Abstract
The diverse functional roles that proteases play in basic biological processes make them essential for virtually all organisms. Not surprisingly, proteolysis is also a critical process required for many aspects of pathogenesis. In particular, obligate intracellular parasites must precisely coordinate proteolytic events during their highly regulated life cycle inside multiple host cell environments. Advances in chemical, proteomic and genetic tools that can be applied to parasite biology have led to an increased understanding of the complex events centrally regulated by proteases. In this review, we outline recent advances in our knowledge of specific proteolytic enzymes in two medically relevant apicomplexan parasites: Plasmodium falciparum and Toxoplasma gondii. Efforts over the last decade have begun to provide a map of key proteotolyic events that are essential for both parasite survival and propagation inside host cells. These advances in our molecular understanding of proteolytic events involved in parasite pathogenesis provide a foundation for the validation of new networks and enzyme targets that could be exploited for therapeutic purposes.
Keywords: apicomplexans, proteases, pathogenesis, Plasmodium, Toxoplasma, malaria, invasion, egress
1. Introduction
The primary function of all proteases is to catalyze the simple process of peptide bond hydrolysis. While this process is the same for all proteases, each family utilizes a different general mechanism to accomplish this goal. Although proteases were originally thought to act as downstream mediators of protein turn-over, we now know that distinct proteolytic processing events can also regulate or initiate key biological processes such as cell death [1], cell cycle progression [2] and cell migration [3]. Given that proteases regulate such basic biological processes, it is not surprising that apicomplexan parasites, with their complex life cycles, also depend on proteolytically regulated processes. Decades of research in parasite biology have begun to define the roles of proteases in all stages of the parasite life cycle. In this review, we will provide a general overview of the current knowledge of proteases found in the apicomplexan parasites Plasmodium spp. and Toxoplasma spp, with special focus on three important cellular events: host-cell invasion, general catabolism and host cell rupture (also known as egress). While the roles of proteases in various aspects of parasite biology have been reviewed in the past [4–7] we will focus on recent discoveries that have contributed to our understanding of critical proteolytic events in these two important human pathogens.
The apicomplexan parasites: Plasmodium and Toxoplasma
The Apicomplexa comprise a phylum of highly diverse eukaryotic protozoa that exists as parasites in animals. Apicomplexans are morphologically characterized by the presence of an eponymous apical complex of organelles consisting of micronemes, rhoptries and dense granules, as well as other less-well characterized secretory organelles such as exonemes. These organelles contain proteins that are secreted around the time of egress and invasion to facilitate parasite motility, host cell adhesion and subsequent remodeling of host cells (see [8] for detailed review). These parasites also have common, yet parasite-specific features such as a chloroplast-like organelle termed the apicoplast that has a role in fatty acid and isoprenoid synthesis [9]. Plasmodium spp., the causative agent of malaria, affects 500 million worldwide and therefore attracts a considerable amount of medical and research attention [10]. Classically at least five species of Plasmodium infect humans: Plasmodium vivax, P. ovale, P. malariae, P. knowlesi and P. falciparum. Each species has a highly regulated life cycle inside the intermediate human host as well as inside the definitive insect vector. In addition all Plasmodium spp. within the intermediate host primarily infect red blood cells. Toxoplasma gondii is related to Plasmodium spp. however a notable difference is that T. gondii infects virtually any nucleated host cell. Up to one third of the entire population of the world is infected by T. gondii [11] and although primary infection by T. gondii is typically subclinical, it can cause serious illness in immuno-compromised patients and lead to fetal death if acquired during pregnancy [12].
Both Plasmodium and Toxoplasma progress through a complicated series of sexual and asexual life cycles where non-replicative extracellular forms of the parasites known as zoites first invade host cells, grow and replicate within these cells, and finally rupture to establish further infections (Figure 1). [13, 14] This variety of molecular events requires the expression of many specialized proteins to coordinate these diverse biochemical activities. Recent efforts have revealed proteases as crucial players in all stages of the parasite life cycle. The roles of various proteases in Plasmodium and Toxoplasma will be reviewed here, but the importance of proteases is not limited to the apicomplexans, and their roles in the progression of other parasitic diseases have been previously reviewed [15].
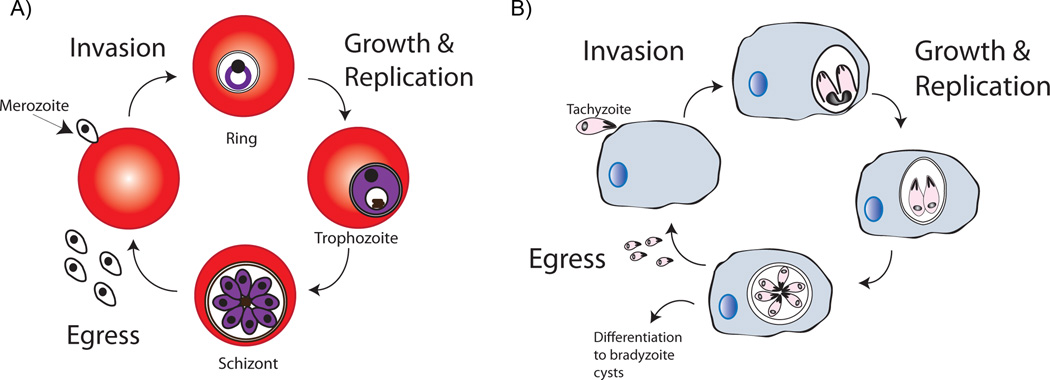
Figure 1.
Asexual life cycle of Plasmodium (A) and Toxoplasma B). During invasion, polar zoite cells randomly establish contact to host cells, followed by reorientation of apical end to host cell membrane. A tight junction forms between the parasite and host membrane, and this junction moves from the apical to the posterior end of the parasite. This results in formation of an invagination which eventually forms the parasitophorous vacuole. This is accompanied by shedding of surface proteins. Once invasion is complete, parasites rapidly grow in size in the intracellular environment, and replicate their genome to prepare for cellular division. When cell division is complete, parasites exit host cells in a process known as egress to invade other cells. In Toxoplasma, chronic infection can occur via transformation of the active tachyzoites to the inactive form of parasites known as bradyzoites which form tissue cysts in hosts.
2. Role of proteases in host-cell invasion
As obligate intracellular parasites, it is vital for the extra-cellular forms of Plasmodium and Toxoplasma to rapidly invade host cells. This process of invasion occurs within a very short time frame because the extracellular forms cannot survive for long periods of time outside host cells. Invasion proceeds following an initial contact by the polar zoite, which reorients such that its apical end contacts the host cell membrane [16, 17]. A tight junction that results from the interaction of multiple receptor proteins forms between the apposed parasite and host membranes, and this junction moves from the apical to the posterior end of the zoite. The active trafficking of the receptor proteins on the parasite surface drags the host cell membrane over the parasite, creating an invagination within the host cell [5, 6]. As invasion proceeds, the zoite must shed the surface proteins initially required for attachment and penetration. Invasion is complete when the parasite is fully engulfed within the parasitophorous vacuole, which seals to create a protective environment where it can subsequently grow and replicate (see Figure 2, Step 1).
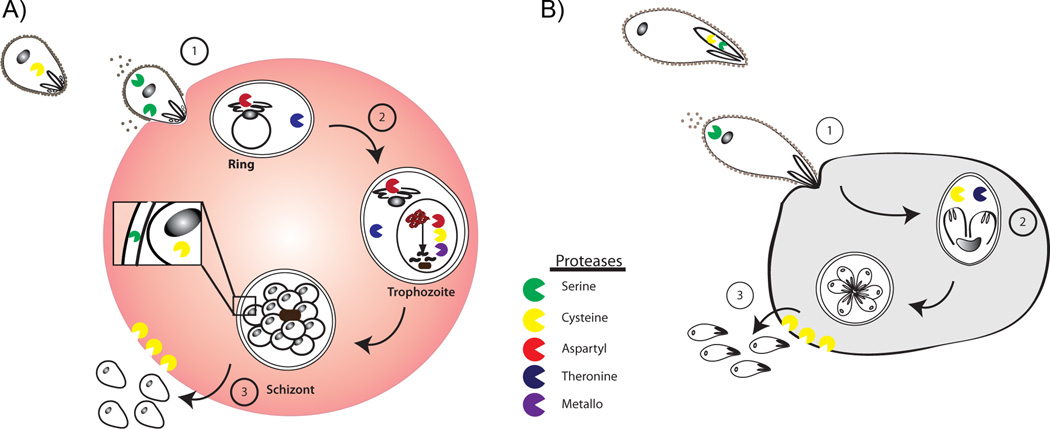
Figure 2.
Schematic depicting the role of proteases in different stages of the parasite life cycle in A) Plasmodium falciparum and B) Toxoplasma gondii. Different classes of proteases are represented in the corresponding color. Proteases are involved in proteolytic maturation of apical organelle proteins prior to invasion (not shown in figure). During invasion (1), the proteins from the apical organelles can facilitate adhesion to the host cell, and these proteins are subsequently shed in a protease-dependent manner. Proteases are also involved in general metabolism (2) to allow growth and replication. After several rounds of cell division, host cells rupture to release parasite zoite cells (3) which then re-invade other host cells and repeat the life cycle.
Many apical surface proteins have been confirmed to be required for successful attachment [7, 18, 19]. In Plasmodium, proteins such as merozoite surface proteins (MSPs), apical membrane antigen 1 (AMA1), rhoptry neck proteins (RONs) and serine repeat antigen (SERA) proteins are thought to play important roles in parasite invasion. In addition, many of these proteins are proteolytically processed either during trafficking to, or arrival at, the parasite surface. However, until recently the identities of the majority of proteases that regulate the maturation and subsequent shedding of the surface proteins during invasion were largely unknown.
