Abstract
Pmel17 is a human amyloid involved in melanin synthesis. A fragment of Pmel17, the repeat domain (RPT) rich in glutamic acids, only forms amyloid under mildly acidic pH. Unlike pathological amyloids, these fibrils dissolve at neutral pH, supporting a reversible aggregation/disaggregation process. Here, we study RPT dissolution using atomic force microscopy and solution-state nuclear magnetic resonance spectroscopy. Our results reveal asymmetric fibril disassembly proceeding in the absence of intermediates. We suggest that fibril unfolding involves multiple deprotonation events resulting in electrostatic charge repulsion and filament dissolution.
Keywords: melanosomes, melanin, NMR, atomic force microscopy
Amyloids are highly-ordered protein aggregates with an unbranched filamentous morphology that have been associated with many human diseases.1 However, emerging evidence suggests that some amyloids are beneficial serving biological roles.2 Pmel17 is a functional amyloid involved in melanin synthesis and deposition, which forms intralumenal filaments in melanosomes, membranous organelles where melanin is synthesized and stored.3 Pmel17 fibrils are proposed to act as scaffolds to which melanin is accumulated and possibly to protect cells by sequestering the reactive chemical intermediates en route to this important pigment.3a,3c
A fragment of Pmel17 (Figure S1), named the repeat (RPT) domain due to its 10 imperfect repeat sequence of proline, serine, threonine, and glutamic acid residues,3b forms amyloid in vitro under mildly acidic conditions (pH 4.5–5.5),4 typical of the melanosomal pH.5 Importantly, these fibrils readily dissolve near neutral pH (≥ 6),4 supporting the requirement of the acidic melanosome pH for amyloid assembly and stability. This striking observation suggests that the intra- and/or intermolecular electrostatic charge repulsion created upon deprotonation of specific carboxylic acids (there are 15 Glu and 1 Asp) is responsible for dissolution of fibrils.4b
This reversibility of RPT aggregation/disaggregation may be a characteristic that defines it as a functional amyloid, since many disease-related amyloids are stable to comparably harsher conditions6 such as chemical denaturation and protease digestion.7 Hence, we hypothesized that melanosomal pH may be a natural regulator of RPT fibril formation and if RPT is released and exposed to the neutral cytosolic environment, filaments would dissociate and thus, maintain their benign nature. To gain further kinetic and structural insights on the RPT fibril disassembly process, we have employed atomic force microscopy (AFM) and solution-state nuclear magnetic resonance (NMR) spectroscopy as ultrastructural and molecular probes, respectively. In particular, we sought to determine whether during this dissolution process, intermediates or oligomers, putative cytotoxic precursors implicated in disease-related amyloids,8 are populated.
To monitor fibril disassembly, preformed RPT fibrils (100 μM in 20 mM sodium acetate buffer with 100 mM NaCl, pH 5.0, 37 °C, agitated at 600 rpm for 4 days) were diluted (final concentration ~5 μM) and deposited on freshly cleaved mica. AFM images were collected under wet buffer conditions and long, straight, and un-branched filaments with an average height of ~4 nm were found (Figure S2). Consistent with our previous work,4a,4b washing these fibrils with pH 5.0 buffer showed no signs of morphological change (Figure 1A) whereas when treated with pH 6.5 buffer, the fibrils begin to dissolve (Figure 1B). By monitoring in real time, the RPT fibril dissolution process reveals fragmentation of larger fibrils (≥ 300 nm) followed by the complete disappearance of the smaller fragments on the order of minutes.
Figure 1.
