Abstract
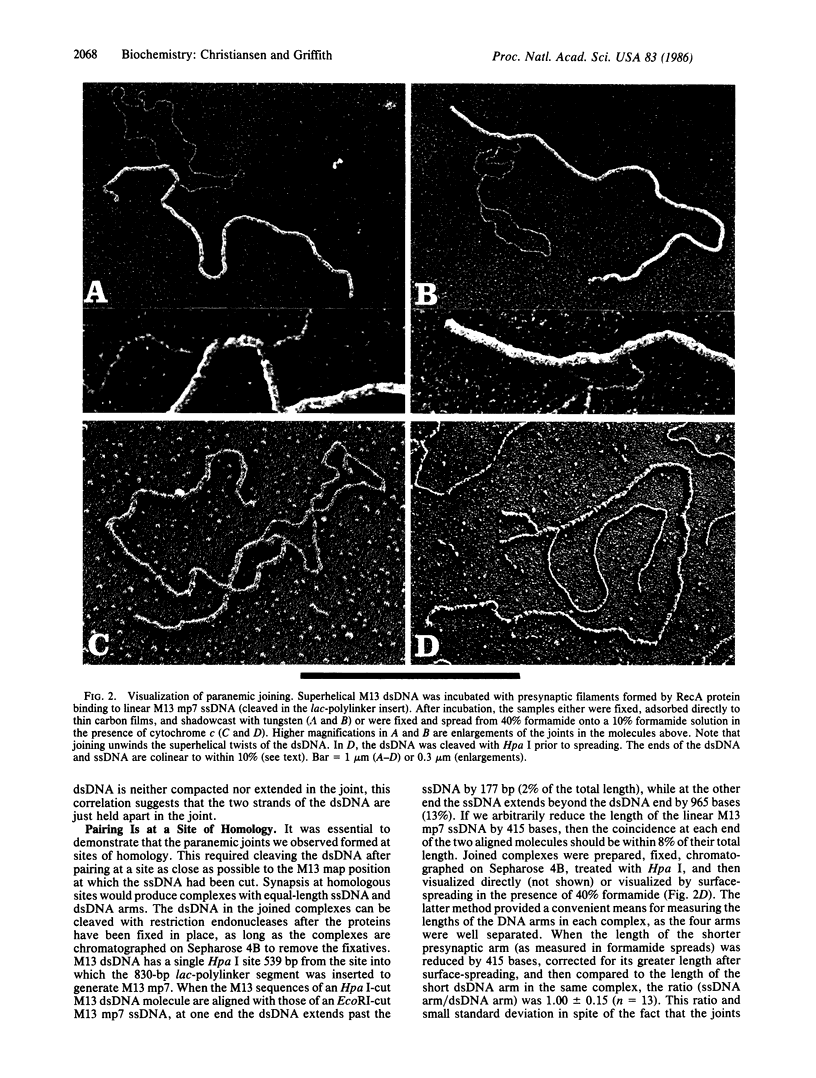
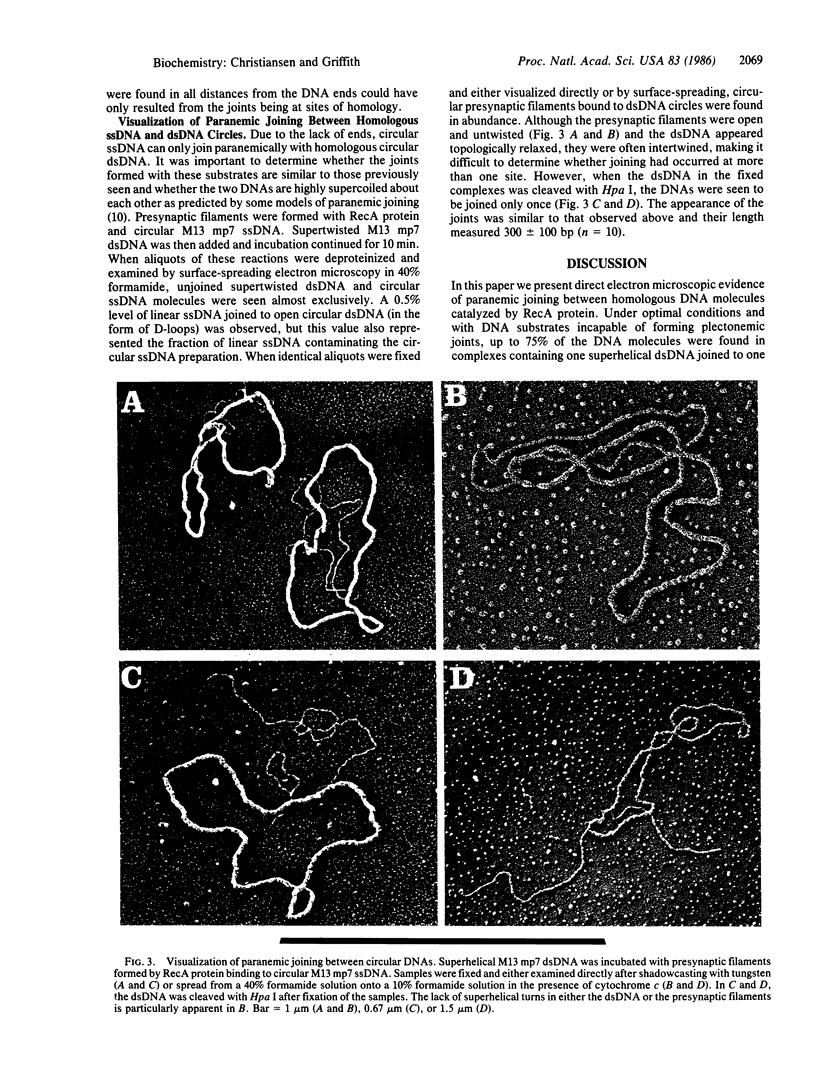
In reactions catalyzed by the RecA protein of Escherichia coli, synapsis between two DNA molecules is believed to occur even in the absence of free homologous DNA ends and to involve a metastable interaction termed paranemic joining. We have used electron microscopic methods to visualize synapse formation between supertwisted M13 double-stranded DNA (dsDNA) and linear M13 mp7 single-stranded DNA (ssDNA) with non-M13 sequences at its ends. These non-M13 sequences block strand invasion and make this pairing equivalent to the joining of two fully circular molecules. We observed a high frequency of joining when the ssDNA was initially assembled into presynaptic filaments with RecA protein. Cleavage of the dsDNA in the joined complexes by Hpa I revealed that the joint was at a site of homology. In these joints, the dsDNA entered the presynaptic filament over a length of 360 +/- 80 base pairs, not visibly altering its ultrastructure, and then dissociated from the filament. Although the dsDNA in the complexes appeared topologically relaxed, deproteinization released supertwisted dsDNA, indicating that the dsDNA was unwound by 34 degrees per base pair in the paranemic joint. When supertwisted M13 dsDNA was paired with circular M13 ssDNA, similar joints were observed and both DNA circles appeared topologically relaxed.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bianchi M., DasGupta C., Radding C. M. Synapsis and the formation of paranemic joints by E. coli RecA protein. Cell. 1983 Oct;34(3):931–939. doi: 10.1016/0092-8674(83)90550-0. [DOI] [PubMed] [Google Scholar]
- Chase J. W., Whittier R. F., Auerbach J., Sancar A., Rupp W. D. Amplification of single-strand DNA binding protein in Escherichia coli. Nucleic Acids Res. 1980 Jul 25;8(14):3215–3227. doi: 10.1093/nar/8.14.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen G., Christiansen C. Heterology of mitochondrial DNA from mammals detected by electron microscopic heteroduplex analyses. Nucleic Acids Res. 1983 Jan 11;11(1):37–56. doi: 10.1093/nar/11.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox M. M., Lehman I. R. Directionality and polarity in recA protein-promoted branch migration. