Thol F, Heuser M, Damm F, Klusmann JH, Reinhardt K, Reinhardt D. DNMT3A mutations are rare in childhood acute myeloid leukemia. Haematologica. 2011;96(8):1238–40. doi: 10.3324/haematol.2011.046839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho PA, Kutny MA, Alonzo TA, Gerbing RB, Joaquin J, Raimondi SC, et al. Leukemic mutations in the methylation-associated genes DNMT3A and IDH2 are rare events in pediatric AML: A report from the Children’s Oncology Group. Pediatr Blood Cancer. 2011;57(2):204–9. doi: 10.1002/pbc.23179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pession A, Rondelli R, Basso G, Rizzari C, Testi AM, Fagioli F, et al. Treatment and long-term results in children with acute myeloid leukaemia treated according to the AIEOP AML protocols. Leukemia. 2005;19(12):2043–53. doi: 10.1038/sj.leu.2403869. [DOI] [PubMed] [Google Scholar]
- 9.Van Vlierberghe P, Patel J, Abdel-Wahab O, Lobry C, Hedvat CV, Balbin M, et al. PHF6 mutations in adult acute myeloid leukemia. Leukemia. 2011;25(1):130–4. doi: 10.1038/leu.2010.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takeuchi S, Matsushita M, Zimmermann M, Ikezoe T, Komatsu N, Seriu T, et al. Clinical significance of aberrant DNA methylation in childhood acute lymphoblastic leukemia. Leuk Res. 2011;35(10):1345–9. doi: 10.1016/j.leukres.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamashita Y, Yuan J, Suetake I, Suzuki H, Ishikawa Y, Choi YL, et al. Array-based genomic resequencing of human leukemia. Oncogene. 2010;29(25):3723–31. doi: 10.1038/onc.2010.117. [DOI] [PubMed] [Google Scholar]
- 12.Shah MY, Licht JD. DNMT3A mutations in acute myeloid leukemia. Nat Genet. 2011;29;43(4):289–90. doi: 10.1038/ng0411-289. [DOI] [PubMed] [Google Scholar]