Abstract
Increased mucosal polyamine levels and ornithine decarboxylase (ODC) activity are associated with an increased risk of colorectal neoplasia, and aspirin treatment reduces risk. Previous studies suggest that a single nucleotide polymorphism (SNP) in the promoter of the ODC gene (rs2302615) may be associated with adenoma risk and/or response to aspirin chemoprevention. However, a comprehensive investigation of common genetic variation in the region of ODC gene is lacking. Using a tag SNP approach, we investigated associations between genotype or haplotype and adenoma risk among a cohort of 792 white non-Hispanic participants in a randomized trial of aspirin. Generalized linear regression was used to compute relative risks (RRs) and 95% confidence intervals (95% CIs) adjusted for age and sex. The false discovery rate was used to account for multiple testing. Interactions terms were used to assess whether genotype modified the effect of aspirin treatment. Of 15 SNPs analyzed, 7 were statistically significantly associated with adenoma risk. However, in multiple SNP regression models, only 2 of these, located downstream of the gene, were independently associated with risk: rs11694911 (1.29 RR, 1.08–1.53 95% CI, P=0.005) and rs2430420 (1.20 RR, 1.03–1.40 95% CI, P=0.022). In addition, there was evidence that rs2430420 and rs28362380 modified the effect of aspirin treatment, whereas the previously investigated SNP, rs2302615, had no statistically significant main effect or interaction with aspirin treatment. Our findings suggest that common genetic variants located downstream (3’) of the ODC gene influence risk of colorectal adenoma and may also impact the efficacy of aspirin chemoprevention.
Keywords: ornithine decarboxylase, polyamines, polymorphism, colorectal adenoma, aspirin, chemoprevention
INTRODUCTION
Colorectal cancer is the second leading cause of cancer death in the United States (1) and is potentially preventable. Modification of diet and lifestyle factors as well as the use of chemoprevention strategies in combination with screening may reduce the burden of this disease (2, 3). Substantial evidence from meta-analyses of randomized clinical trials indicate that aspirin is effective for prevention of colorectal cancer (4) and adenomas (the precursor to most cancers) (5) and potentially cost-effective (6), although its effect may be modest. However, in a recent randomized clinical trial, a combination of the ornithine decarboxylase (ODC) inhibitor difluoromethylornithine (DFMO) and the non-steroidal anti-inflammatory drug (NSAID) sulindac, was remarkably effective in reducing the occurrence of colorectal adenomas: all adenomas were reduced by 70% and advanced lesions by more than 90% (7). ODC is a key regulatory enzyme for the synthesis of polyamines, small highly regulated molecules that are essential for cell growth and for the regulation of numerous processes, including gene expression and ion channel activity (8). ODC catalyzes the first step in polyamine biosynthesis, the conversion of ornithine to putrescine (9). Although the association of increased polyamine synthesis (10, 11) and inflammation (12, 13) with colorectal carcinogenesis has been recognized for some time, the striking results of the DFMO/sulindac trial highlight the potential for an effective combination chemoprevention strategy (14). Given the mandate to use pharmacogenomics to personalize cancer treatment (15) and presumably prevention, it will be important to investigate the potential effect of common genetic variation on such a strategy.
Several previous genetic epidemiological studies of a single nucleotide polymorphism (SNP) in the ODC gene (rs2302615) suggested that the variant allele may be associated with a decreased risk for adenoma recurrence and/or an enhanced response to aspirin use or treatment (16–18). However, in the DFMO/sulindac trial the variant allele appeared to be associated with a reduced response to treatment (19). In addition, it was associated with reduced survival in a cohort of colorectal cancer patients (20). Regardless, a major limitation of this prior work is that only one common SNP in the ODC gene has been investigated to date. The goal of the present study was to extend this work to investigate associations with adenoma risk and response to aspirin chemoprevention of common genetic variation throughout the ODC gene and adjacent chromosomal regions.
MATERIALS AND METHODS
Study Design and Population
We performed a cohort analysis of the association between ODC1 genotypes and colorectal adenoma recurrence among participants in the Aspirin/Folate Polyp Prevention Study, a double-blind, placebo-controlled, randomized clinical trial of aspirin and/or folic acid for the prevention of colorectal adenoma recurrence (21, 22). Human subjects committees at each of the clinical centers approved the study protocol and materials distributed to participants and all participants provided written informed consent. The study design and main findings have been described in detail previously (21, 22). In brief, eligible participants had no history of colorectal cancer or any familial cancer syndrome but had a recent history of one or more histologically confirmed colorectal adenoma and a complete colonoscopy within 3 months prior to enrollment with all polyps removed from the bowel. Subjects, who agreed to avoid NSAID use during their participation in the study, were randomized to aspirin treatment (placebo, 81 or 325 mg daily) and independently to folic acid treatment (placebo or 1 mg daily) in a 3 X 2 factorial design. Aspirin treatment was continued for an average of almost three years (33 months) until a follow-up colonoscopy was performed. The principal outcome of the study was the occurrence of at least one adenoma during randomized treatment. To maximize outcome ascertainment, we included findings from colonoscopies that were performed at least one year after randomization and on or before September 28, 2001, as described in the publication of the main study findings (21). Thus, the actual follow-up period ranged from 19 to 59 months post randomization. A single, blinded, study pathologist provided uniform review of all clinical samples removed from the large bowel.
SNP Selection and Genotyping
To provide comprehensive coverage of the ODC1 gene and adjacent potentially regulatory regions, SNPs within 10 kb upstream and 10 kb downstream of the gene were chosen for genotyping using data from four sources. Two of the sources are publically available databases: the HAPMAP - CEU population (Utah residents with Northern and Western European ancestry from the CEPH collection from NCBI release #36; see (23)) and the NIEHS SNPs (National Institute of Environmental Health Sciences Environmental Genome Project; see (24)). In addition, some SNPs were chosen that were not included in either of the public databases, based on frequency data obtained from more comprehensive genotyping of CEPH samples (N=90) and a sample of 81 participants in the DFMO/sulindac trial (7) [data shared by Drs. Eugene Gerner and Patricia Thompson from the Arizona Cancer Center (Tucson, AZ)]. Because “binning” of SNPs with r2 ≥ 0.8 was inconsistent across the 4 sets of genotyping data, all SNPs with a minor allele frequency (MAF) ≥ 3% were genotyped and tag SNPs that were identified after genotyping of the current population were chosen for analyses (see below).