2.1 Subtilases
The subtilisin-like serine proteases are a class of enzymes present in both P. falciparum and T. gondii. The P. falciparum genome project identified three subtilases but only two, PfSUB1 and PfSUB2, are known to have functional roles in the blood-stages of the parasite life cycle [20]. PfSUB1 has roles in both egress (see section 4.2) and invasion. This protease has a high degree of substrate specificity [21] and its expression peaks in schizont stage [22]. It was initially postulated that PfSUB1might be involved in the shedding of micronemal proteins from the surface of the parasite during invasion. However selective inhibitors of PfSUB1 do not interfere with shedding of surface proteins MSP-1 and AMA-1 [6], but can efficiently block invasion [23]. The effects of PfSUB1 inhibitors were suggested to be the result of a block in processing of surface proteins required for productive invasion. PfSUB1 was subsequently shown to effectively process MSP1/6/7 and that the maturation of these surface proteins was required for successful invasion [24]. A detailed bioinformatic and proteomic analysis has identified additional proteins processed by PfSUB1 that are putatively involved in egress and invasion [25].
While PfSUB1 is involved in maturation of MSP1 before merozoite rupture, PfSUB2 functions as a sheddase during merozoite invasion. PfSUB2 localizes to the micronemes, and is secreted and translocated to the posterior end of parasite surface during invasion [26]. During this translocation, PfSUB2 proteolytically cleaves the ectodomain of MSP1 and AMA1. This processing may disengage adhesion complexes to release the parasite into the parasitophorous vacuole. Evidence also suggests that PfSUB2 is responsible for the proteolytic processing of Plasmodium thrombospondin related apical merozoite protein (PTRAMP), another adhesive protein on the merozoite surface [27]. Similar to MSP1 and AMA1, PTRAMP is shed from the merozoite during erythrocyte invasion.
Given the relatedness of T. gondii and P. falciparum, it is not surprising that subtilisin-like proteases also have important functions in host cell invasion by Toxoplasma [28, 29]. Using a gene disruption strategy to knock out the sub1 gene in tachyzoites, Lagal and co-workers [30] showed that loss of GPI-anchored protease TgSUB1 results in defective surface processing of micronemal proteins MIC2, MIC4 and M2AP. Since secretion of mature forms of these proteins is important for attachment to the host cell, tachyzoites lacking TgSUB1 have a reduced ability to invade. Furthermore, deletion of the sub1 gene results in parasites with a defect in gliding motility, a key process required for invasion of the host. The highly related T. gondii subtilase, TgSUB2, also has been linked to invasion. TgSUB2 is a transmembrane protein that localizes to the rhoptry organelle in tachyzoites [31]. Attempts to disrupt the Tgsub2 gene suggest that it is essential in the T.gondii life cycle. Interestingly, the peptide sequence where autocleavage occurs within the TgSUB2 protein matches that of cleavage sites of other rhoptry protein (e.g. ROP1). This, combined with its known rhoptry localization, is suggestive of a role for TgSUB2 in proteolytic maturation of rhoptry proteins.
2.2 Rhomboid proteases
The Rhomboid family of serine proteases was first discovered in Drosophila [32], and it soon became apparent that these intra-membrane proteases are conserved in both prokaryotic and eukaryotic organisms [33, 34]. The rhomboid proteases have a mechanism of proteolysis that is unique from the soluble serine proteases. Specifically, the protease active site is buried in the plane of the membrane, and proteolysis occurs in, or adjacent to, the transmembrane domain of the substrate [35].
In Toxoplasma, proteomic analysis of invasion-specific cleavage events first hinted at a role for rhomboid proteases in surface protein shedding [36]. Six rhomboid-like genes were identified and five rhomboid proteases, TgROM1–5, were subsequently cloned and characterized [37, 38]. These rhomboid proteases have non-overlapping substrate specificities and localize to different regions of the cell. Of these, TgROM2, TgROM4 and TgROM5 function as sheddases of micronemal adhesin proteins during the process of invasion. Recent work has uncovered a crucial role for TgROM4 in invasion as a result of processing of surface adhesins MIC2, MIC3 and AMA1 [39]. This cleavage of AMA1 from the T. gondii surface during invasion, in turn, triggers a switch of the parasite from an invasive to replicative state [40]. TgROM5, which like TgROM4, is localized to the cell surface of the tachyzoite, cleaves the adhesion protein MIC2 upon its trafficking to the posterior end of the parasite membrane during invasion. The rhomboid proteases in T.gondii thus collectively serve as sheddases that remove adhesion proteins to allow successful host cell invasion. However, it is perhaps worthwhile to note that rhomboid proteases may have functions beyond those required for invasion. For example, TgROM1 has been shown to contribute to intracellular growth of the parasite [41].
Not long after the characterization of TgROMs, rhomboid proteases were identified in P. falciparum and shown to also function in shedding of adhesion proteins during invasion. PfROM1 and PfROM4 are able to cleave a variety of substrates that possess a transmembrane domain, and they are likely to be involved in all invasive stages of P. falciparum [42–44]. PfROM1 has similar substrate preference as the Drosophila Spitz-cleaving rhomboid proteases, while PfROM4 is distinct in that it does not cleave Spitz-type substrate motifs and instead cleaves erythrocyte binding-like (EBL) adhesins [42]. Collectively, these results establish a role for rhomboid proteases in apicomplexan invasion. Further studies will be required to characterize the specific targets of each rhomboid protease in the life cycle of the parasites.
2.3 The cysteine proteases
Although cysteine protease inhibitors have convincingly been shown to block parasite invasion, only a few candidate proteases have been identified. In P. falciparum, the identification of selective inhibitors of falcipain-1, through screening of epoxide inhibitor libraries, provided a tool that implicated this protease in erythrocyte invasion [45]. Unlike the other falcipains that are involved in hemoglobin degradation, falcipain-1 localizes to distinct, granule like structures in newly formed merozoites. However, subsequent gene disruption of falcipain-1 suggests that its role in invasion may be somewhat redundant [46, 47]. The fact that broad-spectrum cysteine protease inhibitors block invasion suggests that yet undiscovered cysteine proteases may function together to ensure successful invasion.
In T. gondii, a cathepsin B homologue TgCPB (also known as toxopain-1 or TgCP1), may play a role in tachyzoite invasion by processing rhoptry proteins [48]. Immunoelectron microscopy using cathepsin B antibodies provides some evidence that TgCPB might be localized to the rhoptries. The use of small molecule inhibitors of cathepsin B have also been shown to disrupt rhoptry protein maturation resulting in abnormal rhoptry development and defects in tachyzoite invasion [49]. However, the specificity of the compounds used in these studies has not been confirmed suggesting that effects could be mediated by other targets. In particular, T. gondii expresses a highly homologous cathepsin L, TgCPL, which has recently been shown to localize to a specialized secretory organelle termed the vacuolar compartment (VAC) where it is believed to play a role in maturation of secretory proteins targeted to the micronemes [50]. In particular, T. gondii deficient for TgCPL showed slower maturation of TgM2AP and TgMIC3 proproteins. As TgM2AP and TgMIC3 are required for efficient host cell invasion, it is not surprising that the TgCPL-deficient strain has reduced ability to invade host cells. Although only a limited number of cysteine proteases have been confirmed to have a role in invasion, recent tools such as the complete genome sequence of P. falciparum and T. gondii, in combination with the use of chemical inhibitors, will hopefully aid in the discovery and characterization of additional important cysteine proteases.
2.4 The metalloprotease Toxolysin 4
The metalloprotease toxolysin 4 contains a signature motif present in insulin-degrading enzymes [51]. This protease localizes to the micronemes, and its secretion coincides with discharge of micronemal contents. Thus, it is possible that toxolysin 4 can function in invasion. However, further studies will be required to assign specific roles to this protease.
3. Proteases in general catabolism
Once invasion has occurred, parasites actively metabolize host cell proteins to generate products required for growth and replication. In Plasmodium, this process has been extensively studied due to its potential as a target pathway for malaria drug-therapy (See Figure 2, Step 2) [52].
3.1 Hemoglobin Degradation in Plasmodium
During the intraerythrocytic stage of the Plasmodium life cycle, the parasite imports host cell hemoglobin and then progressively degrades it to generate key nutrients as the parasite grows. Degradation of hemoglobin provides free amino acids, frees up space for further growth, and also has a possible role in regulating the osmotic status of the cell [53]. This process occurs mainly in the trophozoite stage, during which time hemoglobin is endocytosed into a parasite organelle known as the food vacuole. Once inside this acidic compartment, a cascade of aspartyl and cysteine proteases degrade the hemoglobin to the final amino acid products [54].
The extended family of plasmepsin aspartyl proteases in P. falciparum has been the focus of intensive research mainly due to the parasite-specific nature of these enzymes [55]. Therefore, plasmepsins were initially thought to be ideal targets for development of anti-parasitic drugs. The plasmepsins are a family of aspartyl proteases made up of 4 main members: plasmepsins I, II, IV and histo-aspartic protease [56, 57]. Plasmepsin I and II are thought to initiate hemoglobin degradation, while histo-aspartic protease and plasmepsin IV function in the degradation of peptides generated by the action of upstream members of the family [58–61]. However, the fact that genetic disruption of individual plasmepsins did not result in any difference in P. falciparum morphology suggests that functional redundancy between the family members may make these proteases non-optimal as therapeutic targets [62]. Nevertheless, plasmepsins are important proteases in hemoglobin degradation, especially since a triple or quadruple plasmepsins gene knock-out confers a growth defect in P. falciparum [63].
Other proteases such as the metalloprotease falcilysin [64–66], and the cysteine proteases falcipains 2, 2’ and 3 [67–70] and dipeptidyl aminopeptidase 1 (DPAP1), are also involved in degradation of hemoglobin after it has been cleaved into peptide fragments [71]. Falcipains are thought to function downstream of plasmepsin I and II, and evidence suggests that these proteases are involved in activation of the food vacuole plasmepsins [72]. Gene disruptions as well as studies using small molecule inhibitors of falcipain-2, have confirmed a role for this protease in hemoglobin degradation [73–75]. Furthermore, falcipains may be able to replace the functions of plasmepsins [62] making them promising targets for antimalarials [76].
