Abstract
Rtt109 is a fungal-specific histone acetyltransferase required for modification of histone H3 K9, K27 and K56. These acetylations are associated with nascent H3 and play an integral role in replication- and repair-coupled nucleosome assembly. Rtt109 is unique among acetyltransferases as it is activated by a histone chaperone; either Vps75 or Asf1. Recent biochemical, structural and genetic studies have shed light on the intricacies of this activation. It is now clear that Rtt109-Asf1 acetylates K56, while Rtt109-Vps75 acetylates K9 and K27. This reinforces that Asf1 and Vps75 activate Rtt109 via distinct molecular mechanisms. Structures of Rtt109-Vps75 further imply that Vps75 positions histone H3 in the Rtt109 active site. These structures however raise questions regarding the stoichiometry of the Rtt109-Vps75 complex. This has ramifications for determining the physiological Rtt109 substrate.
Introduction
Histone acetylation is a critical regulatory mechanism of DNA-dependent processes such as transcription, replication and repair. It involves histone acetyl-transferase enzymes (HATs) that transfer an acetyl moiety from acetyl-CoA to the ε-amine group of a histone lysine residue. The cell contains several HATs, for example Gcn5 (KAT2), p300/CBP (KAT3A/B) and Tip60 (KAT5), which between them acetylate different residues on the four core histone proteins [1,2]. A recurrent feature of HATs is their presence in protein complexes with constituents that regulate their activity and substrate selectivity. Regulation prevents uncontrolled histone acetylation causing aberrant DNA metabolic events. The molecular underpinning of such regulation is challenging to decipher as it requires genetic, biochemical and structural approaches. This review will focus on recent developments relating to the HAT, Rtt109 (KAT11), particularly the molecular details of its interaction with and regulation by histone chaperone, Vps75.
Rtt109 Substrates
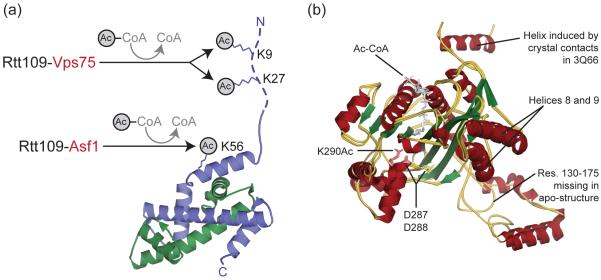
Rtt109 is a fungal-specific HAT historically associated with acetylation of histone H3 K56 (H3 K56Ac) [3-5]. Recent data, however, also establishes H3 K9 and K27 as bona fide Rtt109 targets in yeast [6,7]. Rtt109-dependent K9Ac and K27Ac eluded detection as unlike K56, these residues are also acetylated by Gcn5 [8]. Residues K9 and K27 are in the H3 disordered tail, while residue K56 is in the H3 histone-fold domain (Figure 1a) [9]. As such, Rtt109 cannot acetylate K56 on H3 that is a constituent of the nucleosome [10]. In fact, all Rtt109-dependent acetylations are associated with non-nucleosomal, nascent H3 in yeast [8,11], with K56 acetylation also occurring in multi-cellular eukaryotes [12]. In flies and humans, K56Ac is executed by p300/CBP [12], whose catalytic domain is structurally similar to that of Rtt109 [13]. Rtt109 acetylation of nascent H3 is consistent with a recent study showing Rtt109 activity in the cytoplasm [14]. The genomic instability and sensitivity to genotoxic stress of an Rtt109 knock-out strain, demonstrates the integral role of H3 K9, K27 and K56 acetylation [3-5]. This is reinforced by the more pronounced phenotype of a double Gcn5-Rtt109 knock-out, which completely lacks K9, K27 and K56 acetylation [7]. Rtt109 is thus responsible for H3 acetylations that play an important role in replication- and repair-associated nucleosome assembly. Delineation of HAT-substrate relationships facilitates the dissection of the biological context and mechanistic basis of histone acetylation.
Figure 1. Rtt109 acetylates histone H3 at K9, K27 and K56.
(a) Rtt109-Vps75 acetylates K9 and K27 in the H3 tail (dotted line), while Rtt109-Asf1 acetylates K56 (shown as sticks) in the H3 histone-fold domain. Yeast H3 (blue) and H4 (green) are shown in cartoon (taken from nucleosome PDB 1ID3). (b) Rtt109 adopts a similar conformation when free (apo) or bound by Vps75. Shown is a cartoon superposition of free (PDB 3CZ7) and Vps75-bound Rtt109 (PDB 3Q66 and 3Q33) colored according to secondary structure. The acetyl-CoA and residues K290Ac, D287 and D288 are shown as sticks. Abbreviations: Res. – residues.