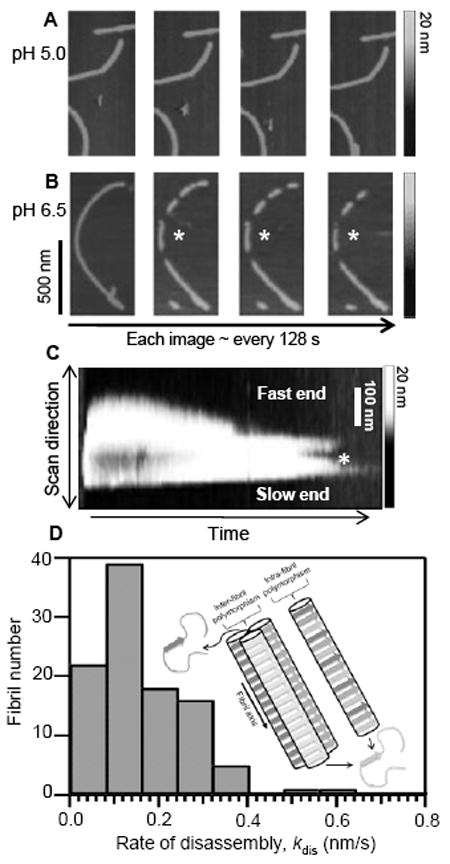
Real-time AFM measurments of RPT fibril dissolution. Fibrils formed at pH 5.0 were washed with (A) pH 5.0 or (B) pH 6.5 sodium acetate buffer and monitored over time (*denotes one disassembly event). Grey scale indicates fibril height. C. Scanning-force kymogram showing a single fibril scanned repetitively along its long axis as a function of scanning time. Ends have been labeled for relative rates (*denotes fragmentation). Grey scale denotes fibril height. Time scale 0–900 s. D. Histogram showing rates of disassembly (kdis) for individual fibrils (n = 102, bin width = 0.08 nm/s, 10 bins). (Inset) Schematic representation of RPT fibril dissolution showing inter/intra fibril polymorphism. With each image taken every 128 s, 42–43 monomers would be released after each scanning cycle.
Using a method called scanning-force kymography (SFK),9 we examined the directionality of fibril disassembly. By repetitively scanning the cantilever tip along the fibril axis, images from a single fibril can be assembled into a kymogram (Figure 1C). A total of 18 individual events were observed using SFK and the rates of disassembly for most fibrils appear to differ at both ends, implying filament directionality, though a few have similar dissociation rates for both ends (Figure S3). A representative kymogram is shown in Figure 1C with a fast- and slow-dissolving end.
While heterogeneous fibril growth has been shown for amyloids such as amylin, Aβ, and the yeast prion PSI+,10 asymmetric disassembly of a functional amyloid has not been demonstrated until now. The observed fragmentation and variation in dissociation rates could be due to structural polymorphism, where a protein can adopt different self-propagating fibril conformations.11 This suggestion is possible for RPT since a recent solid-state NMR report revealed that indeed RPT fibrils are polymorphic.12 Since multiple distinct peptide structures are possible even within a single fibril,13 both intra- and inter-fibril heterogeneity may occur (Figure 1D inset). Another plausible explanation is that there is directional growth of the fibril resulting in distinctive interaction sites at the filament ends.
Despite the presence of heterogeneity, we elected to characterize the kinetics (this rate includes dissolution from both fast and slow ends) by assessing end-to-end filament distances (see Figure 1B). At least one hundred individual dissolution events were counted and an average rate of 0.16 ± 0.13 nm/s was obtained (Figure 1D). Because amyloids adopt a cross-β structure,1 where the β-strands in a β-sheet run perpendicular to the fibril axis at a interstrand distance of approx. 0.47 nm, we estimate that on average it would take ~3 s to remove these β-strands of each monomer from the fibril.
In order to gain residue-specific insight into fibril disassembly, we prepared isotopically labelled RPT for NMR spectroscopy. The 15N HSQC spectrum was first measured for soluble, monomeric RPT at pH 6.5, which provided backbone amide hydrogen and nitrogen resonance assignments for 87 of the 119 non-proline residues, with 29 more being conditionally assigned due to spectral overlap (Figures S4A and C). Spectroscopic features (chemical shift and line width) of all assigned residues were consistent with the protein adopting a flexible random coil conformation and can be classified as an intrinsically disordered polypeptide.14
In the fibrillar state at pH 5.0, no backbone amide resonances were observed for residues 378–444, suggesting that the lack of NMR signal may be due to a large rotational correlation time of the amyloid core (Figures 2, S4B, and D). In contrast, resonances for N-terminal residues, 316–377, remain visible albeit with reduced intensities and broadened line widths (at least twice that of the monomer), consistent with a reduction in motion compared to the monomer, and becoming more immobile as the residues approach the C-terminal region. No chemical shift perturbations were observed (< 0.03 ppm), thus the N-terminal region retains a flexible, unstructured conformation in the amyloid form, nearly indistinguishable from the monomer.
Figure 2.