The cysteine dipeptidyl aminopeptidase 1 (DPAP1) is an exopeptidase in the food vacuole [71]. DPAP1 is a homologue of mammalian cathepsin C that also removes dipeptides from protein substrates. DPAP1 functions in an acidic environment (the food vacuole) and has similar substrate specificity as cathepsin C [77]. Studies using specific inhibitors of DPAP1 have shown that inhibition of this protease leads to an accumulation of parasites at the trophozoite stage and a subsequent decrease in parasitemia [78]. These inhibitors were also tested in a mouse model of malaria and found to reduce parasitemia. Furthermore, by linking DPAP1 inhibitors to a synthetic analog of another effective antimalarial drug, it was possible to enhance killing of parasites in culture [79]. These observations, combined with the essential nature of DPAP1 expression in blood stage parasites [71], suggest that it plays a critical role in parasite survival and is therefore a potentially valuable drug target.
The final stages of hemoglobin degradation are carried out by aminopeptidases that generate single amino acids from cleaved peptides [80]. An additional family of neutral amino-peptidases PfA-M1 and PfM17LAP [81–83] has recently been suggested to degrade peptide substrates that are exported to the cytosol from the food vacuole to generate free amino acids [84–86]. These proteases may also represent promising new targets for the development of anti-malarial drugs, which may be facilitated by the recent structural characterization of members of this family [87].
3.2 Proteases involved in general peptide processing
Proteases that perform essential roles such as protein homeostasis, protein transport and host cell remodeling are also potentially valuable as targets for theraputic intervention. As these processes are required throughout the parasite life cycle, drugs that block the action of these proteases have the potential to be effective at multiple points during an infection.
The proteasome is a large multi-subunit complex that catalyzes the proteolysis of proteins that have been targeted for degradation by ubiquitination. This protease complex is conserved across the eukaryota and is also found in some prokaryotes [88]. The proteasome is involved in regulatory functions such as protein turnover and degradation of misfolded proteins. In mammalian cells, it also regulates cell cycle progression and inflammation pathways [89]. Furthermore, a small molecule proteasome inhibitor has been approved by the FDA for the treatment of multiple myeloma, suggesting that targeting the proteasome could be an effective way to treat other diseases [90].
Initial studies using the natural product lactacystin, which inhibits P. falciparum proteasome at nanomolar concentrations, confirmed that a block in proteasome activity leads to parasite death [91]. However, these initial studies also pointed out the overall toxicity that resulted from inhibition of the host proteasome. More recent studies with several different classes of proteasome inhibitors suggest that the proteasome may be a viable target in Plasmodium spp. [92–94]. Furthermore, at least two proteasome inhibitors – epoxomicin and thiostrepton – also reduce growth of the sexually differentiated Plasmodium gametocyte, suggesting that inhibition of the proteasome could be an effective way to block parasite transmission [95, 96]. Unfortunately, relatively little is known about the biochemical or structural properties of the proteasome from Toxoplasma or Plasmodium and this has hindered the development of parasite-specific inhibitors. Selective inhibitors will almost certainly be required to effectively validate the proteasome as a drug target for malaria and other parasitic diseases.
An additional protease that has been suggested to function in T. gondii in general catabolism is cathepsin C (TgCPC). Three isoforms of this protease have been identified, and TgCP1 is implicated in growth and replication of the parasite [97]. Cathepsin C is an exopeptidase that cleaves dipeptides from the N-terminus of proteins. Treatment with a cathepsin C inhibitor led a reduction in parasitemia and reduced protein degradation in the parasitophorous vacuole (PV). However, additional studies to identify the substrates of cathepsin C will be required to define its role in general peptide degradation in T. gondii.
The recent placement of plasmepsin V in the Plasmodium protein export pathway also opens up new strategies for antimalarial chemotherapy. Unlike plasmepsin I–IV, plasmepsin V is not localized to the food vacuole, but is instead restricted to the endoplasmic reticulum (ER) membrane [98]. Recent work demonstrated the role of this aspartyl protease in cleaving the Plasmodium Export Element (PEXEL) sequence motif, required for the export of PEXEL containing parasite proteins into the erythrocyte cytosol [99, 100]. This export of proteins allows the parasite to remodel the host cell for functions such as adhesion to endothelial cells. Such remodeling could be a mechanism to evade host defenses and therefore inhibition of plasmepsin V could be a strategy for clearing parasites [101, 102].
4. Proteases have central roles in parasite egress
As obligate intracellular parasites, Plasmodium and Toxoplasma spend the majority of their lives within the confines of their host-cell environment. Once replication is complete and several mitotic cycles have taken place, specialized zoite cells are released from the host cell through a lytic process known as egress (See Figure 2, Step 3). Our current understanding of egress is that it is a rapid and tightly regulated process. However, the exact mechanisms that regulate egress remain unclear. In Plasmodium, there are two popular models; the first model proposes that egress occurs by initial degradation of the parasitophorous vacuole membrane (PVM), and then the erythrocyte membrane. The second model proposes the reverse, where the erythrocyte membrane is degraded prior to the release of the merozoites from the PVM [103]. Both models have experimental support, and thus it is difficult to provide a definite mechanism. However, recent advances with video microscopy supports a model whereby the PVM ruptures, followed by the rapid and explosive rupturing of the erythrocyte membrane that is thought to facilitate the dissemination of the daughter zoites from the parent host-cell [104, 105].
Despite our lack of understanding of the precise mechanistic processes underlying egress, proteases appear to play pivotal roles. For example, treatment of P. falciparum infected cells with serine and cysteine protease inhibitors produces a block in egress leading to accumulation of late stage schizonts [106]. However, it is important to note that these experiments used broad-spectrum inhibitors that block proteases implicated in both egress and invasion. Subsequent experiments have identified inhibitors that can specifically block egress, and these have served as useful chemical tools in further exploration of the proteases involved in egress [23, 107]. The following section reviews the role of several proteases that function in Plasmodium egress. Although studies on host-cell rupture in Toxoplasma have uncovered roles for potassium and calcium signaling [108] and the pore-forming perforin proteins [109], virtually nothing is known about proteases that may regulate egress in this organism.
4.1 Serine-repeat antigen (SERA) family
The serine-repeat antigen (SERA) family is group of highly expressed Plasmodium proteins that are characterized by a central papain-like protease domain. Within the family, only SERA 6–8 have the canonical ‘active site’ cysteine residue found in classical papain family proteases, while SERA 1–5 and 9 have a serine in place of this residue [6]. Intriguingly, this family of proteins is not found in related apicomplexan parasites such as Toxoplasma. SERA5 is the most abundant of the SERA family [110] and is found in the parasitophorus vacuole where it is processed around the time of egress. Antibodies against the truncated form of SERA5 and a cyclic peptide directed against the predicted enzyme domain of the protein interferes with its function and cause a reduction in parasite growth, suggesting that SERA5 is vital for Plasmodium egress and/or invasion [110–112]. SERA5 is one of the two SERA genes that cannot be disrupted, further highlighting its importance in the parasite life cycle [113]. As SERA5 possesses a papain-like fold with serine as the active site residue [114], it could potentially function in egress and invasion by proteolytically processing substrates. The recombinant catalytic domain of SERA5 indeed possesses some weak chymotrypsin-like activity and this activity is sensitive to protease inhibitors [115]. However, it is still presently unclear if endogenous SERA5 has proteolytic activity. Crystallographic evidence suggests that the active site in SERA5 is not conducive to substrate binding [114]. As such, although SERA5 may be an important player in egress, any catalytic function remains a mystery. The only SERA protein with a direct link to parasite egress is SERA8. In Plasmodium berghei, disruption of the SERA8 orthologue, ECP1, resulted in a complete block of sporozoite egress from oocytes [116]. Therefore, the SERA proteins may mediate egress in both the sexual and asexual stages of Plasmodium.
4.2 PfSUB1
During maturation, SERA5 is proteolytically cleaved from the full-length form of approximately 126 kDa, through two intermediates before accumulating as a 50 kDa form which is released into culture supernantant during rupture [117]. Investigation of the proteases responsible for the maturation of SERA5 linked the serine protease PfSUB1 and the cysteine protease DPAP3 to this maturation process [23, 107]. In addition to the role of PfSUB1 in priming merozoites during invasion, it has a role in parasite egress. PfSUB1 localizes to a merozoite organelle called the exoneme, and is released into the parasitophorous vacuolar space before egress. Further studies demonstrated that PfSUB1 is responsible for cleaving SERA 4, 5 and 6 during egress. Inhibition of PfSUB1 using a specific inhibitor (MRT 12113) led to significant reduction in egress and invasion. The role of PfSUB1 in processing SERA5 was also confirmed by a chemical screen that identified a chloroisocoumarin (JCP104) that serves as both an inhibitor and probe of PfSUB1 activity [107]. JCP104 specifically blocks schizont rupture further demonstrating that PfSUB1 is a regulator of P. falciparum egress.
4.3 DPAP3
In addition to identifying a small molecule inhibitor of PfSUB1, the small molecule screen by Arastu-Kapur and co-workers identified a cysteine protease inhibitor that specifically blocked host cell rupture [107]. The identified dipeptide vinyl sulfone hit was converted to an active site probe and used to identify the dipeptidyl aminopetidases (DPAPs) as targets of this compound. Further development efforts identified inhibitors that could specifically target the food vacuole specific protease DPAP1 as well as the related family member DPAP3, a protease with unknown functions. These compounds showed that inhibition of DPAP1 resulted in toxicity to the parasite without affecting host cell rupture, while inhibition of DPAP3 specifically blocked parasite release from the host cell. Inhibition of DPAP3 also led to reduction in levels of the mature form of PfSUB1 without causing accumulation of the precursor protein, suggesting that DPAP3 may be required for maturation of PfSUB1. DPAP3 may therefore regulate host cell egress by controlling maturation of secretory proteins required for this process [107].
4.4 Host cell calpain
In addition to parasite derived proteases, a host protease has also recently been implicated in the regulation of parasite egress in both P. falciparum and T. gondii. Specifically, depletion of host cell calpain leads to a complete block in parasite egress [118]. In this study, human calpain-1 activity in erythrocytes was reduced by both chemical inhibition and immunodepletion. The resulting calpain-1 deficient erythrocytes were infected with P. falciparum and a defect in schizont rupture was observed. To confirm the role of calpain in this process, purified calpain was added back to the calpain-depleted host cells and egress was restored. The role of calpain in release of T. gondii tachyzoites was also investigated using siRNA knockdown in U2OS human host cells. Knockdown of both calpain 1 and calpain 2 was achieved by targeting of the regulatory subunit of both calpains. Similar to Plasmodium, calpain knockdown in host-cells blocked T. gondii egress. Therefore, it is likely that host-cell calpain facilitates parasite egress through some type of host cell remodeling, the details of which are yet to be determined.
