Abstract
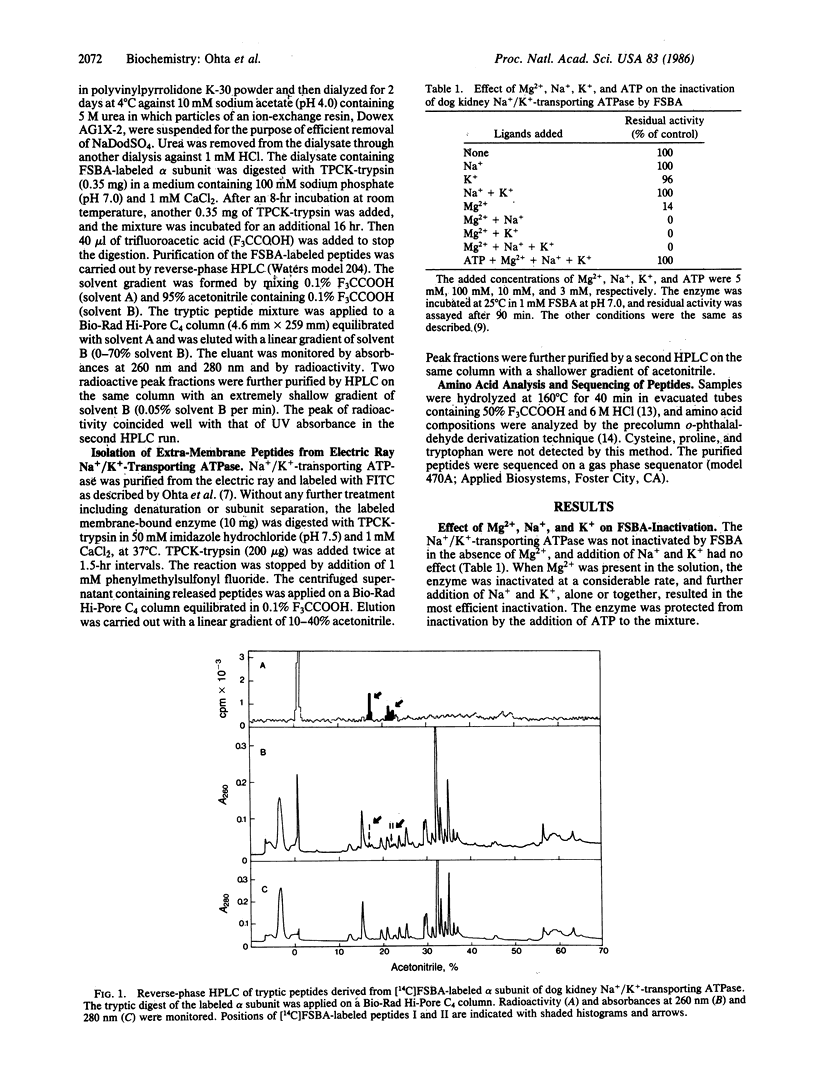
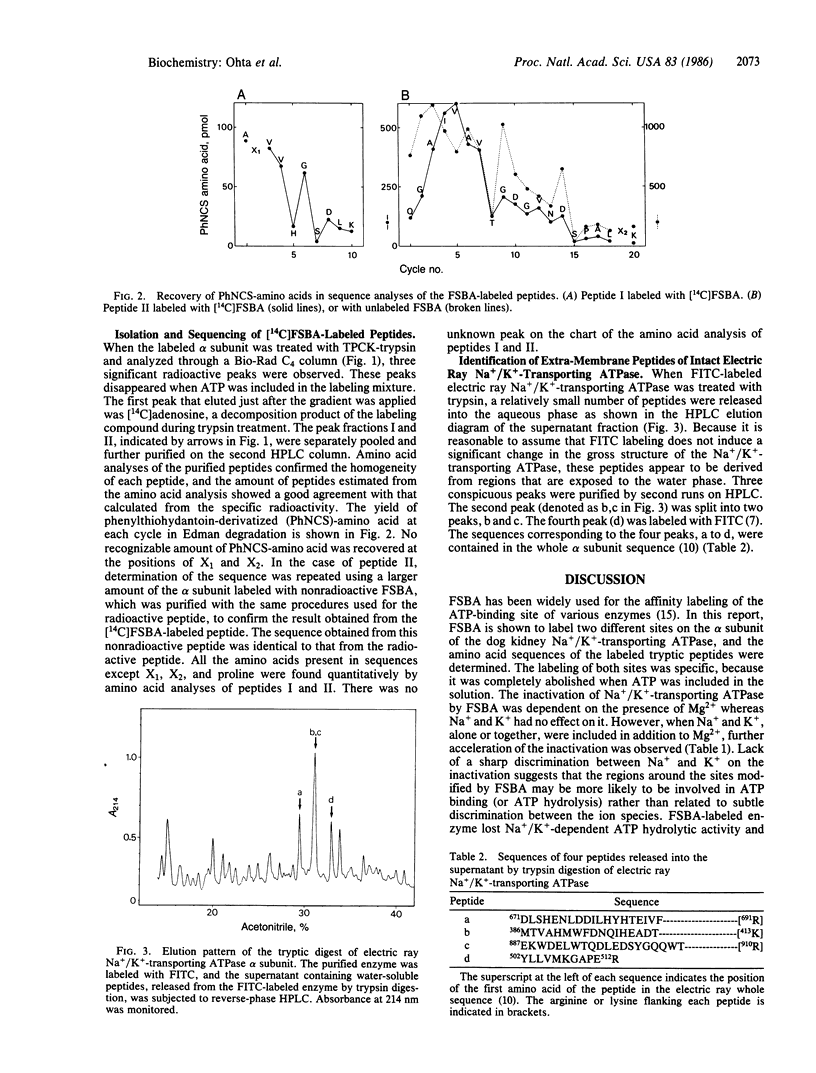
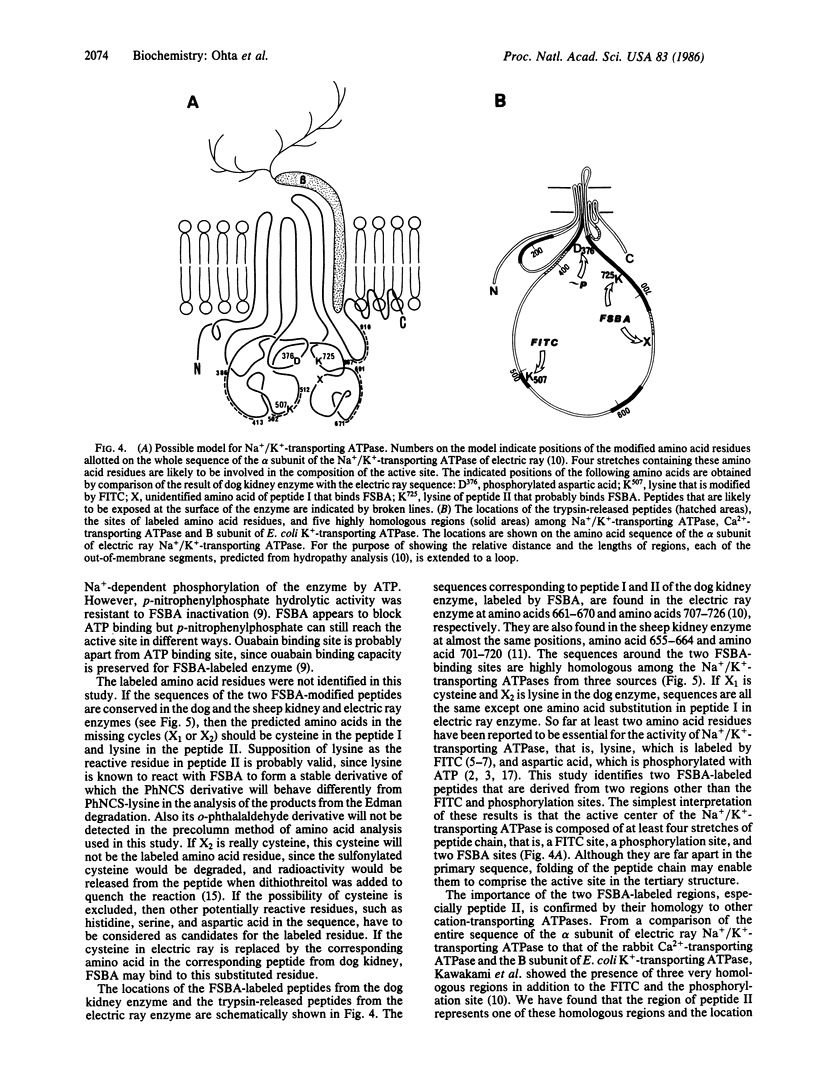
When the dog kidney Na+/K+-transporting ATPase (EC 3.6.1.37, formerly EC 3.6.1.3) was labeled with an ATP analogue, 5'-(p-fluorosulfonyl)benzoyladenosine (FSBA), there was a concomitant loss of ATPase activity. The presence of ATP protected the enzyme from both labeling and inactivation. The ATP-sensitive incorporation of FSBA is associated only with modification of the alpha subunit from which two labeled tryptic peptides were purified and sequenced. To establish any regions of the enzyme protruding from the membrane, the native Na+/K+-transporting ATPase from the electric ray, Torpedo californica, was treated with trypsin; and four peptides, which were released into the water phase, were purified and sequenced. A comparison of the peptide sequences with the deduced amino acid sequences of the DNA coding for the alpha subunit of T. californica and sheep kidney reveal the following. (i) FSBA-labeled peptides from the dog kidney enzyme are located in the central hydrophilic domain and show almost complete sequence homology with the same region in the alpha subunit from the electric ray and sheep kidney. Furthermore, the sequence homology of one of the two labeled peptides can be extended to the sarcoplasmic Ca2+-transporting ATPase and B subunit of Escherichia coli K+-transporting ATPase. (ii) Three trypsin-exposed peptides are found in the central hydrophilic domain, and one peptide is in the hydrophilic segment near the C terminus of the alpha subunit. (iii) The active center of Na+/K+-transporting ATPase is likely to be constructed from at least four different stretches in the primary sequence and, irrespective of the different specificity of cations, the various cation transport ATPases that form phosphorylated enzyme appear to have a common structure at the catalytic site for ATP hydrolysis.