Genomic DNA was isolated as previously described (17, 25). Genotyping was performed by McGill University and Genome Quebec Innovation Center (Montreal, Canada) using Sequenom iPLEX Gold technology according to the manufacturer’s instructions (Sequenom, San Diego, CA) using a single panel. The oligos used are available upon request. Of a total of 31 SNPs selected for genotyping, 2 were in GC rich regions and failed the initial validation step and therefore were not genotyped (rs2302616 and rs28742580). The remaining 29 SNPs had call rates ranging from 98.14 to 100% (median = 99.73%) and were in Hardy Weinberg equilibrium (P>0.05 using a χ2 test for the comparison between observed and expected genotype counts among white non-Hispanic subjects with no adenoma recurrence). Concordance rates among 44 blinded duplicate samples was 100% for all SNPs except for one (rs13000913), which had one error for a concordance rate of 97.7%. The sample success rate was 99.0%; samples that could not be called at more than 3 of the 29 SNPs were deemed to have failed and were dropped from the analysis dataset. Of the successful samples, 93.2% could be called at all SNPs and another 5.8% could be called at all but one SNP. Of the 29 SNPs that were successfully genotyped, 13 were excluded from the analyses because they were in high linkage disequilibrium (r2 ≥ 0.8) with a tag SNP selected using Haploview Tagger (26), and 1 was excluded because it had a MAF less than 3% in our dataset. The excluded SNPs are: rs2463463, rs4669584, rs28362433, rs28362422, rs2357550, rs1405948, rs3752661, rs2302613, rs2302614, rs12616336, rs7608353, rs7558559, rs2009741, and rs6432097. Thus, a total of 15 tag SNPs are included in the statistical analyses presented here along with rs2302615, which we genotyped previously (17). Notably, rs2302615 is not in high linkage disequilibrium (r2 ≥ 0.8) with any of the newly genotyped SNPs and thus meets the criteria for inclusion in these analyses as an independent tag SNP.
Statistical Analysis
Of the total of 920 participants from the Aspirin/Folate Polyp Prevention Study with both genetic data and trial end-point data available for this analysis, 128 (13.9%) reported a race/ethnicity other than non-Hispanic white. Genotype was statistically significantly associated with race for 8 of the 15 tag SNPs analyzed in our dataset (not shown) and race/ethnicity may be associated with outcome [see (25)]. Therefore, we limited our analyses to participants self-identified as “white, not of Hispanic origin” (N=792) because we did not have adequate numbers of other racial/ethnic groups to appropriately address population stratification (see Figure 1). The outcome assessed in these analyses was the recurrence of one or more adenomas during randomized aspirin treatment. Because the asymptomatic nature of this outcome prohibited time-to-event analysis, findings were assessed at follow-up colonoscopy. Risk ratios and 95% confidence intervals used to estimate the association between genotype and adenoma recurrence were calculated with an underdispersed generalized linear regression model using the Poisson distribution as an approximation to the binomial family. Minimally adjusted (for age and sex) relative risks are shown in the tables. Several levels of genotype association analyses were performed, as described below.
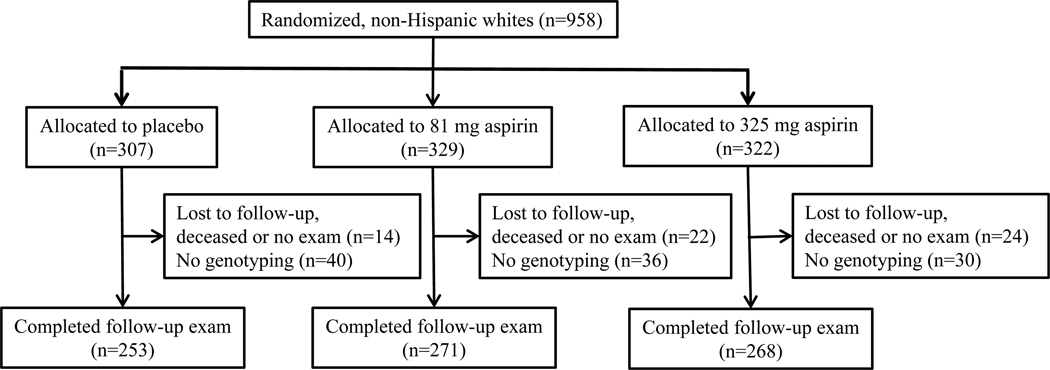
Figure 1.
Flow diagram showing the numbers of participants from the Aspirin/Folate Polyp Prevention Study that were included in this secondary analysis involving ODC genotyping. The non-Hispanic whites shown here represent 85% of the total randomized population (n=1121).
For SNP level analyses, genotypes were included in the regression equation one at a time using an additive genetic model, providing per-allele relative risks and Wald test p-values for the tables. To account for multiple statistical testing of 15 SNPs, false discovery rate (FDR) q values were calculated (27) using the R statistical package (see (28)). Using this FDR method we control the proportion of type I errors made rather than the probability of making even a single type I error (i.e. 5% probability with the classical Bonferroni method of multiple testing correction, which assumes independence of tests and thus is not appropriate for genetic analyses within and around a single gene). We used an FDR threshold of 20%, which has been suggested for candidate gene studies (29), such that up to 20% of the declared discoveries are expected to be false. In secondary analyses of statistically significantly associated SNPs, genetic models (additive, dominant and recessive) were examined for best fit using maximum likelihood-based statistics.
For haplotype level analyses, used to assess the combined effect of correlated SNPs, linkage disequilibrium blocks were defined with Haploview (26) using the default algorithm based on confidence bounds on D’ (30) (see Figure 2). Phased haplotype pairs and probabilities were estimated with Powermarker v3.25 (31) using the EM algorithm (32). Generalized linear regression was used to estimate the haplotype association with risk for adenoma recurrence taking haplotype uncertainty (probability) into account. The haplotype was modeled as a continuous variable wherein the number of copies of each allele was multiplied by its probability to obtain a continuous variable. The most common haplotype was used as the reference group and omitted from the model. For each haplotype, the model provides an estimate of the risk associated with each additional copy of the specified haplotype. The most frequent haplotypes (with frequencies above 3%) were analyzed individually, and the remaining rare haplotypes were pooled. Wald test p values were calculated for each individual haplotype and a likelihood ratio test p-value was calculated for the joint contribution of all haplotypes to the model.
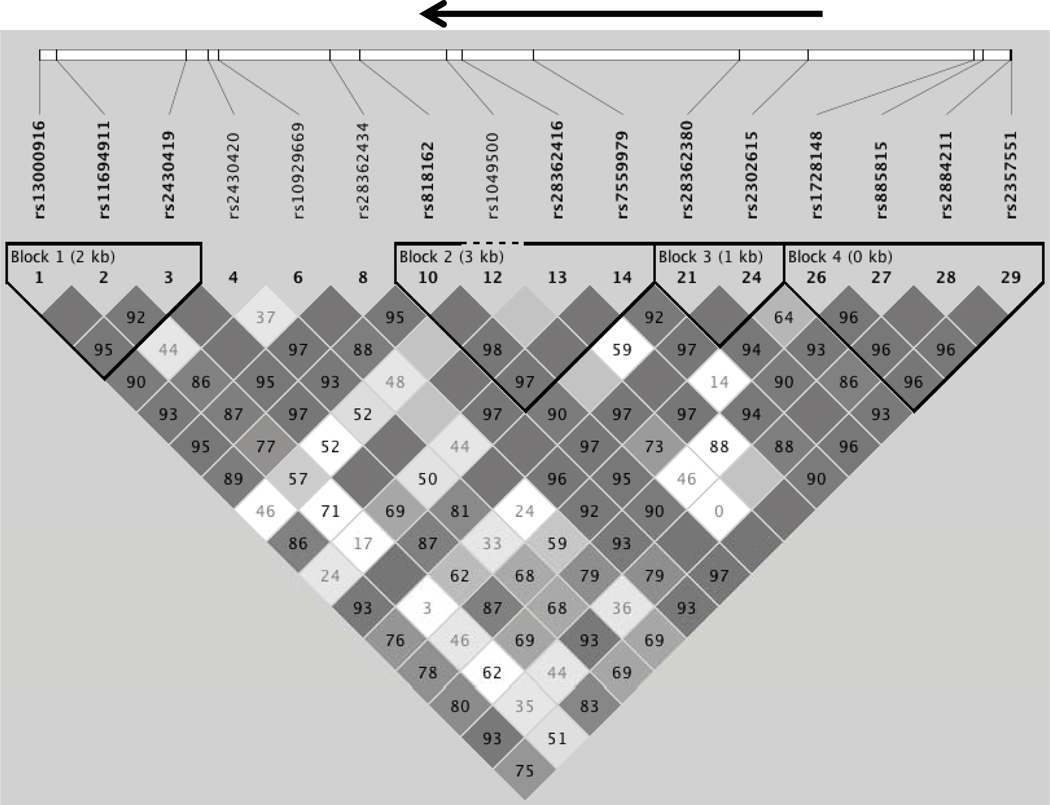
Figure 2.