5. Proteases as targets for anti-malarials
Proteases are attractive targets for drug development because the presence of a well-defined active site facilitates interactions with small molecules, and thus many classes of small molecule proteases inhibitors have been developed [119]. Our understanding of the biology of specific proteases in the intra-erythrocytic stages during malaria infection has advanced greatly in the past decade and this information will likely facilitate the design of specific protease inhibitors as anti-parasitic agents (Table 1). Furthermore, the rapid emergence of drug resistance in Plasmodium spp. suggests an urgent need for new anti-malarial compounds. Current approved drugs focus mainly on targeting processes in the apicoplast or food vacuole, which are responsible for parasite growth and metabolism [52]. Proteases involved in hemoglobin metabolism are the most commonly targeted proteases in anti-malarial drugs currently under development, yet it may not be the most optimal pathway due to highly redundant mechanisms and the ability of the parasite to actively export compounds out of the food vacuole. While much effort has been directed towards developing lead compounds to target falcipains and plasmepsins, it is important to note that redundancies exist between these two families of proteases [62, 63] and hence effective therapy would most likely require targeting of both. Recent work has highlighted the potential of DPAP1 as a target for drug development efforts [78]. The inability to disrupt the DPAP1 gene highlights its vital role in parasite catabolism. Furthermore a hybrid of a DPAP1 inhibitor and an artemisinin analogue was able to effectively reduce parasite growth at low nano-molar concentrations with low cell toxicity [79]. Such a hybrid drug is less likely to induce resistance since each fragment of the compound uses distinct mechanisms for parasite killing.
Table 1.
List of partially characterized proteases organized by mechanism of proteolysis in Toxoplasma gondii and Plasmodium falciparum. Cited references are representative papers from studies of each protease listed.
| Protease Class | Protease | Apicomplexan species |
Implicated function in asexual stage |
Protease function and localization |
Literature |
|---|---|---|---|---|---|
| Aspartyl | Plasmepsins I–IV | Plasmodium falciparum | Growth and Replication | Hemoglobin degradation in food vacuole | [55], [60] |
| Plasmepsin V | Plasmodium falciparum | Growth and Replication | PEXEL processing for export into the erythrocytic cytosol; localized to the ER | [99], [100] | |
| Cysteine | TgCPB/Toxopain-1 | Toxoplasma gondii | Invasion | Rhoptry protein processing; localized to the rhoptries | [48] |
| TgCPCs | Toxoplasma gondii | Growth and Replication | Cleavage of dipeptides from N-terminal in cytosol, exact functions unknown | [97] | |
| TgCPL | Toxoplasma gondii | Invasion | Maturation of micronemal proteins; localized in the vacuolar compartment | [50] | |
| PfDPAP1 | Plasmodium falciparum | Growth and Replication | Involved in later stages of hemoglobin degradation in food vacuole | [77], [78] | |
| PfDPAP3 | Plasmodium falciparum | Egress | Facilitates PfSUB1 maturation in apical end of merozoites (unpublished)l | [107] | |
| Falcipain-1 | Plasmodium falciparum | Invasion | Exact functions unknown; localized to apical end of merozoites | [45], [46] | |
| Falcipain-2,2',3 | Plasmodium falciparum | Growth and Replication | Hemoglobin degradation in food vacuole | [69], [70], [74] | |
| Metalloprotease | Falcilysin | Plasmodium falciparum | Growth and Replication | Involved in later stages of hemoglobin degradation in food vacuole | [64], [66] |
| PfM1AAP and PfM17LAP | Plasmodium falciparum | Growth and Replication | Involved in terminal stages of hemoglobin degradation in food vacuole | [80], [82] | |
| Toxolysin 4 | Toxoplasma gondii | Invasion/Egress | Exact function unknown; localized to micronemes | [51] | |
| Serine | PfSUB1 | Plasmodium falciparum | Invasion and egress | Processing of merozoite surface and parasitophorous vacuole (PV) constituents prior to egress; localized to the exonemes | [23], [24], [107] |
| PfSUB2 | Plasmodium falciparum | Invasion | Surface protein sheddase;localized to the micronemes | [26] | |
| TgSUB1 | Toxoplasma gondii | Invasion | Processes micronemal proteins involved in host cell attachment; localized to the micronemes | [30] | |
| TgSUB2 | Toxoplasma gondii | Invasion | Maturation of rhoptry proteins; localized to the rhoptries | [31] | |
| PfSERA5 | Plasmodium falciparum | Egress | Potential role in egress; localized to the PV | [110], [114] | |
| TgROM2,4,5 | Toxoplasma gondii | Invasion | Surface protein sheddase;localized to tachyzoite plasma membrane | [38] | |
| PfROM1 and 4 | Plasmodium falciparum | Invasion | Surface protein sheddase;localized to merozoite plasma membrane | [42] | |
| Threonine | PfHslV | Plasmodium falciparum | Growth and Replication | Possible function as peptidase, but targets unknown; localized to mitochondria | [141], [142] |
| Proteasome | Plasmodium and Toxoplasma | Growth and Replication | General protein degradation; Regulates protein turnover; localization unknown | [94], [140] | |
Other proteases involved in Plasmodium egress and invasion are gaining attention as potential targets for therapy [103]. Arguably, egress and invasion are attractive processes for inhibitor development because Plasmodium spp. are obligate intracellular parasites that are particularly vulnerable outside their host cells [7]. Our lack of understanding of specific players in these processes is probably the reason for slow development of compounds that target egress and invasion. While there have been a recent series of published inhibitors that target DPAP3 and PfSUB1 [23, 107], these compounds are generally not optimal drugs and significant additional efforts will be required to convert these lead compounds into more suitable therapeutic agents.
6. Tools to facilitate dissection of apicomplexan parasite proteases
The post-genomic era offers exciting prospects to begin to further define apicomplexan biology. Genome sequences [120–122] and transcriptome profiling [123, 124] of P. falciparum and T. gondii enable genetic validation of protease activities and increase the rate of discovery of novel proteases with important functions. While T. gondii is a relatively genetically tractable organism [125], P. falciparum remains challenging to study using classical genetic methods. Low transfection efficiency [126, 127], coupled with the lack of RNAi machinery [128, 129] make standard genetic tools untenable. Fortunately, recent advances in chemical genetics [107, 130] and systems biology are helping to advance the study of basic parasite biology.
One particularly exciting area of development is the use of small molecules to conditionally regulate protein stability. This technique makes use of ligands that can stabilize a protein domain known as the destabilizing domain (DD), which is otherwise degraded in the absence of a stabilizing small molecule [131]. Recent success in P. falciparum has allowed the conditional knockdown of falcipain-2 [132], parasite calpain [133], calcium dependent-protein kinase 5 [134] and a proteasome cap regulatory subunit Rpn6 [135]. In P. falciparum, the DD system has therefore proven to be more generally applicable than the previously reported tetracycline-inducible transactivator system [136]. Moreover, the DD system allows the rapid temporal control of protein levels and thus enables the study of protein function at precise stages of the parasite life cycle.
The use of activity-based probes (ABPs) as a forward chemical genetic tool [137] also represents an exciting method to study parasite protease biology [78, 107, 130]. Using inhibitor scaffolds tagged with a reporter, the activity of proteases can be monitored in intact parasites. This technique can also be applied effectively to identify selective substrates that can then be used for high throughput screens of protease targets [138]. In addition, various groups have harnessed mass spectrometry (MS) as a tool to assess global proteomic changes in P. falciparum [25, 139]. These approaches allow for a more global mapping of proteolytic events, and could lead to the identification of essential pathways that are controlled by proteases. The regulators of these pathways can then be prioritized as targets for small molecule therapies.
Conclusion
In this review, we have described the roles of various proteases throughout the life cycles of Plasmodium and Toxoplasma. Although we have highlighted many proteases that have been the focus of significant study over the past decade, we also chose to draw attention to proteases where further investigation is required to better understand their function in the parasite. With the help of novel genetic and chemical tools, coupled with systems-based analysis, the functions of poorly understood and novel proteases can hopefully be defined in the near future. As proteases represent very attractive targets in drug development, further studies will facilitate discovery of novel drugs against malaria and toxoplasmosis that will hopefully overcome emerging resistance to current drug therapies.
Highlights.
Discussion of Proteases as important mediators of pathogenesis in parasites
Outline of proteases in Plasmodium falciparum and Toxoplasma gondii, and their roles in invasion, growth, replication and egress from host cells.
Outline of advances in chemical genetics and systems biology that have advanced our understanding of the roles of proteases in parasite biology.
Discussion of how new information on proteases in parasite pathogens could lead to new anti-parasitic drug targets.