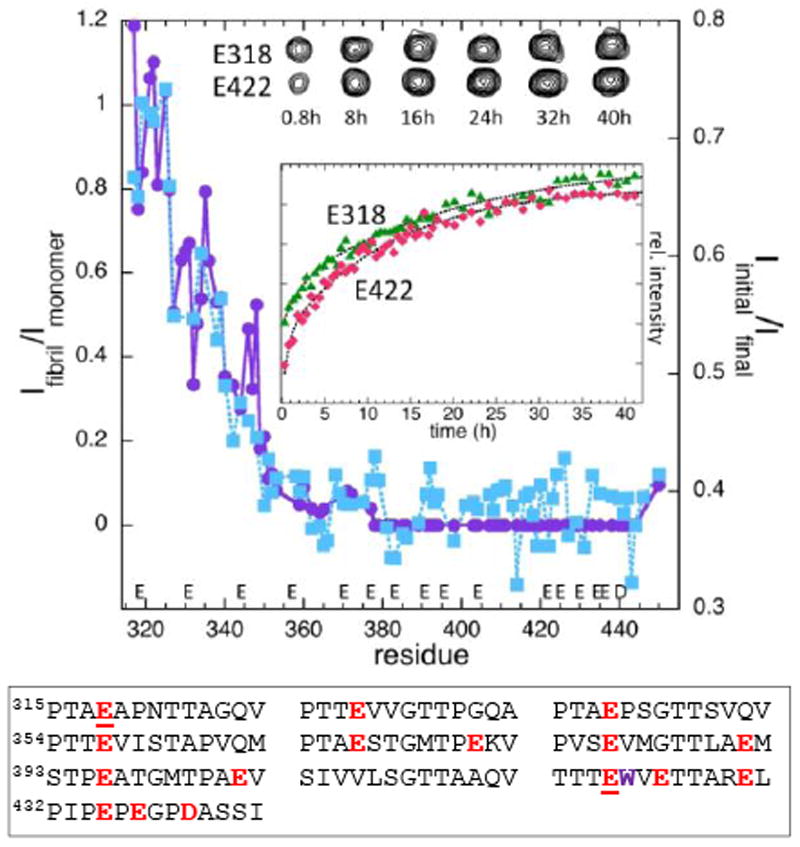
Comparison of NMR amide resonance intensities for RPT fibril dissolution (right axis) and fibrillar vs. monomer RPT (left axis). Intensity ratios (Ifibril/Imonomer, purple circles and Iinitial/Ifinal, blue squares) are plotted as a function of residue number for the fibril (Ifibril, pH 5), monomer (Imonomer, pH 6.5), initial (Iinitial) and final (Ifinal) time point of the disassembly process at pH 6.5. (Inset) Disassembly kinetics monitored by E318 (green triangles) and E422 (red diamonds) with corresponding resonance peaks at selected times shown above. Acidic residues in RPT are colored red with E318 and E422 underlined and W423 colored purple.
Based on these data, we propose that the RPT fibril core is contained within the C-terminal repeats (residues 378–444). This is consistent with our prior fluorescence study using W423 in repeat 9, which also indicated a key role for the RPT C-terminal region in fibril assembly.4b The N-terminal intensity reduction is consistent with restricted motion due to being tethered to the much larger fibril. However, it cannot be ruled out that some reduction could be due to the incorporation of a subpopulation of the N-terminal residues into the amyloid.12
To map the RPT disassembly process in detail, we resuspended preformed fibrils in pH 6.5 buffer and recorded a 15N HSQC spectrum every 30 min up to 42 h at 22 °C. The first spectrum showed 35% recovery of the intensity observed for the monomer spectrum. Subsequent spectra showed a slower, exponential recovery with an average time constant (τ) of 12 h for all residues. The first spectrum appears to be a sum of fibril and monomer spectra, with no evidence of intermediates during the first 30 min. This is shown in Figure 2 by comparing the intensity ratios of the initial and final spectra to those of the fibril and monomer spectra, which clearly follow the same trend within experimental error.
Though kinetics for every assigned residue were examined, we paid special attention to the Glu backbone amides (Figure S5) due to their implications in the pH dependent dissolution process (vide supra). Figure 2 inset displays a representative example, showing backbone amide kinetics of E318 and E422, with E318 positioned outside and E422 within the putative fibril core.4b It is clear that there are no significant differences amongst the different glutamates suggesting that fibril unfolding is not a local but rather global process involving multiple deprotonation events. Consistent with the spectral data, no intermediate states are apparent as evidenced by the comparable rates, suggesting such states are avoided or they are beyond our detection.15
The absence of stable oligomeric states during RPT fibril disassembly was also verified using size exclusion chromatography (SEC). RPT fibrils were resuspended in pH 6.5 buffer and directly applied to a pre-equilibrated column. The resulting chromatogram showed two peaks, one of the monomer and the other eluting in the void volume, suggestive of small fibrils with a mass >600 kDa (Figure S6). By pre-incubating RPT at pH 6.5 for various lengths of time, it is clear that more monomer is released when the fibrils are exposed for longer time periods, in accord with the NMR results (Figure S6). Our current data demonstrate that RPT fibril dissociation is fully reversible since nearly all of the NMR monomer signal intensity is recovered and no apparent stable oligomers are identified by SEC. Nevertheless, further work utilizing other site-specific and highly sensitive probes (e.g. Förster energy transfer) is necessary to delineate the presence of small population of oligomers.