The linkage disequilibrium (LD) block structure of the 19.33 kB genotyped region is shown as determined using Haploview (26). The heavy black arrow on top shows the location of the ODC gene (transcribed region). D’ values are shown for each pairwise comparison (dark blocks without values are in complete LD, while lighter boxes without values are “inconclusive”) (26).
For gene level analyses, a multiple SNP test was used to assess which SNPs in this chromosomal region were independently associated with risk of adenoma recurrence in an exploratory data-driven analysis. Starting with a composite regression equation containing all 16 tag SNPs, a step-down selection process was used in which SNP variables were removed from the equation one at a time in order of decreasing likelihood ratio test p-values until only SNPs with p-values <0.05 remained. A step-up approach gave the same results. A global multiple degrees of freedom likelihood ratio test was used to assess the statistical significance of the joint contribution of all independently associated SNPs to the model. In addition, a composite genetic risk score was created for independently associated SNPs by summing the number of risk genotypes over these loci.
We also evaluated whether aspirin treatment interacted with ODC genotypes to modify associations with adenoma risk using interaction terms in the single SNP regression models and Wald tests. Stratified analyses (by aspirin treatment group or by genotype) were used to obtain stratum specific estimates of relative risk and confidence intervals. We did not account for multiple testing in these analyses as we had limited power.
Analyses that included study treatment with aspirin or folate were conducted according to the intention-to-treat principle. All statistical tests were two-sided and considered significant at a value of P < 0.05, except as otherwise indicated above. Stata (version 10) was used for all analyses, except as described above.
RESULTS
Demographic and other selected characteristics of non-Hispanic white participants from the Aspirin/Folate Polyp Prevention Study with genotypeand outcome data that were included in the present analysis (see Figure 1) are presented in Table 1. Among the 792 participants, 370 (46.7%) had a recurrence of one or more colorectal adenomas during follow-up. The mean age was almost 58 years and the majority of participants were males (64%). Approximately 39% of participants had a family history of colorectal cancer among first-degree relatives. On average, participants were followed for 32.8 months from randomization to their follow-up colonoscopy. As observed in the full analyses of the trial (21, 22), individuals who were randomized to 81 mg/day of aspirin were less likely to have a recurrence compared with those randomized to the placebo arm (P=0.04), whereas treatment with 325 mg/day aspirin (P=0.83) or 1 mg/day folate (P=0.51) was not significantly associated with the outcome. In addition, the main effect of aspirin in this subset of 792 participants (0.77 RR, 0.63–0.94 95% CI, P=0.009 for 81 mg aspirin and 0.99 RR, 0.82–1.19 95% CI, P=0.89 for 325 mg aspirin) was similar to that for the entire cohort (21).
Table 1.
Selected characteristics of the study populationa
| Characteristics | Total | No Adenoma |
Adenoma Recurrence |
Pb |
|---|---|---|---|---|
| No. of subjects (%) | 792 (100) | 422 (53.3) | 370 (46.7) | |
| Mean age at enrollment ± SD, y | 57.83 ± 9.53 | 56.4 ± 9.5 | 59.4 ± 9.3 | <0.001 |
| Sex, No. (%) | ||||
| Male | 507 (64.0) | 250 (59.2) | 257 (69.5) | 0.003 |
| Female | 285 (36.0) | 172 (40.8) | 113 (30.5) | |
| Family history of colorectal cancer, No. (%)c | ||||
| No | 397 (60.9) | 218 (61.8) | 179 (59.9) | 0.24 |
| Yes | 255 (39.1) | 135 (38.2) | 120 (40.1) | |
| Follow-up time (mean ± SD), mod | 32.8 ± 3.6 | 32.6 ± 3.1 | 33.0 ± 4.1 | 0.14 |
| Aspirin treatment group, No. (%) | ||||
| Placebo | 253 (31.9) | 128 (30.3) | 125 (33.8) | |
| 81 mg Aspirin | 271 (34.2) | 161 (38.2) | 110 (29.7) | 0.04 |
| 325 mg Aspirin | 268 (33.8) | 133 (31.5) | 135 (36.5) | 0.83 |
| Folate treatment group, No. (%) | ||||
| Placebo | 358 (45.2) | 196 (46.5) | 162 (43.8) | |
| 1 mg Folate | 367 (46.3) | 192 (45.5) | 175 (47.3) | 0.51 |
| Not randomized to folate | 67 (8.5) | 34 (8.1) | 33 (8.9) | 0.55 |
Only participants self-identifying as “white, not of Hispanic origin” were included.
Tests for comparison between group with no adenoma and group with adenoma recurrence using two-sample t test for continuous variables and Pearson χ2 test for categorical variables.
Family History data is missing for 140 subjects.
Months between randomization and follow-up colonoscopy.
We first examined associations of common SNPs in the vicinity of the ODC1 gene with risk of adenoma recurrence in single SNP analyses (Table 2). The region analyzed encompassed about 23 kb in total: including approximately 7 kb upstream (5’) and 8 kb downstream (3’) of the 8 kb transcribed region of the ODC1 gene. In Table 2, minor allele frequencies (MAFs) and gene locations are shown for the SNPs that were included in the statistical analyses, including 15 newly genotyped SNPs and rs2302615, which was genotyped previously (17). Results for seven SNPs were statistically significant at a value of P < 0.05. Of these, five were associated with increased risk: rs11694911 (1.29 RR, 1.10–1.51 95% CI), rs2430420 (1.17 RR, 1.05–1.31 95% CI), rs10929669 (1.22 RR, 1.04–1.43 95% CI), rs1049500 (1.38 RR, 1.10–1.73 95% CI), rs2357551 (1.13 RR, 1.01–1.27 95% CI); and two were associated with decreased risk: rs13000916 (0.89 RR, 0.80–0.99 95% CI), rs818162 (0.86 RR, 0.76–0.97 95% CI) of adenoma recurrence. After accounting for multiple comparisons using a 20% false discovery rate threshold (Q < 0.2), all seven associations were still statistically significant. The previously investigated SNP, rs2302615, was not associated with risk when the analysis was restricted to non-Hispanic whites, in agreement with our previous results for all participants (17). Although an additive genetic model was used for all SNPs in the analyses shown in Table 2, other models (i.e. dominant or recessive) provided a better fit for several of the SNPs with a significant association suggesting larger effect sizes (data not shown).
Table 2.