Acknowledgement
We thank Dr. J. Boothroyd for helpful discussions. This work was funded by a Burroughs Wellcome New Investigator in Pathogenesis Grant (to MB) and NIH grants R01 AI078947 and R21 AI088541 (to MB). H.L is supported by the NSS-Ph.D Scholarship from Agency for Science, Technology and Research Singapore.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cohen GM. Caspases: the executioners of apoptosis. Biochemical Journal. 1997;326:1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li SJ, Hochstrasser M. A new protease required for cell-cycle progression in yeast. Nature. 1999;398:246–251. doi: 10.1038/18457. [DOI] [PubMed] [Google Scholar]
- 3.Huttenlocher A, Palecek SP, Lu Q, Zhang WL, Mellgren RL, Lauffenburger DA, Ginsberg MH, Horwitz AF. Regulation of cell migration by the calcium-dependent protease calpain. Journal of Biological Chemistry. 1997;272:32719–32722. doi: 10.1074/jbc.272.52.32719. [DOI] [PubMed] [Google Scholar]
- 4.Klemba M, Goldberg DE. Biological roles of proteases in parasitic protozoa. Annual Review of Biochemistry. 2002;71:275–305. doi: 10.1146/annurev.biochem.71.090501.145453. [DOI] [PubMed] [Google Scholar]
- 5.Kim K. Role of proteases in host cell invasion by Toxoplasma gondii and other apicomplexa. Acta Tropica. 2004;91:69–81. doi: 10.1016/j.actatropica.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 6.Blackman MJ. Proteases in host cell invasion by the malaria parasite. Cellular Microbiology. 2004;6:893–903. doi: 10.1111/j.1462-5822.2004.00437.x. [DOI] [PubMed] [Google Scholar]
- 7.Cowman AF, Crabb BS. Invasion of red blood cells by malaria parasites. Cell. 2006;124:755–766. doi: 10.1016/j.cell.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Blackman MJ, Bannister LH. Apical organelles of Apicomplexa: biology and isolation by subcellular fractionation. Molecular and Biochemical Parasitology. 2001;117:11–25. doi: 10.1016/s0166-6851(01)00328-0. [DOI] [PubMed] [Google Scholar]
- 9.Walter RF, McFadden GI. The apicoplast: A review of the derived plastid of apicomplexan parasites. Current Issues in Molecular Biology. 2005;7:57–79. [PubMed] [Google Scholar]
- 10.Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tenter AM, Heckeroth AR, Weiss LM. Toxoplasma gondii: from animals to humans. International Journal for Parasitology. 2000;30:1217–1258. doi: 10.1016/s0020-7519(00)00124-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montoya JG, Liesenfeld O. Toxoplasmosis. Lancet. 2004;363:1965–1976. doi: 10.1016/S0140-6736(04)16412-X. [DOI] [PubMed] [Google Scholar]
- 13.Black MW, Boothroyd JC. Lytic cycle of Toxoplasma gondii. Microbiology and Molecular Biology Reviews. 2000;64 doi: 10.1128/mmbr.64.3.607-623.2000. 607-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Striepen B, Jordan CN, Reiff S, van Dooren GG. Building the perfect parasite: Cell division in Apicomplexa. Plos Pathogens. 2007;3:691–698. doi: 10.1371/journal.ppat.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKerrow JH, Caffrey C, Kelly B, Loke P, Sajid M. Proteases in parasitic diseases. Annual Review of Pathology-Mechanisms of Disease. 2006;1:497–536. doi: 10.1146/annurev.pathol.1.110304.100151. [DOI] [PubMed] [Google Scholar]
- 16.Smith JE. A UBIQUITOUS INTRACELLULAR PARASITE - THE CELLULAR BIOLOGY OF TOXOPLASMA-GONDII. International Journal for Parasitology. 1995;25:1301–1309. doi: 10.1016/0020-7519(95)00067-c. [DOI] [PubMed] [Google Scholar]
- 17.Soldati D, Foth BJ, Cowman AF. Molecular and functional aspects of parasite invasion. Trends in Parasitology. 2004;20:567–574. doi: 10.1016/j.pt.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 18.Soldati D, Dubremetz JF, Lebrun M. Microneme proteins: structural and functional requirements to promote adhesion and invasion by the apicomplexan parasite Toxoplasma gondii. International Journal for Parasitology. 2001;31:1293–1302. doi: 10.1016/s0020-7519(01)00257-0. [DOI] [PubMed] [Google Scholar]
- 19.Huynh MH, Harper JM, Carruthers VB. Preparing for an invasion: charting the pathway of adhesion proteins to Toxoplasma micronemes. Parasitol Res. 2006;98:389–395. doi: 10.1007/s00436-005-0062-2. [DOI] [PubMed] [Google Scholar]
- 20.Withers-Martinez C, Jean L, Blackman MJ. Subtilisin-like proteases of the malaria parasite. Molecular Microbiology. 2004;53:55–63. doi: 10.1111/j.1365-2958.2004.04144.x. [DOI] [PubMed] [Google Scholar]
- 21.Sajid M, Withers-Martinez C, Blackman MJ. Maturation and specificity of Plasmodium falciparum subtilisin-like protease-1, a malaria merozoite subtilisin-like serine protease. Journal of Biological Chemistry. 2000;275:631–641. doi: 10.1074/jbc.275.1.631. [DOI] [PubMed] [Google Scholar]
- 22.Blackman MJ, Fujioka H, Stafford WHL, Sajid M, Clough B, Fleck SL, Aikawa M, Grainger M, Hackett F. A subtilisin-like protein in secretory organelles of Plasmodium falciparum merozoites. Journal of Biological Chemistry. 1998;273:23398–23409. doi: 10.1074/jbc.273.36.23398. [DOI] [PubMed] [Google Scholar]
- 23.Yeoh S, O'Donnell RA, Koussis K, Dluzewski AR, Ansell KH, Osborne SA, Hackett F, Withers-Martinez C, Mitchell GH, Bannister LH, Bryans JS, Kettleborough CA, Blackman MJ. Subcellular discharge of a serine protease mediates release of invasive malaria parasites from host erythrocytes. Cell. 2007;131:1072–1083. doi: 10.1016/j.cell.2007.10.049. [DOI] [PubMed] [Google Scholar]
- 24.Koussis K, Withers-Martinez C, Yeoh S, Child M, Hackett F, Knuepfer E, Juliano L, Woehlbier U, Bujard H, Blackman MJ. A multifunctional serine protease primes the malaria parasite for red blood cell invasion. Embo Journal. 2009;28:725–735. doi: 10.1038/emboj.2009.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silmon de Monerri NC, Flynn HR, Campos MG, Hackett F, Koussis K, Withers-Martinez C, Skehel JM, Blackman MJ. Global Identification of Multiple Substrates for Plasmodium falciparum SUB1,an Essential Malarial Processing Protease. Infect Immun. 2011;79:1086–1097. doi: 10.1128/IAI.00902-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris PK, Yeoh S, Dluzewski AR, O'Donnell RA, Withers-Martinez C, Hackett F, Bannister LH, Mitchell GH, Blackman MJ. Molecular identification of a malaria merozoite surface sheddase. Plos Pathogens. 2005;1:241–251. doi: 10.1371/journal.ppat.0010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Green JL, Hinds L, Grainger M, Knueffer E, Holder AA. Plasmodium thrombospondin related apical merozoite protein (PTRAMP) is shed from the surface of merozoites by PfSUB2 upon invasion of erythrocytes. Molecular and Biochemical Parasitology. 2006;150:114–117. doi: 10.1016/j.molbiopara.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 28.Miller SA, Binder EM, Blackman MJ, Carruthers VB, Kim K. A conserved subtilisin-like protein TgSUB1 in microneme organelles of Toxoplasma gondii. Journal of Biological Chemistry. 2001;276:45341–45348. doi: 10.1074/jbc.M106665200. [DOI] [PubMed] [Google Scholar]
- 29.Binder EM, Lagal V, Kim K. The prodomain of Toxoplasma gondii GPI-anchored subtilase TgSUB1 mediates its targeting to micronemes. Traffic. 2008;9:1485–1496. doi: 10.1111/j.1600-0854.2008.00774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lagal V, Binder EM, Huynh MH, Kafsack BF, Harris PK, Diez R, Chen D, Cole RN, Carruthers VB, Kim K. Toxoplasma gondii protease TgSUB1 is required for cell surface processing of micronemal adhesive complexes and efficient adhesion of tachyzoites. Cell Microbiol. 2010;12:1792–1808. doi: 10.1111/j.1462-5822.2010.01509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller SA, Thathy V, Ajioka JW, Blackman MJ, Kim K. TgSUB2 is a Toxoplasma gondii rhoptry organelle processing proteinase. Mol Microbiol. 2003;49:883–894. doi: 10.1046/j.1365-2958.2003.03604.x. [DOI] [PubMed] [Google Scholar]
- 32.Urban S, Lee JR, Freeman M. Drosophila Rhomboid-1 defines a family of putative intramembrane serine proteases. Cell. 2001;107:173–182. doi: 10.1016/s0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- 33.Urban S, Freeman M. Intramembrane proteolysis controls diverse signalling pathways throughout evolution. Current Opinion in Genetics & Development. 2002;12:512–518. doi: 10.1016/s0959-437x(02)00334-9. [DOI] [PubMed] [Google Scholar]
- 34.Urban S, Schlieper D, Freeman M. Conservation of intramembrane proteolytic activity and substrate specificity in prokaryotic and eukaryotic rhomboids. Current Biology. 2002;12:1507–1512. doi: 10.1016/s0960-9822(02)01092-8. [DOI] [PubMed] [Google Scholar]
- 35.Freeman M. Rhomboid Proteases and their Biological Functions. Annual Review of Genetics. 2008;42:191–210. doi: 10.1146/annurev.genet.42.110807.091628. [DOI] [PubMed] [Google Scholar]
- 36.Zhou XW, Blackman MJ, Howell SA, Carruthers VB. Proteomic analysis of cleavage events reveals a dynamic two-step mechanism for proteolysis of a key parasite adhesive complex. Molecular & Cellular Proteomics. 2004;3:565–576. doi: 10.1074/mcp.M300123-MCP200. [DOI] [PubMed] [Google Scholar]