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6018–6022. doi: 10.1073/pnas.78.10.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox M. M., Lehman I. R. recA protein-promoted DNA strand exchange. Stable complexes of recA protein and single-stranded DNA formed in the presence of ATP and single-stranded DNA binding protein. J Biol Chem. 1982 Jul 25;257(14):8523–8532. [PubMed] [Google Scholar]
- Cunningham R. P., Wu A. M., Shibata T., DasGupta C., Radding C. M. Homologous pairing and topological linkage of DNA molecules by combined action of E. coli RecA protein and topoisomerase I. Cell. 1981 Apr;24(1):213–223. doi: 10.1016/0092-8674(81)90517-1. [DOI] [PubMed] [Google Scholar]
- DasGupta C., Shibata T., Cunningham R. P., Radding C. M. The topology of homologous pairing promoted by RecA protein. Cell. 1980 Nov;22(2 Pt 2):437–446. doi: 10.1016/0092-8674(80)90354-2. [DOI] [PubMed] [Google Scholar]
- Flory J., Tsang S. S., Muniyappa K. Isolation and visualization of active presynaptic filaments of recA protein and single-stranded DNA. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7026–7030. doi: 10.1073/pnas.81.22.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J. D., Christiansen G. Electron microscope visualization of chromatin and other DNA-protein complexes. Annu Rev Biophys Bioeng. 1978;7:19–35. doi: 10.1146/annurev.bb.07.060178.000315. [DOI] [PubMed] [Google Scholar]
- Griffith J. D. DNA structure: evidence from electron microscopy. Science. 1978 Aug 11;201(4355):525–527. doi: 10.1126/science.663672. [DOI] [PubMed] [Google Scholar]
- Griffith J. D., Harris L. D., Register J., 3rd Visualization of SSB-ssDNA complexes active in the assembly of stable RecA-DNA filaments. Cold Spring Harb Symp Quant Biol. 1984;49:553–559. doi: 10.1101/sqb.1984.049.01.062. [DOI] [PubMed] [Google Scholar]
- Griffith J., Shores C. G. RecA protein rapidly crystallizes in the presence of spermidine: a valuable step in its purification and physical characterization. Biochemistry. 1985 Jan 1;24(1):158–162. doi: 10.1021/bi00322a022. [DOI] [PubMed] [Google Scholar]
- Howard-Flanders P., West S. C., Stasiak A. Role of RecA protein spiral filaments in genetic recombination. Nature. 1984 May 17;309(5965):215–219. doi: 10.1038/309215a0. [DOI] [PubMed] [Google Scholar]