Associations of ODC genotypes with risk of adenoma recurrence, Aspirin/Folate Polyp Prevention Study, 1994–2001
| SNP | Positiona | Location | Call rate | PHWE | MAF | RR (95% CI)c | P | Qd |
|---|---|---|---|---|---|---|---|---|
| rs13000916 | 10490304 | 3’ flanking | 0.995 | 0.09 | 0.449 | 0.89 (0.80–0.99) | 0.031 | 0.07 |
| rs11694911 | 10490649 | 3’flanking | 0.991 | 0.96 | 0.106 | 1.29 (1.10–1.51) | 0.002 | 0.027 |
| rs2430419 | 10493239 | 3’flanking | 1.000 | 0.59 | 0.247 | 1.09 (0.97–1.24) | 0.15 | 0.27 |
| rs2430420 | 10493677 | 3’flanking | 1.000 | 0.86 | 0.324 | 1.17 (1.05–1.31) | 0.005 | 0.027 |
| rs10929669 | 10493883 | 3’flanking | 0.997 | 0.61 | 0.115 | 1.22 (1.04–1.43) | 0.017 | 0.054 |
| rs28362434 | 10496095 | 3’flanking | 1.000 | 0.82 | 0.183 | 0.94 (0.81–1.08) | 0.36 | 0.52 |
| rs818162 | 10496675 | 3’flanking | 1.000 | 0.44 | 0.302 | 0.86 (0.76–0.97) | 0.012 | 0.048 |
| rs1049500 | 10498418 | Exon 12b | 1.000 | 0.57 | 0.041 | 1.38 (1.10–1.73) | 0.005 | 0.027 |
| rs28362416 | 10498720 | Intron 11 | 0.996 | 0.10 | 0.067 | 0.85 (0.67–1.07) | 0.18 | 0.29 |
| rs7559979 | 10500130 | Intron 8 | 0.985 | 0.61 | 0.347 | 1.05 (0.93–1.17) | 0.44 | 0.59 |
| rs28362380 | 10504231 | Intron 1 | 0.999 | 0.83 | 0.103 | 1.04 (0.88–1.23) | 0.67 | 0.77 |
| rs2302615e | 10505589 | Intron 1 | 0.998 | 0.50 | 0.254 | 1.02 (0.90–1.15) | 0.80 | |
| rs1728148 | 10508908 | 5’ flanking | 0.999 | 0.79 | 0.451 | 0.92 (0.83–1.02) | 0.12 | 0.24 |
| rs885815 | 10509079 | 5’ flanking | 0.992 | 0.32 | 0.214 | 0.96 (0.84–1.09) | 0.51 | 0.63 |
| rs2884211 | 10509623 | 5’ flanking | 1.000 | 0.70 | 0.077 | 1.01 (0.83–1.23) | 0.89 | 0.94 |
| rs2357551 | 10509631 | 5’ flanking | 1.000 | 0.73 | 0.342 | 1.13 (1.01–1.27) | 0.029 | 0.07 |
Abbreviations: SNP, single nucleotide polymorphism; MAF, minor allele frequency; HWE, Hardy-Weinberg equilibrium; RR, relative risk; CI, confidence interval.
Location on chromosome 2p25 according to NCBI Human Genome Map Build 36.
The substitution is synonymous.
Per allele relative risk and Wald test p-value using an additive genetic model adjusted for age and sex
False-discovery rate Q-values (27).
Previously genotyped SNP, not included in multiple testing correction (n=765)(17).
We also examined the association of adenoma recurrence with common haplotypes found within 4 linkage blocks identified in this segment of the chromosome (see Table 3) to assess the combined effect of correlated SNPs. In the first block, which contains 3 SNPs, a haplotype (GTG) found in 10.6% of the study population (8.6% controls, 13.0% cases) was associated with a 33% increased risk (1.33 RR, 1.12–1.57 95% CI, P=0.001) of adenoma recurrence (per copy of the haplotype) compared to the most common haplotype in this block (TCG). Another haplotype (GCA), occurring in 24% of the study population (23.1% controls, 25.5% cases), was associated with a 14% increased risk that was borderline statistically significant (1.14 RR, 1.00–1.30 95% CI, P=0.06). A test of the overall association of genetic variation in block 1 with adenoma recurrence was statistically significant (P=0.011). In the second block, which contains 4 SNPs, one haplotype (CCCT) found in 23.3% of individuals (25.2% controls, 21.4% cases) was associated with a 15% decreased risk (0.85 RR, 0.73–0.98 95% CI, P=0.029) and another (GTCC) found in 4.1% of individuals (2.7% controls, 5.7% cases) was associated with a 27% increased risk (1.27 RR, 1.01–1.61 95% CI, P=0.044) compared to the most common haplotype (GCCT). Overall, genetic variation in this block was also statistically significantly associated with adenoma recurrence (P=0.012). Genetic variation in block 3 was not associated with the outcome (P=0.63). For the fourth block, one haplotype (CAAA) found in 26.1% of individuals (23.8% controls, 28.9% cases) was associated with a 17% increased risk (1.17 RR, 1.02–1.33 95% CI, P=0.021) per copy of the haplotype compared to the most common haplotype in this block (GAAG), but the test of overall association was not statistically significant (P=0.19).
Table 3.
Association of ODC haplotypes with risk for adenoma recurrence, Aspirin/Folate Polyp Prevention Study, 1994–2001
| Haplotype | Frequency (%) Controls/Cases |
RR (95% CI) | Pa | Pb |
|---|---|---|---|---|
| Block 1c | ||||
| TCG | 47.2 / 41.6 | 1.00 (reference) | 0.011 | |
| GCA | 23.1 / 25.5 | 1.14 (1.00–1.30) | 0.06 | |
| GCG | 21.0 / 19.2 | 1.02 (0.88–1.17) | 0.84 | |
| GTG | 8.5 / 13.0 | 1.33 (1.12–1.57) | 0.001 | |
| Rare | 0.2 / 0.7 | 1.41 (0.70–2.81) | 0.34 | |
| Block 2d | ||||
| GCCT | 33.3 / 37.3 | 1.00 (reference) | 0.012 | |
| GCCC | 31.0 / 29.7 | 0.91 (0.80–1.05) | 0.20 | |
| CCCT | 25.2 / 21.4 | 0.85 (0.73–0.98) | 0.029 | |
| CCTT | 7.3 / 5.8 | 0.82 (0.64–1.04) | 0.11 | |
| GTCC | 2.7 / 5.7 | 1.27 (1.01–1.61) | 0.044 | |
| Rare | 0.4 / 0.1 | 0.55 (0.13–2.32) | 0.41 | |
| Block 3e | ||||
| TG | 66.1 / 64.3 | 1.00 (reference) | 0.63 | |
| TA | 23.7 / 25.3 | 1.05 (0.93–1.20) | 0.41 | |
| CG | 10.2 / 10.4 | 1.06 (0.89–1.27) | 0.50 | |
| Block 4f | ||||
| GAAG | 46.4 / 41.9 | 1.00 (reference) | 0.19 | |
| CAAA | 23.8 / 28.9 | 1.17 (1.02–1.33) | 0.021 | |
| CGAG | 21.4 / 20.1 | 1.02 (0.88–1.18) | 0.81 | |
| CAGA | 7.3 / 7.4 | 1.06 (0.86–1.31) | 0.57 | |
| Rare | 0.9 / 1.6 | 1.27 (0.86–1.31) | 0.33 |
Abbreviations: RR, relative risk; CI, confidence interval
Per haplotype relative risk and Wald test P-value adjusted for age and sex.