- 37.Dowse TJ, Pascall JC, Brown KD, Soldati D. Apicomplexan rhomboids have a potential role in microneme protein cleavage during host cell invasion. International Journal for Parasitology. 2005;35:747–756. doi: 10.1016/j.ijpara.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 38.Brossier F, Jewett TJ, Sibley LD, Urban S. A spatially localized rhomboid protease cleaves cell surface adhesins essential for invasion by Toxoplasma. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:4146–4151. doi: 10.1073/pnas.0407918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buguliskis JS, Brossier F, Shuman J, Sibley LD. Rhomboid 4 (ROM4) Affects the Processing of Surface Adhesins and Facilitates Host Cell Invasion by Toxoplasma gondii. Plos Pathogens. 2010;6 doi: 10.1371/journal.ppat.1000858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santos JM, Ferguson DJP, Blackman MJ, Soldati-Favre D. Intramembrane Cleavage of AMA1 Triggers Toxoplasma to Switch from an Invasive to a Replicative Mode. Science. 2011;331:473–477. doi: 10.1126/science.1199284. [DOI] [PubMed] [Google Scholar]
- 41.Brossier F, Starnes GL, Beatty WL, Sibley LD. Microneme rhomboid protease TgROM1 is required for efficient intracellular growth of Toxoplasma gondii. Eukaryotic Cell. 2008;7:664–674. doi: 10.1128/EC.00331-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baker RP, Wijetilaka R, Urban S. Two Plasmodium rhomboid proteases preferentially cleave different adhesins implicated in all invasive stages of malaria. Plos Pathogens. 2006;2:922–932. doi: 10.1371/journal.ppat.0020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Donnell RA, Hackett F, Howell SA, Treeck M, Struck N, Krnajski Z, Withers-Martinez C, Gilberger TW, Blackman MJ. Intramembrane proteolysis mediates shedding of a key adhesin during erythrocyte invasion by the malaria parasite. Journal of Cell Biology. 2006;174:1023–1033. doi: 10.1083/jcb.200604136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Srinivasan P, Coppens I, Jacobs-Lorena M. Distinct Roles of Plasmodium Rhomboid 1 in Parasite Development and Malaria Pathogenesis. Plos Pathogens. 2009;5 doi: 10.1371/journal.ppat.1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greenbaum DC, Baruch A, Grainger M, Bozdech Z, Medzihradszky KF, Engel J, DeRisi J, Holder AA, Bogyo M. A role for the protease falcipain 1 in host cell invasion by the human malaria parasite. Science. 2002;298:2002–2006. doi: 10.1126/science.1077426. [DOI] [PubMed] [Google Scholar]
- 46.Sijwali PS, Kato K, Seydel KB, Gut J, Lehman J, Klemba M, Goldberg DE, Miller LH, Rosenthal PJ. Plasmodium falciparum cysteine protease falcipain-1 is not essential in erythrocytic stage malaria parasites. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8721–8726. doi: 10.1073/pnas.0402738101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eksi S, Czesny B, Greenbaum DC, Bogyo M, Williamson KC. Targeted disruption of Plasmodium falciparum cysteine protease falcipain 1 reduces oocyst production not erythrocytic stage growth. Molecular Microbiology. 2004;53:243–250. doi: 10.1111/j.1365-2958.2004.04108.x. [DOI] [PubMed] [Google Scholar]
- 48.Que XC, Ngo HA, Lawton J, Gray M, Liu Q, Engel J, Brinen L, Ghosh P, Joiner KA, Reed SL. The cathepsin B of Toxoplasma gondii toxopain-1is critical for parasite invasion and rhoptry protein processing. Journal of Biological Chemistry. 2002;277:25791–25797. doi: 10.1074/jbc.M202659200. [DOI] [PubMed] [Google Scholar]
- 49.Que XC, Wunderlich A, Joiner KA, Reed SL. Toxopain-1 is critical for infection in a novel chicken embryo model of congenital toxoplasmosis. Infection and Immunity. 2004;72:2915–2921. doi: 10.1128/IAI.72.5.2915-2921.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parussini F, Coppens I, Shah PP, Diamond SL, Carruthers VB. Cathepsin L occupies a vacuolar compartment and is a protein maturase within the endo/exocytic system of Toxoplasma gondii. Molecular Microbiology. 2010;76:1340–1357. doi: 10.1111/j.1365-2958.2010.07181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laliberte J, Carruthers VB. Toxoplasma gondii toxolysin 4 is an extensively processed putative metalloproteinase secreted from micronemes. Mol Biochem Parasitol. 2011;177:49–56. doi: 10.1016/j.molbiopara.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kappe SH, Vaughan AM, Boddey JA, Cowman AF. That was then but this is now: malaria research in the time of an eradication agenda. Science. 2010;328:862–866. doi: 10.1126/science.1184785. [DOI] [PubMed] [Google Scholar]
- 53.Goldberg DE. Hemoglobin degradation, in: Malaria: Drugs. Disease and Post-Genomic Biology. 2005;295:275–291. doi: 10.1007/3-540-29088-5_11. [DOI] [PubMed] [Google Scholar]
- 54.Francis SE, Sullivan DJ, Goldberg DE. Hemoglobin metabolism in the malaria parasite Plasmodium falciparum. Annual Review of Microbiology. 1997;51:97–123. doi: 10.1146/annurev.micro.51.1.97. [DOI] [PubMed] [Google Scholar]
- 55.Coombs GH, Goldberg DE, Klemba M, Berry C, Kay J, Mottram JC. Aspartic proteases of Plasmodium falciparum and other parasitic protozoa as drug targets. Trends in Parasitology. 2001;17:532–537. doi: 10.1016/s1471-4922(01)02037-2. [DOI] [PubMed] [Google Scholar]
- 56.Francis SE, Banerjee R, Goldberg DE. Biosynthesis and maturation of the malaria aspartic hemoglobinases plasmepsins I and II. Journal of Biological Chemistry. 1997;272:14961–14968. doi: 10.1074/jbc.272.23.14961. [DOI] [PubMed] [Google Scholar]
- 57.Liu J, Gluzman IY, Drew ME, Goldberg DE. The role of Plasmodium falciparum food vacuole plasmepsins. Journal of Biological Chemistry. 2005;280:1432–1437. doi: 10.1074/jbc.M409740200. [DOI] [PubMed] [Google Scholar]
- 58.Luker KE, Francis SE, Gluzman IY, Goldberg DE. Kinetic analysis of plasmepsins I and II,aspartic proteases of the Plasmodium falciparum digestive vacuole. Molecular and Biochemical Parasitology. 1996;79:71–78. doi: 10.1016/0166-6851(96)02651-5. [DOI] [PubMed] [Google Scholar]
- 59.Silva AM, Lee AY, Gulnik SV, Majer P, Collins J, Bhat TN, Collins PJ, Cachau RE, Luker KE, Gluzman IY, Francis SE, Oksman A, Goldberg DE, Erickson JW. Structure and inhibition of plasmepsin II, a hemoglobin-degrading enzyme from Plasmodium falciparum. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:10034–10039. doi: 10.1073/pnas.93.19.10034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Banerjee R, Liu J, Beatty W, Pelosof L, Klemba M, Goldberg DE. Four plasmepsins are active in the Plasmodium falciparum food vacuole including a protease with an active-site histidine. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:990–995. doi: 10.1073/pnas.022630099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dame JB, Yowell CA, Omara-Opyene L, Carlton JM, Cooper RA, Li T. Plasmepsin 4, the food vacuole aspartic proteinase found in all Plasmodium spp. infecting man. Molecular and Biochemical Parasitology. 2003;130:1–12. doi: 10.1016/s0166-6851(03)00137-3. [DOI] [PubMed] [Google Scholar]
- 62.Liu J, Istvan ES, Gluzman IY, Gross J, Goldberg DE. Plasmodium falciparum ensures its amino acid supply with multiple acquisition pathways and redundant proteolytic enzyme systems. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8840–8845. doi: 10.1073/pnas.0601876103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bonilla JA, Moura PA, Bonilla TD, Yowell CA, Fidock DA, Dame JB. Effects on growth hemoglobin metabolism and paralogous gene expression resulting from disruption of genes encoding the digestive vacuole plasmepsins of Plasmodium falciparum. International Journal for Parasitology. 2007;37:317–327. doi: 10.1016/j.ijpara.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 64.Eggleson KK, Duffin KL, Goldberg DE. Identification and characterization of falcilysin a metallopeptidase involved in hemoglobin catabolism within the malaria parasite Plasmodium falciparum. Journal of Biological Chemistry. 1999;274:32411–32417. doi: 10.1074/jbc.274.45.32411. [DOI] [PubMed] [Google Scholar]
- 65.Murata CE, Goldberg DE. Plasmodium falciparum falcilysin: an unprocessed food vacuole enzyme. Molecular and Biochemical Parasitology. 2003;129:123–126. doi: 10.1016/s0166-6851(03)00098-7. [DOI] [PubMed] [Google Scholar]
- 66.Ralph SA. Subcellular multitasking - multiple destinations and roles for the Plasmodium falcilysin protease. Molecular Microbiology. 2007;63:309–313. doi: 10.1111/j.1365-2958.2006.05528.x. [DOI] [PubMed] [Google Scholar]
- 67.Salas F, Fichmann J, Lee GK, Scott MD, Rosenthal PJ. FUNCTIONAL EXPRESSION OF FALCIPAIN A PLASMODIUM-FALCIPARUM CYSTEINE PROTEINASE SUPPORTS ITS ROLE AS A MALARIAL HEMOGLOBINASE. Infection and Immunity. 1995;63:2120–2125. doi: 10.1128/iai.63.6.2120-2125.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sijwali PS, Shenai BR, Gut J, Singh A, Rosenthal PJ. Expression and characterization of the Plasmodium falciparum haemoglobinase falcipain-3. Biochemical Journal. 2001;360:481–489. doi: 10.1042/0264-6021:3600481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sabnis YA, Desai PV, Rosenthal PJ, Avery MA. Probing the structure of falcipain-3 a cysteine protease from Plasmodium falciparum: Comparative protein modeling and docking studies. Protein Science. 2003;12:501–509. doi: 10.1110/ps.0228103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Singh N, Sijwali PS, Pandey KC, Rosenthal PJ. Plasmodium falciparum: Biochemical characterization of the cysteine protease falcipain-2 '. Experimental Parasitology. 2006;112:187–192. doi: 10.1016/j.exppara.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 71.Klemba M, Gluzman I, Goldberg DE. A Plasmodium falciparum dipeptidyl aminopeptidase I participates in vacuolar hemoglobin degradation. Journal of Biological Chemistry. 2004;279:43000–43007. doi: 10.1074/jbc.M408123200. [DOI] [PubMed] [Google Scholar]