In summary, we report new structural insights into RPT fibril dissolution. At the microscopic level, fibrils often fragmented before their complete disappearance. In addition, asymmetric dissolution rates were observed. On the residue level, solution state NMR data identified the RPT amyloid core in the C-terminal region. Moreover, spectral and kinetics data indicate that intermediate states are not populated during dissolution. Since amyloid toxicity has been associated with oligomeric or pre-fibrillar species that manifest during aggregation and potentially causing membrane disruption,16 we suggest that in the case of the RPT domain, despite being encapsulated in melanosomes, membranous organelles, these events are mitigated because of its reversible and pH dependent amyloid formation.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program of the National Institutes of Health, National Heart, Lung, and Blood Institute. We thank K.-N. Hu for technical assistance and N. Tjandra for use of the 800 MHz NMR spectrometer.
Footnotes
Supporting Information. Materials and methods and Figures S1-S6. This material is free of charge online at http://pubs.acs.org.
References
- 1.(a) Shewmaker F, McGlinchey RP, Wickner RB. J Biol Chem. 2011;286:16533–16540. doi: 10.1074/jbc.R111.227108. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Chiti F, Dobson CM. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]; (c) Jahn TR, Radford SE. Arch Biochem Biophys. 2008;469:100–117. doi: 10.1016/j.abb.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Fowler DM, Koulov AV, Balch WE, Kelly JW. Trends Biochem Sci. 2007;32:217–224. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]; (b) Greenwald J, Riek R. Structure. 2010;18:1244–1260. doi: 10.1016/j.str.2010.08.009. [DOI] [PubMed] [Google Scholar]; (c) Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]; (d) Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Balguerie A, Dos Reis S, Ritter C, Chaignepain S, Coulary-Salin B, Forge V, Bathany K, Lascu I, Schmitter JM, Riek R, Saupe SJ. Embo J. 2003;22:2071–2081. doi: 10.1093/emboj/cdg213. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) McGlinchey RP, Yap TL, Lee JC. Phys Chem Chem Phys. 2011 doi: 10.1039/C1031CP21376H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Berson JF, Harper DC, Tenza D, Raposo G, Marks MS. Mol Biol Cell. 2001;12:3451–3464. doi: 10.1091/mbc.12.11.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Hoashi T, Muller J, Vieira WD, Rouzaud F, Kikuchi K, Tamaki K, Hearing VJ. J Biol Chem. 2006;281:21198–21208. doi: 10.1074/jbc.M601643200. [DOI] [PubMed] [Google Scholar]; (c) Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW. PLoS Biol. 2006;4:100–107. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Raposo G, Marks MS. Nat Rev Mol Cell Biol. 2007;8:786–797. doi: 10.1038/nrm2258. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Hurbain I, Geerts WJC, Boudier T, Marco S, Verkleij AJ, Marks MS, Raposo G. Proc Natl Acad Sci U S A. 2008;105:19726–19731. doi: 10.1073/pnas.0803488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) McGlinchey RP, Shewmaker F, McPhie P, Monterroso B, Thurber K, Wickner RB. Proc Natl Acad Sci U S A. 2009;106:13731–13736. doi: 10.1073/pnas.0906509106. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Pfefferkorn CM, McGlinchey RP, Lee JC. Proc Natl Acad Sci U S A. 2010;107:21447–21452. doi: 10.1073/pnas.1006424107. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) McGlinchey RP, Shewmaker F, Hu KN, McPhie P, Tycko R, Wickner RB. J Biol Chem. 2011;286:8385–8393. doi: 10.1074/jbc.M110.197152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhatnagar V, Anjaiah S, Puri N, Darshanam BNA, Ramaiah A. Arch Biochem Biophys. 1993;307:183–192. doi: 10.1006/abbi.1993.1577. [DOI] [PubMed] [Google Scholar]