Likelihood ratio test P-values for joint test of all haplotypes in the block.
Block 1 includes rs13000916, rs11694911 and rs2430419.
Block 2 includes rs818162, rs1049500, rs28362416 and rs7559979.
Block 3 includes rs28362380 and rs2302615.
Block 4 includes rs1728148, rs885815, rs2884211 and rs2357551.
Although the tag SNPs analyzed here were not in strong linkage disequilibrium (by definition r2<0.8), some correlation still exists between them. Thus, in addition to the single SNP and haplotype analyses described above, a multiple SNP analysis was used to assess which SNPs in this chromosomal region were independently associated with risk of adenoma recurrence. Starting with all SNPs in Table 2, those that were not statistically significantly associated with risk were successively removed from a composite regression model, until only 2 SNPs remained showing statistically significant independent associations with risk: rs11694911 (1.29 RR, 1.08–1.53 95% CI, P=0.005) and rs2430420 (1.20 RR, 1.03–1.40 95% CI, P=0.022) using dominant genetic models (which provided a better fit than additive or recessive models). The joint contribution of these two SNPs to adenoma risk was highly statistically significant (P=0.0003) and the linkage disequilibrium between them was very low (r2 = 0.05). In addition, there was no evidence for an interaction between the two SNPs: their combined effect was essentially the sum of their excess risks. Thus, having at least one risk allele at both loci (15% of the study population) was associated with a 53% increased risk (1.53 RR, 1.24–1.90 95% CI, P<0.001) whereas having at least one risk allele at only one loci (45% of the study population) was associated with a 24% increased risk (1.24 RR, 1.12–1.38 95% CI, P<0.001) compared to having no risk alleles at either loci (40% of the study population, referent group).
Finally, we evaluated whether there was evidence for an interaction between ODC genotypes and aspirin treatment on risk for adenoma recurrence. Table 4 shows the association of each genotype with adenoma risk stratified by aspirin treatment group. Two SNPs (rs2430420 and rs28362380) had nominally statistically significant interactions, although these findings were not corrected for multiple testing. For rs2430420, which had an independent statistically significant main effect as described above, the variant allele was not associated with risk in the placebo group, but was associated with an increased risk of 21% (1.21 RR, 0.98–1.49 95% CI) and 38% (1.38 RR, 1.15–1.66 95% CI) per allele in the 81 and 325 mg aspirin treatment groups, respectively. Conversely, when the effect of aspirin was stratified by rs2430420 genotype (Table 5), 81 mg aspirin treatment appeared to be protective among wild type homozygotes, with a risk reduction of 32% (RR 0.68, 0.50–0.94 95% CI) compared to placebo, but not among heterozygotes/variant homozygotes (RR 0.95, 0.75–1.20 95% CI). Notably, there was no evidence for interaction of aspirin treatment with genotype at the other SNP that was independently associated with risk (rs11694911). However, for rs28362380, which did not have a main effect on risk overall (see Table 2), each variant allele was associated with a 25% risk reduction in the placebo group (0.75 RR, 0.53–1.04 95% CI), a 39% risk increase in the 81 mg/day aspirin treatment group (1.39 RR, 1.02–1.87 95% CI), but virtually no change in risk in the 325 mg/day aspirin treatment group (1.03 RR, 0.80–1.35 95% CI) (Table 4). Conversely, when the effect of aspirin was stratified by genotype (Table 5), 81 mg aspirin treatment was associated with a 25% risk reduction in wild type homozyzgotes (0.75 RR, 0.61–0.92 95% CI) compared to placebo, but not among heterozygotes/variant homozygotes (1.32 RR, 0.85–2.06 95% CI).
Table 4.
Association of ODC genotypes with risk of adenoma recurrence stratified by aspirin treatment group, Aspirin/Folate Polyp Prevention Study, 1994–2001
| RR (95% CI)a | ||||
|---|---|---|---|---|
| SNP | Placebo | 81 mg/day Aspirin | 325 mg/day Aspirin | Pint |
| rs13000916 | 0.89 (0.75–1.07) | 0.94 (0.76–1.15) | 0.84 (0.71–1.00) | 0.76 |
| rs11694911 | 1.40 (1.06–1.85) | 1.24 (0.92–1.67) | 1.31 (1.00–1.70) | 0.82 |
| rs2430419 | 0.91 (0.73–1.12) | 1.13 (0.89–1.42) | 1.27 (1.05–1.55) | 0.08 |
| rs2430420 | 0.99 (0.8201.20) | 1.21 (0.98–1.49) | 1.38 (1.15–1.66) | 0.05 |
| rs10929669 | 1.26 (0.94–1.69) | 1.23 (0.92–1.66) | 1.21 (0.94–1.56) | 0.97 |
| rs28362434 | 1.14 (0.91–1.44) | 0.83 (0.62–1.11) | 0.82 (0.65–1.04) | 0.10 |
| rs818162 | 0.94 (0.77–1.14) | 0.77 (0.61–0.98) | 0.84 (0.70–1.03) | 0.45 |
| rs1049500 | 1.42 (0.99–2.05) | 1.53 (1.03–2.27) | 1.16 (0.75–1.80) | 0.63 |
| rs28362416 | 0.63 (0.41–0.97) | 0.79 (0.51–1.25) | 1.19 (0.84–1.69) | 0.08 |
| rs7559979 | 1.01 (0.84–1.22) | 1.15 (0.92–1.45) | 0.98 (0.82–1.18) | 0.50 |
| rs28362380 | 0.75 (0.53–1.04) | 1.39 (1.02–1.87) | 1.03 (0.80–1.35) | 0.03 |
| rs2302615b | 1.11 (0.91–1.36) | 0.94 (0.73–1.21) | 0.97 (0.79–1.19) | 0.56 |
| rs1728148 | 0.88 (0.73–1.06) | 0.94 (0.76–1.15) | 0.93 (0.78–1.10) | 0.91 |
| rs885815 | 1.13 (0.91–1.40) | 0.86 (0.65–1.13) | 0.86 (0.69–1.09) | 0.19 |
| rs2884211 | 0.95 (0.68–1.34) | 1.06 (0.74–1.52) | 1.06 (0.76–1.47) | 0.89 |
| rs2357551 | 1.05 (0.86–1.28) | 1.15 (0.93–1.43) | 1.22 (1.02–1.46) | 0.56 |
Abbreviations: SNP, single nucleotide polymorphism; RR, relative risk; CI, confidence interval; Pint = P for interaction (between aspirin treatment and genotype, modeled additively).
Per allele relative risk using an additive genetic model adjusted for age and sex.
Previously genotyped SNP (17)
Table 5.