- 72.Drew ME, Banerjee R, Uffman EW, Gilbertson S, Rosenthal PJ, Goldberg DE. Plasmodium food vacuole plasmepsins are activated by falcipains. Journal of Biological Chemistry. 2008;283:12870–12876. doi: 10.1074/jbc.M708949200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rosenthal PJ, Olson JE, Lee GK, Palmer JT, Klaus JL, Rasnick D. Antimalarial effects of vinyl sulfone cysteine proteinase inhibitors. Antimicrobial Agents and Chemotherapy. 1996;40:1600–1603. doi: 10.1128/aac.40.7.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shenai BR, Sijwali PS, Singh A, Rosenthal PJ. Characterization of native recombinant falcipain-2 a principal trophozoite cysteine protease and essential hemoglobinase of Plasmodium falciparum. Journal of Biological Chemistry. 2000;275:29000–29010. doi: 10.1074/jbc.M004459200. [DOI] [PubMed] [Google Scholar]
- 75.Sijwali PS, Rosenthal PJ. Gene disruption confirms a critical role for the cysteine protease falcipain-2 in hemoglobin hydrolysis by Plasmodium falciparum. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4384–4389. doi: 10.1073/pnas.0307720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kerr ID, Lee JH, Farady CJ, Marion R, Rickert M, Sajid M, Pandey KC, Caffrey CR, Legac J, Hansell E, McKerrow JH, Craik CS, Rosenthal PJ, Brinen LS. Vinyl Sulfones as Antiparasitic Agents and a Structural Basis for Drug Design. Journal of Biological Chemistry. 2009;284:25697–25703. doi: 10.1074/jbc.M109.014340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang F, Krai P, Deu E, Bibb B, Lauritzen C, Pedersen J, Bogyo M, Klemba M. Biochemical characterization of Plasmodium falciparum dipeptidyl aminopeptidase 1. Molecular and Biochemical Parasitology. 2011;175:10–20. doi: 10.1016/j.molbiopara.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Deu E, Leyva MJ, Albrow VE, Rice MJ, Ellman JA, Bogyo M. Functional Studies of Plasmodium falciparum Dipeptidyl Aminopeptidase I Using Small Molecule Inhibitors and Active Site Probes. Chemistry & Biology. 2010;17:808–819. doi: 10.1016/j.chembiol.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mahajan SS, Deu E, Lauterwasser EM, Leyva MJ, Ellman JA, Bogyo M, Renslo AR. A fragmenting hybrid approach for targeted delivery of multiple therapeutic agents to the malaria parasite. ChemMedChem. 2011;6:415–419. doi: 10.1002/cmdc.201100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dalal S, Klemba M. Roles for two aminopeptidases in vacuolar hemoglobin catabolism in Plasmodium falciparum. Journal of Biological Chemistry. 2007;282:35978–35987. doi: 10.1074/jbc.M703643200. [DOI] [PubMed] [Google Scholar]
- 81.Gavigan CS, Dalton JP, Bell A. The role of aminopeptidases in haemoglobin degradation in Plasmodium falciparum-infected erythrocytes. Molecular and Biochemical Parasitology. 2001;117:37–48. doi: 10.1016/s0166-6851(01)00327-9. [DOI] [PubMed] [Google Scholar]
- 82.Stack CM, Lowther J, Cunningham E, Donnelly S, Gardiner DL, Trenholme KR, Skinner-Adams TS, Teuscher F, Grembecka J, Mucha A, Kafarski P, Lua L, Bell A, Dalton JP. Characterization of the Plasmodium falciparum M17 leucyl aminopeptidase - A protease involved in amino acid regulation with potential for antimalarial drug development. Journal of Biological Chemistry. 2007;282:2069–2080. doi: 10.1074/jbc.M609251200. [DOI] [PubMed] [Google Scholar]
- 83.Skinner-Adams TS, Stack CM, Trenholme KR, Brown CL, Grembecka J, Lowther J, Mucha A, Drag M, Kafarski P, McGowan S, Whisstock JC, Gardiner DL, Dalton JP. Plasmodium falciparum neutral aminopeptidases: new targets for anti-malarials. Trends in Biochemical Sciences. 2010;35:53–61. doi: 10.1016/j.tibs.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 84.Allary M, Schrevel J, Florent I. Propertiesstage-dependent expression and localization of Plasmodium falciprum M1 family zinc-aminopeptidase. Parasitology. 2002;125:1–10. doi: 10.1017/s0031182002001828. [DOI] [PubMed] [Google Scholar]
- 85.McGowan S, Porter CJ, Lowther J, Stack CM, Golding SJ, Skinner-Adams TS, Trenholme KR, Teuscher F, Donnelly SM, Grembecka J, Mucha A, Kafarski P, DeGori R, Buckle AM, Gardiner DL, Whisstock JC, Dalton JP. Structural basis for the inhibition of the essential Plasmodium falciparum M1 neutral aminopeptidase. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2537–2542. doi: 10.1073/pnas.0807398106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Azimzadeh O, Sow C, Geze M, Nyalwidhe J, Florent I. Plasmodium falciparum PfA-M1 aminopeptidase is trafficked via the parasitophorous vacuole and marginally delivered to the food vacuole. Malaria Journal. 2010;9 doi: 10.1186/1475-2875-9-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McGowan S, Oellig CA, Birru WA, Caradoc-Davies TT, Stack CM, Lowther J, Skinner-Adams T, Mucha A, Kafarski P, Grembecka J, Trenholme KR, Buckle AM, Gardiner DL, Dalton JP, Whisstock JC. Structure of the Plasmodium falciparum M17 aminopeptidase and significance for the design of drugs targeting the neutral exopeptidases. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:2449–2454. doi: 10.1073/pnas.0911813107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 89.Borissenko L, Groll M. 20S proteasome and its inhibitors: crystallographic knowledge for drug development. Chem Rev. 2007;107:687–717. doi: 10.1021/cr0502504. [DOI] [PubMed] [Google Scholar]
- 90.Richardson PG, Mitsiades C, Hideshima T, Anderson KC. Bortezomib: Proteasome inhibition as an effective anticancer therapy. Annu. Rev. Med. 2006;57:33–47. doi: 10.1146/annurev.med.57.042905.122625. [DOI] [PubMed] [Google Scholar]
- 91.Gantt SM, Myung JM, Briones MRS, Li WD, Corey EJ, Omura S, Nussenzweig V, Sinnis P. Proteasome inhibitors block development of Plasmodium spp. Antimicrobial Agents and Chemotherapy. 1998;42:2731–2738. doi: 10.1128/aac.42.10.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lindenthal C, Weich N, Chia YS, Heussler V, Klinkert MQ. The proteasome inhibitor MLN-273 blocks exoerythrocytic and erythrocytic development of Plasmodium parasites. Parasitology. 2005;131:37–44. doi: 10.1017/s003118200500747x. [DOI] [PubMed] [Google Scholar]
- 93.Hatabu T, Hagiwara M, Taguchi N, Kiyozawa M, Suzuki M, Kano S, Sato K. Plasmodium falciparum: The fungal metabolite gliotoxin inhibits proteasome proteolytic activity and exerts a plasmodicidal effect on Plasmodium falciparum. Experimental Parasitology. 2006;112:179–183. doi: 10.1016/j.exppara.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 94.Kreidenweiss A, Kremsner PG, Mordmuller B. Comprehensive study of proteasome inhibitors against Plasmodium falciparum laboratory strains and field isolates from Gabon. Malaria Journal. 2008;7 doi: 10.1186/1475-2875-7-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Czesny B, Goshu S, Cook JL, Williamson KC. The Proteasome Inhibitor Epoxomicin Has Potent Plasmodium falciparum Gametocytocidal Activity. Antimicrobial Agents and Chemotherapy. 2009;53:4080–4085. doi: 10.1128/AAC.00088-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Aminake MN, Schoof S, Sologub L, Leubner M, Kirschner M, Arndt HD, Pradel G. Thiostrepton derivatives exhibit antimalarial and gametocytocidal activity by dually targeting parasite proteasome and apicoplast. Antimicrob Agents Chemother. 2011 doi: 10.1128/AAC.01096-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Que XC, Engel JC, Ferguson D, Wunderlich A, Tomavo S, Reed SL. Cathepsin Cs are key for the intracellular survival of the protozoan parasite Toxoplasma gondii. Journal of Biological Chemistry. 2007;282:4994–5003. doi: 10.1074/jbc.M606764200. [DOI] [PubMed] [Google Scholar]
- 98.Klemba M, Goldberg DE, Characterization of plasmepsin V. a membrane-bound aspartic protease homolog in the endoplasmic reticulum of Plasmodium falciparum. Molecular and Biochemical Parasitology. 2005;143:183–191. doi: 10.1016/j.molbiopara.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 99.Boddey JA, Hodder AN, Gunther S, Gilson PR, Patsiouras H, Kapp EA, Pearce JA, de Koning-Ward TF, Simpson RJ, Crabb BS, Cowman AF. An aspartyl protease directs malaria effector proteins to the host cell. Nature. 2010;463:627–U652. doi: 10.1038/nature08728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Russo I, Babbitt S, Muralidharan V, Butler T, Oksman A, Goldberg DE. Plasmepsin V licenses Plasmodium proteins for export into the host erythrocyte. Nature. 2010;463:632–636. doi: 10.1038/nature08726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Haase S, de Koning-Ward TF. New insights into protein export in malaria parasites. Cellular Microbiology. 2010;12:580–587. doi: 10.1111/j.1462-5822.2010.01455.x. [DOI] [PubMed] [Google Scholar]
- 102.Goldberg DE, Cowman AF. Moving in and renovating: exporting proteins from Plasmodium into host erythrocytes. Nature Reviews Microbiology. 2010;8:617–621. doi: 10.1038/nrmicro2420. [DOI] [PubMed] [Google Scholar]
- 103.Blackman MJ. Malarial proteases and host cell egress: an 'emerging' cascade. Cellular Microbiology. 2008;10:1925–1934. doi: 10.1111/j.1462-5822.2008.01176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gilson PR, Crabb BS. Morphology and kinetics of the three distinct phases of red blood cell invasion by Plasmodium falciparum merozoites. Int J Parasitol. 2009;39:91–96. doi: 10.1016/j.ijpara.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 105.Glushakova S, Yin D, Li T, Zimmerberg J. Membrane transformation during malaria parasite release from human red blood cells. Curr Biol. 2005;15:1645–1650. doi: 10.1016/j.cub.2005.07.067. [DOI] [PubMed] [Google Scholar]
- 106.Lyon JA, Haynes JD. Plasmodium falciparum antigens synthesized by schizonts and stabilized at the merozoite surface when schizonts mature in the presence of protease inhibitors. J Immunol. 1986;136:2245–2251. [PubMed] [Google Scholar]