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Colman R. F. Affinity labeling of purine nucleotide sites in proteins. Annu Rev Biochem. 1983;52:67–91. doi: 10.1146/annurev.bi.52.070183.000435. [DOI] [PubMed] [Google Scholar]
- Cooper J. B., Winter C. G. 5'-p-fluorosulfonylbenzoyladenosine as an ATP site affinity probe for Na+, K+-ATPase. J Supramol Struct. 1980;13(2):165–174. doi: 10.1002/jss.400130204. [DOI] [PubMed] [Google Scholar]
- Farley R. A., Tran C. M., Carilli C. T., Hawke D., Shively J. E. The amino acid sequence of a fluorescein-labeled peptide from the active site of (Na,K)-ATPase. J Biol Chem. 1984 Aug 10;259(15):9532–9535. [PubMed] [Google Scholar]
- Hesse J. E., Wieczorek L., Altendorf K., Reicin A. S., Dorus E., Epstein W. Sequence homology between two membrane transport ATPases, the Kdp-ATPase of Escherichia coli and the Ca2+-ATPase of sarcoplasmic reticulum. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4746–4750. doi: 10.1073/pnas.81.15.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen P. L. Mechanism of the Na+, K+ pump. Protein structure and conformations of the pure (Na+ +K+)-ATPase. Biochim Biophys Acta. 1982 Aug 11;694(1):27–68. doi: 10.1016/0304-4157(82)90013-2. [DOI] [PubMed] [Google Scholar]
- Kawakami K., Noguchi S., Noda M., Takahashi H., Ohta T., Kawamura M., Nojima H., Nagano K., Hirose T., Inayama S. Primary structure of the alpha-subunit of Torpedo californica (Na+ + K+)ATPase deduced from cDNA sequence. Nature. 1985 Aug 22;316(6030):733–736. doi: 10.1038/316733a0. [DOI] [PubMed] [Google Scholar]