- 6.(a) β2 microglobulin fibrils associated with dialysis related amyloidosis have been demonstrated to exhibit highly pH sensitive fibril stability. Upon treatment with neutral or basic solution filaments formed at very low pH (2 to 3) can dissociate and reform folded monomers. [Google Scholar]; (b) Platt GW, Radford SE. FEBS Lett. 2009;583:2623–2629. doi: 10.1016/j.febslet.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Yamamoto S, Hasegawa K, Yamaguchi I, Goto Y, Gejyo F, Naiki H. BBA-Proteins Proteomics. 2005;1753:34–43. doi: 10.1016/j.bbapap.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 7.Murphy RM. Annu Rev Biomed Eng. 2002;4:155–174. doi: 10.1146/annurev.bioeng.4.092801.094202. [DOI] [PubMed] [Google Scholar]
- 8.(a) Roychaudhuri R, Yang M, Hoshi MM, Teplow DB. J Biol Chem. 2009;284:4749–4753. doi: 10.1074/jbc.R800036200. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kostka M, Hogen T, Danzer KM, Levin J, Habeck M, Wirth A, Wagner R, Glabe CG, Finger S, Heinzelmann U, Garidel P, Duan W, Ross CA, Kretzschmar H, Giese A. J Biol Chem. 2008;283:10992–11003. doi: 10.1074/jbc.M709634200. [DOI] [PubMed] [Google Scholar]; (c) Stefani M. Curr Protein Pept Sci. 2010;11:343–354. doi: 10.2174/138920310791330631. [DOI] [PubMed] [Google Scholar]; (d) Uversky VN. FEBS J. 2010;277:2940–2953. doi: 10.1111/j.1742-4658.2010.07721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Sakono M, Zako T. FEBS J. 2010;277:1348–1358. doi: 10.1111/j.1742-4658.2010.07568.x. [DOI] [PubMed] [Google Scholar]
- 9.Kellermayer MSZ, Karsai A, Benke M, Soos K, Penke B. Proc Natl Acad Sci U S A. 2008;105:141–144. doi: 10.1073/pnas.0704305105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.(a) Goldsbury C, Kistler J, Aebi U, Arvinte T, Cooper GJS. J Mol Biol. 1999;285:33–39. doi: 10.1006/jmbi.1998.2299. [DOI] [PubMed] [Google Scholar]; (b) Blackley HKL, Sanders GHW, Davies MC, Roberts CJ, Tendler SJB, Wilkinson MJ. J Mol Biol. 2000;298:833–840. doi: 10.1006/jmbi.2000.3711. [DOI] [PubMed] [Google Scholar]; (c) DePace AH, Weissman JS. Nat Struct Biol. 2002;9:389–396. doi: 10.1038/nsb786. [DOI] [PubMed] [Google Scholar]
- 11.(a) Petkova AT, Leapman RD, Guo ZH, Yau WM, Mattson MP, Tycko R. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]; (b) Pedersen JS, Andersen CB, Otzen DE. FEBS J. 2010;277:4591–4601. doi: 10.1111/j.1742-4658.2010.07888.x. [DOI] [PubMed] [Google Scholar]; (c) Giraldo R. Chem Bio Chem. 2010;11:2347–2357. doi: 10.1002/cbic.201000412. [DOI] [PubMed] [Google Scholar]; (d) Kodali R, Williams AD, Chemuru S, Wetzel R. J Mol Biol. 2010;401:503–517. doi: 10.1016/j.jmb.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu KN, McGlinchey RP, Tycko R, Wickner RB. Biophys J. 2011;101:2242–2250. doi: 10.1016/j.bpj.2011.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewandowski JR, van der Wel PCA, Rigney M, Grigorieff N, Griffin RG. J Am Chem Soc. 2011;133:14686–14698. doi: 10.1021/ja203736z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.(a) Mittag T, Forman-Kay JD. Curr Opin Struct Biol. 2007;17:3–14. doi: 10.1016/j.sbi.2007.01.009. [DOI] [PubMed] [Google Scholar]; (b) Wishart DS, Sykes BD. J Biomol NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 15.Since the NMR signals constitute only the monomer and the flexible random coil region of the fibril core, if intermediate states are manifested in the fibrillar or immobile polypeptide region during the disassembly process they would not be detectable.
- 16.Uversky VN, Eliezer D. Curr Protein Pept Sci. 2009;10:483–499. doi: 10.2174/138920309789351921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