- 107.Arastu-Kapur S, Ponder EL, Fonovic UP, Yeoh S, Yuan F, Fonovic M, Grainger M, Phillips CI, Powers JC, Bogyo M. Identification of proteases that regulate erythrocyte rupture by the malaria parasite Plasmodium falciparum. Nature Chemical Biology. 2008;4:203–213. doi: 10.1038/nchembio.70. [DOI] [PubMed] [Google Scholar]
- 108.Lavine MD, Arrizabalaga G. Invasion and egress by the obligate intracellular parasite Toxoplasma gondii: potential targets for the development of new antiparasitic drugs. Curr Pharm Des. 2007;13:641–651. doi: 10.2174/138161207780162854. [DOI] [PubMed] [Google Scholar]
- 109.Kafsack BF, Pena JD, Coppens I, Ravindran S, Boothroyd JC, Carruthers VB. Rapid membrane disruption by a perforin-like protein facilitates parasite exit from host cells. Science. 2009;323:530–533. doi: 10.1126/science.1165740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Aoki S, Li J, Itagaki S, Okech BA, Egwang TG, Matsuoka H, Palacpac NM, Mitamura T, Horii T. Serine repeat antigen (SERA5) is predominantly expressed among the SERA multigene family of Plasmodium falciparum and the acquired antibody titers correlate with serum inhibition of the parasite growth. J Biol Chem. 2002;277:47533–47540. doi: 10.1074/jbc.M207145200. [DOI] [PubMed] [Google Scholar]
- 111.Pang XL, Mitamura T, Horii T. Antibodies reactive with the N-terminal domain of Plasmodium falciparum serine repeat antigen inhibit cell proliferation by agglutinating merozoites and schizonts. Infect Immun. 1999;67:1821–1827. doi: 10.1128/iai.67.4.1821-1827.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fairlie WD, Spurck TP, McCoubrie JE, Gilson PR, Miller SK, McFadden GI, Malby R, Crabb BS, Hodder AN. Inhibition of malaria parasite development by a cyclic peptide that targets the vital parasite protein SERA5. Infect Immun. 2008;76:4332–4344. doi: 10.1128/IAI.00278-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.McCoubrie JE, Miller SK, Sargeant T, Good RT, Hodder AN, Speed TP, de Koning-Ward TF, Crabb BS. Evidence for a common role for the serine-type Plasmodium falciparum serine repeat antigen proteases: implications for vaccine and drug design. Infect Immun. 2007;75:5565–5574. doi: 10.1128/IAI.00405-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hodder AN, Malby RL, Clarke OB, Fairlie WD, Colman PM, Crabb BS, Smith BJ. Structural insights into the protease-like antigen Plasmodium falciparum SERA5 and its noncanonical active-site serine. J Mol Biol. 2009;392:154–165. doi: 10.1016/j.jmb.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 115.Hodder AN, Drew DR, Epa VC, Delorenzi M, Bourgon R, Miller SK, Moritz RL, Frecklington DF, Simpson RJ, Speed TP, Pike RN, Crabb BS. Enzymic phylogenetic and structural characterization of the unusual papain-like protease domain of Plasmodium falciparum SERA5. J Biol Chem. 2003;278:48169–48177. doi: 10.1074/jbc.M306755200. [DOI] [PubMed] [Google Scholar]
- 116.Aly AS, Matuschewski K. A malarial cysteine protease is necessary for Plasmodium sporozoite egress from oocysts. J Exp Med. 2005;202:225–230. doi: 10.1084/jem.20050545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li J, Mitamura T, Fox BA, Bzik DJ, Horii T. Differential localization of processed fragments of Plasmodium falciparum serine repeat antigen and further processing of its N-terminal 47 kDa fragment. Parasitol Int. 2002;51:343–352. doi: 10.1016/s1383-5769(02)00042-9. [DOI] [PubMed] [Google Scholar]
- 118.Chandramohanadas R, Davis PH, Beiting DP, Harbut MB, Darling C, Velmourougane G, Lee MY, Greer PA, Roos DS, Greenbaum DC. Apicomplexan parasites co-opt host calpains to facilitate their escape from infected cells. Science. 2009;324:794–797. doi: 10.1126/science.1171085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Powers JC, Asgian JL, Ekici OD, James KE. Irreversible inhibitors of serine cysteine and threonine proteases. Chem Rev. 2002;102:4639–4750. doi: 10.1021/cr010182v. [DOI] [PubMed] [Google Scholar]
- 120.Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DMA, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Boothroyd JC. Toxoplasma gondii: 25 years and 25 major advances for the field. Int J Parasitol. 2009;39:935–946. doi: 10.1016/j.ijpara.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gopalakrishnan AM, Lopez-Estrano C. Comparative analysis of stage specific gene regulation of apicomplexan parasites: Plasmodium falciparum and Toxoplasma gondii. Infect Disord Drug Targets. 2010;10:303–311. doi: 10.2174/187152610791591593. [DOI] [PubMed] [Google Scholar]
- 123.Bozdech Z, Llinas M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003;1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hu G, Cabrera A, Kono M, Mok S, Chaal BK, Haase S, Engelberg K, Cheemadan S, Spielmann T, Preiser PR, Gilberger TW, Bozdech Z. Transcriptional profiling of growth perturbations of the human malaria parasite Plasmodium falciparum. Nat Biotechnol. 2010;28:91–98. doi: 10.1038/nbt.1597. [DOI] [PubMed] [Google Scholar]
- 125.Meissner M, Breinich MS, Gilson PR, Crabb BS. Molecular genetic tools in Toxoplasma and Plasmodium: achievements and future needs. Current Opinion in Microbiology. 2007;10:349–356. doi: 10.1016/j.mib.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 126.van Dijk MR, Waters AP, Janse CJ. Stable transfection of malaria parasite blood stages. Science. 1995;268 :1358–1362. doi: 10.1126/science.7761856. [DOI] [PubMed] [Google Scholar]
- 127.Wu Y, Sifri CD, Lei HH, Su XZ, Wellems TE. Transfection of Plasmodium falciparum within human red blood cells. Proc Natl Acad Sci U S A. 1995;92:973–977. doi: 10.1073/pnas.92.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Aravind L, Iyer LM, Wellems TE, Miller LH. Plasmodium biology: genomic gleanings. Cell. 2003;115:771–785. doi: 10.1016/s0092-8674(03)01023-7. [DOI] [PubMed] [Google Scholar]
- 129.Blackman MJ. RNAi in protozoan parasites: what hope for the Apicomplexa? Protist. 2003;154:177–180. doi: 10.1078/143446103322166482. [DOI] [PubMed] [Google Scholar]
- 130.Greenbaum DC. Is chemcal genetics the new frontier for malaria biology? Trends in Pharmacological Sciences. 2008;29:51–56. doi: 10.1016/j.tips.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 131.Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ, A rapid reversible. and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Armstrong CM, Goldberg DE. An FKBP destabilization domain modulates protein levels in Plasmodium falciparum. Nat Methods. 2007;4:1007–1009. doi: 10.1038/nmeth1132. [DOI] [PubMed] [Google Scholar]
- 133.Russo I, Oksman A, Vaupel B, Goldberg DE. A calpain unique to alveolates is essential in Plasmodium falciparum and its knockdown reveals an involvement in pre-S-phase development. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1554–1559. doi: 10.1073/pnas.0806926106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dvorin JD, Martyn DC, Patel SD, Grimley JS, Collins CR, Hopp CS, Bright AT, Westenberger S, Winzeler E, Blackman MJ, Baker DA, Wandless TJ, Duraisingh MT. A plant-like kinase in Plasmodium falciparum regulates parasite egress from erythrocytes. Science. 2010;328:910–912. doi: 10.1126/science.1188191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Muralidharan V, Oksman A, Iwamoto M, Wandless TJ, Goldberg DE. Asparagine repeat function in a Plasmodium falciparum protein assessed via a regulatable fluorescent affinity tag. Proc Natl Acad Sci U S A. 2011;108:4411–4416. doi: 10.1073/pnas.1018449108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Meissner M, Krejany E, Gilson PR, de Koning-Ward TF, Soldati D, Crabb BS. Tetracycline analogue-regulated transgene expression in Plasmodium falciparum blood stages using Toxoplasma gondii transactivators. Proc Natl Acad Sci U S A. 2005;102:2980–2985. doi: 10.1073/pnas.0500112102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sadaghiani AM, Verhelst SHL, Bogyo M. Tagging and detection strategies for activity-based proteomics. Curr. Opin. Chem. Biol. 2007;11:20–28. doi: 10.1016/j.cbpa.2006.11.030. [DOI] [PubMed] [Google Scholar]
- 138.Deu E, Yang Z, Wang F, Klemba M, Bogyo M. Use of activity-based probes to develop high throughput screening assays that can be performed in complex cell extracts. Plos One. 2010) e;5:11985. doi: 10.1371/journal.pone.0011985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bowyer PW, Simon GM, Cravatt BF, Bogyo M. Global profiling of proteolysis during rupture of P. falciparum from the host erythrocyte. Mol Cell Proteomics. 2010 doi: 10.1074/mcp.M110.001636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tschan S, Mordmuller B, Kun JF. Threonine peptidases as drug targets against malaria. Expert Opin Ther Targets. 2011 doi: 10.1517/14728222.2011.555399. [DOI] [PubMed] [Google Scholar]
- 141.Mordmuller B, Fendel R, Kreidenweiss A, Gille C, Hurwitz R, Metzger WG, Kun JFJ, Lamkemeyer T, Nordheim A, Kremsner PG. Plasmodia express two threonine-peptidase complexes during asexual development. Molecular and Biochemical Parasitology. 2006;148:79–85. doi: 10.1016/j.molbiopara.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 142.Ramasamy G, Gupta D, Mohmmed A, Chauhan VS. Characterization and localization of Plasmodium falciparum homolog of prokaryotic C1pQ/Hs1V protease. Molecular and Biochemical Parasitology. 2007;152:139–148. doi: 10.1016/j.molbiopara.2007.01.002. [DOI] [PubMed] [Google Scholar]