Association of aspirin treatment with risk of adenoma recurrence stratified by ODC genotypes, Aspirin/Folate Polyp Prevention Study, 1994–2001
| [# controls / # cases] RR (95% CI) | ||||||||
|---|---|---|---|---|---|---|---|---|
| SNP | Placebo | 81 mg/day Aspirin | 325 mg/day Aspirin | Pint | ||||
| rs2430420 | ||||||||
| GG | [58/58] | 1.0 (referent) | [76/40] | 0.68 (0.50–0.94) | [75/50] | 0.81 (0.61–1.09) | ||
| GA/AA | [70/67] | 1.0 (referent) | [85/70] | 0.95 (0.75–1.20) | [58/85] | 1.22 (0.98–1.53) | 0.06 | |
| rs28362380 | ||||||||
| TT | [98/106] | 1.0 (referent) | [136/84] | 0.75 (0.61–0.92) | [105/110] | 0.97 (0.80–1.18) | ||
| TC/CC | [30/19] | 1.0 (referent) | [24/26] | 1.32 (0.85–2.06) | [28/25] | 1.24 (0.80–1.94) | 0.06 | |
| rs2302615a | ||||||||
| GG | [72/59] | 1.0 (referent) | [78/65] | 0.97 (0.75–1.25) | [71/76] | 1.11 (0.87–1.42) | ||
| AG/AA | [49/64] | 1.0 (referent) | [75/44] | 0.65 (0.47–0.88) | [54/58] | 0.96 (0.73–1.26) | 0.25 | |
Abbreviations: SNP, single nucleotide polymorphism; RR, relative risk; CI, confidence interval; Pint = P for interaction (between aspirin treatment and genotype, modeled dominantly).
Previously genotyped SNP(17).
Lastly, although we previously reported a statistically significant interaction between rs2302615 and aspirin treatment (17), the interaction was not statistically significant in the current analyses which differed from the prior analyses in that they were restricted to non-Hispanic whites, were adjusted for age and sex and analyzed the aspirin treatment groups separately (Tables 4 and 5). However, when we precisely mimicked the previous analysis by assessing the combined aspirin treatment effect (81 and 325 mg doses together) stratified by genotype using a dominant genetic model, the interaction was still not statistically significant although the results were similar (not shown): specifically, in the current analysis, as in the prior analysis, there was no risk reduction in the wild type homozygotes (1.04 RR, 0.83–1.30 95% CI), while a 17% risk reduction was observed among heterozygotes/variant homozygotes (0.83 RR, 0.66–1.04 95% CI, Pint=0.15) that was only slightly smaller in magnitude than that reported previously (23% risk reduction, 0.77 RR, 0.63–0.95 95% CI, Pint=0.04, see (17)).
DISCUSSION
We observed that several common genetic variants in or near the ODC gene are associated with risk of colorectal adenoma recurrence among non-Hispanic white participants in a randomized aspirin trial. After adjustment for multiple comparisons, significant associations between adenoma recurrence and genotype at seven tag SNPs remained using an additive genetic model, including 5 downstream (rs13000916, rs11694911, rs2430420, rs10929669, rs818162), 1 upstream (rs2357551) and 1 within the transcribed region (exon 12) of the ODC gene (rs1049500). In addition, common haplotypes in three out of four haplotype blocks were significantly associated with adenoma risk, as was overall variation in two of the blocks. However, there was no evidence for combined allelic effects within a block since the haplotype effect sizes were similar to that seen in the single SNP analyses. In a composite (multiple SNP) analysis, only two of the SNPs were independently associated with risk: having at least one variant allele was associated with increased risks of 20% for rs2430420 (1.20 RR, 1.03–1.40 95% CI, P=0.022) and 29% for rs11694911 (1.29 RR, 1.08–1.53 95% CI, P=0.005). In addition, in the 15% of the population with at least one variant allele at both loci, risk was increased by 53% compared to individuals without any variant alleles at these two loci (1.53 RR, 1.24–1.90 95% CI, P<0.001). There was also evidence for an interaction between rs2430420 genotype and aspirin treatment since genotype was only associated with increased risk in aspirin treated participants. Interestingly, another SNP (rs28362380) without a main effect on risk, also appeared to interact with aspirin treatment.
These results provide additional support for the importance of the polyamine pathway in the development of colorectal neoplasia, since common genetic variation in a potential regulatory region just downstream of the ODC gene is associated with risk of adenoma recurrence. Polyamines may play an important role in etiology of colorectal carcinogenesis since DFMO treatment (in combination with sulindac) was highly effective in a chemoprevention trial (7). ODC gene variants have not been identified in GWAS of colorectal cancer performed to date even though the effect sizes and minor allele frequencies seen here are of the same order of magnitude as for the SNPs detected by these studies (33, 34). It is possible that an intermediate outcome, such as adenoma, may show stronger associations with genotype than cancer if the genotype effect is related to early events in adenoma initiation or progression. Thus, it may be worthwhile to perform GWAS of colorectal adenomas, as opposed to cancer, in future studies in order to identify (additional) chemoprevention targets. However, it is also possible that the true genotype effect sizes are smaller than we see here and, therefore, it will be important to replicate these findings in other studies.
Whether the SNPs (rs2430420 and rs11694911) identified in this study are themselves causal, or are in linkage disequilibrium with unmeasured causal variants is not known. However our results suggest that there are at least two causal variants near the ODC gene that independently impact risk and whose effects are additive with each other. These results highlight the potential importance of genetic variation in non-coding regions, which is likely to be involved in the regulation of gene expression, and the gaps in our knowledge regarding mechanisms for such effects. These findings are consistent with the results of numerous GWAS studies performed to date, where the vast majority (about 80%) of “hits” (disease- or trait-associated SNPs) were from non-coding introns and intergenic regions (35). To our knowledge, the function of these SNPs (rs2430420 and rs11694911) has not been investigated and potential gene regulatory mechanisms are not known. Notably, no SNPs were in linkage disequilibrium (r2≥0.8) with these two SNPs in our dataset of 28 other SNPs genotyped within 23 kb of the ODC gene. Also, no SNPs in the HAPMAP CEU dataset of 41 SNPs genotyped within 50 kb of the ODC gene were in linkage disequilibrium with rs11694911 (note that rs2430420 is not in the HAPMAP dataset and so could not be included in this type of analysis).
The mechanism by which common genetic variants in or near the ODC gene may affect the efficacy of aspirin chemoprevention is also not known. However, it has been suggested previously that aspirin, in addition to inhibiting the prostagladin pathway, may also induce spermidine/spermine-N-acetyltransferase (SSAT) activity (16), which catalyzes the first step in polyamine catabolism or excretion from the cell (36, 37). Aspirin and some other NSAIDs, including sulindac, have been shown to stimulate polyamine catabolism in colon cancer cells via induction of SSAT activity (38, 39). It has been suggested that polyamines may play a role in the relationship between inflammation and carcinogenesis (40, 41). Interestingly, in our randomized aspirin trial, 81 mg/day aspirin was effective in reducing the recurrence of colorectal adenoma whereas 325 mg/day was not (21). The reason for this is not clear, but may relate to differential pro- and anti-carcinogenic effects of aspirin at different doses, which could involve differential effects on the polyamine pathway, although this is speculation. Our results hint that the interaction between ODC genotype and aspirin may differ by aspirin dose, which could lend support to this idea and could be explored in future studies.
