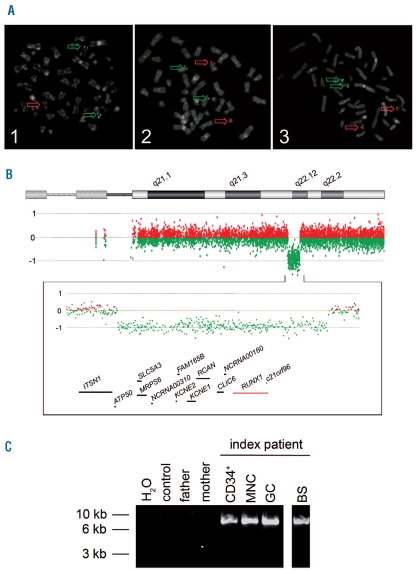
Figure 1.
(A) ETV6/RUNX1 metaphase fluorescence in situ hybridization (FISH). Metaphase plates of PHA-stimulated peripheral blood cells of the index patient (1) and his parents (2, 3) were investigated using specific probes for RUNX1 (LSI AML1, 21q22, red) and ETV6 (LSI TEL, 12p13, green) (Vysis, Abott, LSI ETV6(TEL)/RUNX1(AML1) ES dual color translocation probe set, Wiesbaden, Germany). While metaphases of the index patient (1) displayed one red and two green signals indicative of a loss of one RUNX1 allele, the metaphases of the parents (2, 3) showed two red and two green signals. Interphase FISH analysis of 100 nuclei displayed the submicroscopic loss of one RUNX1 allele in 93% of cell nuclei investigated. (B) High resolution oligo array comparative genomic hybridization (aCGH) of the index patient. DNA of peripheral blood cells was analyzed using a 400k oligo array following the manufacturer’s instructions (Agilent Technologies, Boeblingen, Germany). In comparison to a healthy control sample, aCGH displayed a 1.6 Mb deletion in the long arm of a chromosome 21 of the index patient which is probably due to a de novo rearrangement: arr 21q22.11q22.12(35,304,856–36,864,010)x1 dn. Below the ideogram of chromosome 21 from 21pter to 21qter, the aberration states based on normalized log2 transformed fluorescent intensity ratios are shown indicating the heterozygous interstitial deletion. Further on, an enlarged view of the aberrant region is given (chromosome 21: 34,912,522–37,106,477; 2.19Mb) displaying genes located within the region lost as well as some of the neighboring genes. The RUNX1 locus is highlighted in red. (C) Breakpoint spanning long-distance PCR. Using primers located within the last aCGH probes before and after the deletion detected, a breakpoint spanning PCR product of approximately 9 kb was generated in the index patient (lanes 5–8) while no product was seen in peripheral blood DNA samples of his parents (lane 3, 4) and a healthy control (lane 2). Analyses of DNA of several peripheral blood cell populations of the index patient displayed the deleted allele in CD34+ cells (CD34+, lane 5), cells of a mononuclear (MNC, lane 6), and a granulocytic cell fraction (GC, lane 7). Finally, in DNA of a buccal swab of the patient, the PCR product could also be amplified indicating the germline origin of the submicroscopic deletion in 21q (BS, lane 8). The identified de novo deletion is probably the result of a recombination between two L1PA2 long interspersed nuclear elements (LINE) that lie in the breakpoint region and display a sequence identity of 97%.