- Kawamura M., Ohta T., Nagano K. Effect of reducing agents on the solubilization of renal sodium and potassium dependent ATPase with detergent. J Biochem. 1980 May;87(5):1327–1333. doi: 10.1093/oxfordjournals.jbchem.a132871. [DOI] [PubMed] [Google Scholar]
- Kirley T. L., Wallick E. T., Lane L. K. The amino acid sequence of the fluorescein isothiocyanate reactive site of lamb and rat kidney Na+- and K+-dependent ATPase. Biochem Biophys Res Commun. 1984 Dec 14;125(2):767–773. doi: 10.1016/0006-291x(84)90605-3. [DOI] [PubMed] [Google Scholar]
- MacLennan D. H., Brandl C. J., Korczak B., Green N. M. Amino-acid sequence of a Ca2+ + Mg2+-dependent ATPase from rabbit muscle sarcoplasmic reticulum, deduced from its complementary DNA sequence. Nature. 1985 Aug 22;316(6030):696–700. doi: 10.1038/316696a0. [DOI] [PubMed] [Google Scholar]
- Ohta T., Morohashi M., Kawamura M., Yoshida M. The amino acid sequence of the fluorescein-labeled peptides of electric ray and brine shrimp (Na,K)-ATPase. Biochem Biophys Res Commun. 1985 Jul 16;130(1):221–228. doi: 10.1016/0006-291x(85)90405-x. [DOI] [PubMed] [Google Scholar]
- Post R. L., Kume S. Evidence for an aspartyl phosphate residue at the active site of sodium and potassium ion transport adenosine triphosphatase. J Biol Chem. 1973 Oct 25;248(20):6993–7000. [PubMed] [Google Scholar]
- Shull G. E., Schwartz A., Lingrel J. B. Amino-acid sequence of the catalytic subunit of the (Na+ + K+)ATPase deduced from a complementary DNA. Nature. 1985 Aug 22;316(6030):691–695. doi: 10.1038/316691a0. [DOI] [PubMed] [Google Scholar]
- Walderhaug M. O., Post R. L., Saccomani G., Leonard R. T., Briskin D. P. Structural relatedness of three ion-transport adenosine triphosphatases around their active sites of phosphorylation. J Biol Chem. 1985 Mar 25;260(6):3852–3859. [PubMed] [Google Scholar]