Previous work has focused only on a SNP (rs2302615) in the promoter region of the ODC gene, near binding sites for transcription factors, that appears to affect transcriptional activity (16, 42, 43). Among participants in the Wheat Bran Fiber Trial, individuals homozygous for the variant allele had an approximately 50% reduced risk of colorectal adenoma recurrence and appeared to have an enhanced risk reduction in response to aspirin use compared to individuals that were homozygous wild type, although these associations did not reach statistical significance (16). Similar results were found among participants in the United Kingdom Colorectal Adenoma Prevention Trial (18). However, in our previous analysis of all subjects in the Aspirin/Folate Polyp Prevention Study (regardless of race/ethnicity), we found that genotype was not associated with a main effect on risk of adenoma recurrence (17) consistent with the current analysis of non-Hispanic whites. Although we previously reported a statistically significant interaction between genotype and aspirin treatment in an unadjusted analysis using all subjects (17), the interaction is no longer statistically significant after adjusting for age and sex and including only non-Hispanic whites in the current analysis (see Table 5). The main difference was the adjustment for age, as the magnitude of the effect was not attenuated and the interaction was still significant after adjustment for sex and restriction to non-Hispanic whites. Notably, a published meta-analysis analysis of these three studies (16–18) used raw (unadjusted) data (18). In light of our findings, a more rigorous analysis of this SNP (rs2302615) may be warranted. Finally, it is worth noting that this previously studied SNP is not correlated with the either of the two SNPs (rs2430420 and rs11694911) identified here to be independently associated with adenoma risk (r2=0 and 0.01, respectively).
There are some limitations to the current analyses. Due to the size of the study population, we had limited power, especially for investigating interactions with aspirin treatment. Also, due to the limited sample size and power, we did not investigate associations with advanced lesions (which occur much more rarely) or include minorities (individuals with a race or ethnicity other than non-Hispanic whites). In addition, the study was performed on individuals with a history of adenoma who may be at increased risk relative to the general population undergoing screening or surveillance colonoscopy, potentially limiting the generalizability of the findings. However, since adenoma prevalence in the general (middle-aged or older) population is high, likely between 25–50% (44, 45), this is unlikely to be a major limitation. Finally, multiple comparisons were conducted, which increases the likelihood that some of our findings may be due to chance. Although the analyses of the SNP main effects were adjusted for multiple comparisons (and were still statistically significant) the tests for interactions with aspirin were not. Thus, we can be less certain that the interactions discovered are not due to chance. However, this concern is mitigated to some extent by the fact that these tests were chosen a priori, based on previous studies and a biological rationale, and not as part of a data-dredging exercise.
This study also had several notable strengths. A single study pathologist reviewed all lesions from trial participants, ensuring uniform endpoint ascertainment. Data on environmental exposures and subject characteristics were collected in a detailed and uniform manner at the time of participant enrollment. Aspirin treatment was randomly assigned, thereby ensuring uniform exposure and minimizing concerns about differences between the treated and placebo groups, which could confound the results. Subject compliance with study procedures was excellent (21), including pill taking and avoidance of outside use of the study agents. Finally, a comprehensive tag SNP approach was used to capture common genetic variation throughout the ODC gene and adjacent chromosomal regions. This was important for capturing genetic variation in potential regulatory regions that are likely to influence gene expression through effects on transcriptional or translation efficiency.
This work suggests several potential lines of investigation for future studies. First, these findings need to be replicated in other populations and with larger samples sizes. It would be especially useful to investigate these effects in other populations with well-characterized or randomized aspirin treatment with different doses, as well as in studies of colorectal cancer. If these results are replicated, it will be important to try to identify the causal SNPs by exploring the functionality of the SNPs identified here, especially rs2430420 and rs11694911, and by sequencing this chromosomal region to ascertain unmeasured or rare variants in linkage disequilibrium with these SNPs that may themselves be the causal variants. In addition, given the strong evidence basis for both aspirin and DFMO in colorectal chemoprevention, it will be valuable to genotype these SNPs in individuals that are treated with these agents in future clinical studies in order to assess their impact on efficacy. Finally, given the strong association of genetic variation in this region with race and ethnicity, it will also be worthwhile to investigate these effects in minority populations, especially African Americans and Hispanics.
Acknowledgements
The authors are grateful to the co-investigators, study coordinators and participants in the Aspirin/Folate Polyp Prevention Study who made this research possible and to Bayer for providing the aspirin and placebo tablets for the clinical trial. In addition, we especially wish to thank Drs. Eugene Gerner and Patricia Thompson from the Arizona Cancer Center (Tucson, AZ) for sharing genotype data obtained from their National Cancer Institute SPORE grant (Specialized Program Research Excellence in Gastrointestinal Cancer, CA95060, E.W. Gerner).
Grant Support
National Cancer Institute (CA59005, J.A. Baron); and a grant from the Norris Cotton Cancer Center (E.L. Barry).
REFERENCES
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Chan AT, Giovannucci EL. Primary prevention of colorectal cancer. Gastroenterology. 2010;138:e10. doi: 10.1053/j.gastro.2010.01.057. 2029–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackburn EH, Tlsty TD, Lippman SM. Unprecedented opportunities and promise for cancer prevention research. Cancer Prev Res (Phila) 2010;3:394–402. doi: 10.1158/1940-6207.CAPR-10-0051. [DOI] [PubMed] [Google Scholar]
- 4.Rothwell PM, Wilson M, Elwin CE, Norrving B, Algra A, Warlow CP, et al. Long-term effect of aspirin on colorectal cancer incidence and mortality: 20-year follow-up of five randomised trials. Lancet. 2010;376:1741–1750. doi: 10.1016/S0140-6736(10)61543-7. [DOI] [PubMed] [Google Scholar]
- 5.Cole BF, Logan RF, Halabi S, Benamouzig R, Sandler RS, Grainge MJ, et al. Aspirin for the chemoprevention of colorectal adenomas: meta-analysis of the randomized trials. J Natl Cancer Inst. 2009;101:256–266. doi: 10.1093/jnci/djn485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper K, Squires H, Carroll C, Papaioannou D, Booth A, Logan RF, et al. Chemoprevention of colorectal cancer: systematic review and economic evaluation. Health Technol Assess. 2010;14:1–206. doi: 10.3310/hta14320. [DOI] [PubMed] [Google Scholar]