- Kahn R., Cunningham R. P., DasGupta C., Radding C. M. Polarity of heteroduplex formation promoted by Escherichia coli recA protein. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4786–4790. doi: 10.1073/pnas.78.8.4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn R., Radding C. M. Separation of the presynaptic and synaptic phases of homologous pairing promoted by recA protein. J Biol Chem. 1984 Jun 25;259(12):7495–7503. [PubMed] [Google Scholar]
- Kmiec E. B., Holloman W. K. Synapsis promoted by Ustilago rec1 protein. Cell. 1984 Mar;36(3):593–598. doi: 10.1016/0092-8674(84)90338-6. [DOI] [PubMed] [Google Scholar]
- Kmiec E., Kroeger P., Holliday R., Holloman W. Homologous pairing promoted by Ustilago RecI protein. Cold Spring Harb Symp Quant Biol. 1984;49:675–682. doi: 10.1101/sqb.1984.049.01.076. [DOI] [PubMed] [Google Scholar]
- McEntee K., Weinstock G. M., Lehman I. R. Initiation of general recombination catalyzed in vitro by the recA protein of Escherichia coli. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2615–2619. doi: 10.1073/pnas.76.6.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddles P. W., Lehman I. R. The formation of paranemic and plectonemic joints between DNA molecules by the recA and single-stranded DNA-binding proteins of Escherichia coli. J Biol Chem. 1985 Jan 10;260(1):165–169. [PubMed] [Google Scholar]
- Shibata T., DasGupta C., Cunningham R. P., Radding C. M. Purified Escherichia coli recA protein catalyzes homologous pairing of superhelical DNA and single-stranded fragments. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1638–1642. doi: 10.1073/pnas.76.4.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperrazza J. M., Register J. C., 3rd, Griffith J. Electron microscopy can be used to measure DNA supertwisting. Gene. 1984 Nov;31(1-3):17–22. doi: 10.1016/0378-1119(84)90190-2. [DOI] [PubMed] [Google Scholar]
- Wang J. C. Helical repeat of DNA in solution. Proc Natl Acad Sci U S A. 1979 Jan;76(1):200–203. doi: 10.1073/pnas.76.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A. M., Bianchi M., DasGupta C., Radding C. M. Unwinding associated with synapsis of DNA molecules by recA protein. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1256–1260. doi: 10.1073/pnas.80.5.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]