- 7.Meyskens FL, Jr, McLaren CE, Pelot D, Fujikawa-Brooks S, Carpenter PM, Hawk E, et al. Difluoromethylornithine plus sulindac for the prevention of sporadic colorectal adenomas: a randomized placebo-controlled, double-blind trial. Cancer Prev Res (Phila Pa) 2008;1:32–38. doi: 10.1158/1940-6207.CAPR-08-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pegg AE. Mammalian polyamine metabolism and function. IUBMB Life. 2009;61:880–894. doi: 10.1002/iub.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pegg AE. Regulation of ornithine decarboxylase. J Biol Chem. 2006;281:14529–14532. doi: 10.1074/jbc.R500031200. [DOI] [PubMed] [Google Scholar]
- 10.Casero RA, Jr, Marton LJ. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat Rev Drug Discov. 2007;6:373–390. doi: 10.1038/nrd2243. [DOI] [PubMed] [Google Scholar]
- 11.Gerner EW, Meyskens FL., Jr Polyamines and cancer: old molecules, new understanding. Nat Rev Cancer. 2004;4:781–792. doi: 10.1038/nrc1454. [DOI] [PubMed] [Google Scholar]
- 12.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 14.Sporn MB, Hong WK. Clinical prevention of recurrence of colorectal adenomas by the combination of difluoromethylornithine and sulindac: an important milestone. Cancer Prev Res (Phila Pa) 2008;1:9–11. doi: 10.1158/1940-6207.CAPR-08-0049. [DOI] [PubMed] [Google Scholar]
- 15.Freedman AN, Sansbury LB, Figg WD, Potosky AL, Weiss Smith SR, Khoury MJ, et al. Cancer pharmacogenomics and pharmacoepidemiology: setting a research agenda to accelerate translation. J Natl Cancer Inst. 2010;102:1698–1705. doi: 10.1093/jnci/djq390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez ME, O'Brien TG, Fultz KE, Babbar N, Yerushalmi H, Qu N, et al. Pronounced reduction in adenoma recurrence associated with aspirin use and a polymorphism in the ornithine decarboxylase gene. Proc Natl Acad Sci U S A. 2003;100:7859–7864. doi: 10.1073/pnas.1332465100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barry EL, Baron JA, Bhat S, Grau MV, Burke CA, Sandler RS, et al. Ornithine decarboxylase polymorphism modification of response to aspirin treatment for colorectal adenoma prevention. J Natl Cancer Inst. 2006;98:1494–1500. doi: 10.1093/jnci/djj398. [DOI] [PubMed] [Google Scholar]
- 18.Hubner RA, Muir KR, Liu JF, Logan RF, Grainge MJ, Houlston RS. Ornithine decarboxylase G316A genotype is prognostic for colorectal adenoma recurrence and predicts efficacy of aspirin chemoprevention. Clin Cancer Res. 2008;14:2303–2309. doi: 10.1158/1078-0432.CCR-07-4599. [DOI] [PubMed] [Google Scholar]
- 19.Zell JA, McLaren CE, Chen WP, Thompson PA, Gerner EW, Meyskens FL. Ornithine Decarboxylase-1 Polymorphism, Chemoprevention With Eflornithine and Sulindac, and Outcomes Among Colorectal Adenoma Patients. J Natl Cancer Inst. 2010 doi: 10.1093/jnci/djq325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zell JA, Ziogas A, Ignatenko N, Honda J, Qu N, Bobbs AS, et al. Associations of a polymorphism in the ornithine decarboxylase gene with colorectal cancer survival. Clin Cancer Res. 2009;15:6208–6216. doi: 10.1158/1078-0432.CCR-09-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baron JA, Cole BF, Sandler RS, Haile RW, Ahnen D, Bresalier R, et al. A randomized trial of aspirin to prevent colorectal adenomas. N Engl J Med. 2003;348:891–899. doi: 10.1056/NEJMoa021735. [DOI] [PubMed] [Google Scholar]
- 22.Cole BF, Baron JA, Sandler RS, Haile RW, Ahnen DJ, Bresalier RS, et al. Folic acid for the prevention of colorectal adenomas: a randomized clinical trial. JAMA. 2007;297:2351–2359. doi: 10.1001/jama.297.21.2351. [DOI] [PubMed] [Google Scholar]
- 23. http://hapmap.ncbi.nlm.nih.gov.
- 24. http://egp.gs.washington.edu.
- 25.Barry EL, Sansbury LB, Grau MV, Ali IU, Tsang S, Munroe DJ, et al. Cyclooxygenase-2 polymorphisms, aspirin treatment, and risk for colorectal adenoma recurrence--data from a randomized clinical trial. Cancer Epidemiol Biomarkers Prev. 2009;18:2726–2733. doi: 10.1158/1055-9965.EPI-09-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 27.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- 28. http://www.R-project.org.
- 29.Smith NL, Hindorff LA, Heckbert SR, Lemaitre RN, Marciante KD, Rice K, et al. Association of genetic variations with nonfatal venous thrombosis in postmenopausal women. JAMA. 2007;297:489–498. doi: 10.1001/jama.297.5.489. [DOI] [PubMed] [Google Scholar]
- 30.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 31.Liu K, Muse SV. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21:2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- 32.Excoffier L, Slatkin M. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol. 1995;12:921–927. doi: 10.1093/oxfordjournals.molbev.a040269. [DOI] [PubMed] [Google Scholar]
- 33.Tenesa A, Dunlop MG. New insights into the aetiology of colorectal cancer from genome-wide association studies. Nat Rev Genet. 2009;10:353–358. doi: 10.1038/nrg2574. [DOI] [PubMed] [Google Scholar]
- 34.Houlston RS, Cheadle J, Dobbins SE, Tenesa A, Jones AM, Howarth K, et al. Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat Genet. 2010;42:973–977. doi: 10.1038/ng.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010;363:166–176. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- 36.Pegg AE. Spermidine/spermine-N(1)-acetyltransferase: a key metabolic regulator. Am J Physiol Endocrinol Metab. 2008;294:E995–E1010. doi: 10.1152/ajpendo.90217.2008. [DOI] [PubMed] [Google Scholar]
- 37.Casero RA, Pegg AE. Polyamine catabolism and disease. Biochem J. 2009;421:323–338. doi: 10.1042/BJ20090598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Babbar N, Ignatenko NA, Casero RA, Jr, Gerner EW. Cyclooxygenase-independent induction of apoptosis by sulindac sulfone is mediated by polyamines in colon cancer. J Biol Chem. 2003;278:47762–47775. doi: 10.1074/jbc.M307265200. [DOI] [PubMed] [Google Scholar]
- 39.Babbar N, Gerner EW, Casero RA., Jr Induction of spermidine/spermine N1-acetyltransferase (SSAT) by aspirin in Caco-2 colon cancer cells. Biochem J. 2006;394:317–324. doi: 10.1042/BJ20051298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Babbar N, Murray-Stewart T, Casero RA., Jr Inflammation and polyamine catabolism: the good, the bad and the ugly. Biochem Soc Trans. 2007;35:300–304. doi: 10.1042/BST0350300. [DOI] [PubMed] [Google Scholar]
- 41.Gerner EW, Meyskens FL., Jr Combination chemoprevention for colon cancer targeting polyamine synthesis and inflammation. Clin Cancer Res. 2009;15:758–761. doi: 10.1158/1078-0432.CCR-08-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walhout AJ, van der Vliet PC, Timmers HT. Sequences flanking the E-box contribute to cooperative binding by c-Myc/Max heterodimers to adjacent binding sites. Biochim Biophys Acta. 1998;1397:189–201. doi: 10.1016/s0167-4781(97)00227-3. [DOI] [PubMed] [Google Scholar]
- 43.Guo Y, Harris RB, Rosson D, Boorman D, O'Brien TG. Functional analysis of human ornithine decarboxylase alleles. Cancer Res. 2000;60:6314–6317. [PubMed] [Google Scholar]
- 44.Neugut AI, Jacobson JS, Rella VA. Prevalence and incidence of colorectal adenomas and cancer in asymptomatic persons. Gastrointest Endosc Clin N Am. 1997;7:387–399. [PubMed] [Google Scholar]
- 45.Kahi CJ, Anderson JC, Waxman I, Kessler WR, Imperiale TF, Li X, et al. High-definition chromocolonoscopy vs. high-definition white light colonoscopy for average-risk colorectal cancer screening. Am J Gastroenterol. 2010;105:1301–1307. doi: 10.1038/ajg.2010.51. [DOI] [PubMed] [Google Scholar]