Rtt109 Activation - Vps75 versus Asf1
A well-established and intriguing feature of Rtt109 is its stimulation by binding partners Vps75 and Asf1 [4,10]. Vps75 and Asf1 are structurally unrelated histone chaperones that interact with histone (H3-H4)2 tetramer and H3-H4 dimer, respectively [15,16]. The interactions between Vps75/Asf1 and H3-H4 have nanomolar affinity, ten times stronger than that between Rtt109 and H3-H4 [17,18]. However, both Vps75 and Asf1 enhance the catalytic activity of Rtt109, while having little effect on Rtt109 histone binding. They individually increase kcat 100-fold, but don’t drastically affect KM for acetyl-coA or H3 substrate [10,19,20]. This 100-fold increase transforms Rtt109 activity from negligible to comparable with that of other HATs [2]. Despite this similarity, Vps75 and Asf1 likely induce Rtt109 activity differently. This assertion is based on several key differences between Rtt109-Vps75 and Rtt109-Asf1. Rtt109-Vps75 can acetylate H3-H4, H3 alone or a H3 tail peptide [19], while Rtt109-Asf1 is only active towards H3-H4 [10]. This is consistent with the Asf1-H3-H4 crystal structure, which reveals direct contacts between Asf1 and H3, and Asf1 and H4 [16]. Vps75 forms a tight complex with Rtt109 [21], while Asf1 interacts transiently [10]. A tight Rtt109-Vps75 complex is indicated by co-purification of Rtt109 and Vps75 from yeast and bacteria, as well as the Vps75 concentration used to reach maximum Rtt109 catalytic efficiency [21]. Rtt109 mutants that abolish the Rtt109-Vps75 interaction can also have no effect on Rtt109-Asf1 activity [20,22].
Distinct Vps75 and Asf1 stimulation of Rtt109 is further evidenced by recent acetylation profiles of deletion strains. These profiles are based on western blots, as well as mass spectrometry data [14,22]. In particular, Asf1 deletion abolishes K56Ac, slightly reduces K9Ac and maintains K27Ac [4-6], while Vps75 deletion maintains K56Ac, but reduces K9Ac and K27Ac [14,22]. Concomitant deletion of Vps75 and Gcn5 completely abolishes both K9Ac and K27Ac [14,22]. These data suggest that Rtt109-Asf1 is responsible for K56Ac, while Rtt109-Vps75 and Gcn5 are responsible for K9Ac and K27Ac (Figure 1a). Unlike the substrate selectivity of Rtt109-Asf1, which has been established for several years [10], that of Rtt109-Vps75 is a recent hypothesis. Further support for this hypothesis comes from a series of well-designed kinetic assays using Rtt109-Vps75 and H3 mutants [22]. These assays reveal that Rtt109-Vps75 exhibits wild-type catalytic efficiency toward H3-K27R/K56R (only K9 available for acetylation), and reduced catalytic efficiency toward any H3-K9R-containing mutant [22]. Quantitative mass spectrometry of products obtained with a wild-type H3-H4 substrate, also confirms Rtt109-Vps75 preference for K9Ac [20,22]. In vivo, Vps75 mutants with weakened but not abolished Rtt109 binding show a greater reduction in K27Ac than K9Ac [14]. Why a preference for K27Ac is not observed, and why K56Ac has frequently been observed in vitro [10,20], remain to be determined. The varied influence of Vps75 and Asf1 on Rtt109 substrate selectivity reinforces their differential modes of Rtt109 activation.
Rtt109 Catalytic Mechanism
Determining how histone chaperones activate Rtt109 ultimately requires insight into Rtt109 catalytic mechanism. Such insight has been gleaned from biochemical analyses [20,21,23] performed in the wake of the three Rtt109 crystal structures (Figure 1b) [19,24,25]. The structures of Rtt109 revealed an overall fold similar to the p300/CBP HAT domain, albeit with a non-conserved active site [19,24,26]. The structures also uncovered an acetylated lysine residue (K290) in the vicinity of the acetyl-CoA binding pocket [19,24,26]. An outstanding question related to the role of D287 or D288 as the active site base, as in other HATs the active site base is typically a glutamate [2]. For catalysis to occur in the context of Rtt109-Vps75, the active site base is required to deprotonate a group with a pKa of 8.5, most likely the ε-amine of the substrate lysine [21]. Contrary to predictions, recent data suggests that neither D287 or D288 are the active site base and that the decreased activity of D287/D288 mutants results from reduced acetyl-CoA binding, possibly due to the absence of K290Ac (see next paragraph) [21]. The possibility that Vps75 contributes the active site base has also been entertained, although this is not likely as Rtt109 alone and Rtt109-Vps75 have similar kcat pH profiles [20]. The intricacies of the Rtt109 active site thus continue to evade elucidation.
Another question raised by the Rtt109 structures was if Rtt109 K290Ac was an enzyme-acyl intermediate indicative of a ping-pong mechanism [24]. This is not the case as simultaneously published data established K290Ac as the product of Rtt109 auto-acetylation [19,26]. Enzymatic studies have since verified that Rtt109-Vps75 adopts a sequential mechanism whereby the binding of acetyl-CoA and histone H3 substrate can occur in any order, and do not influence each other [21]. Moreover, the rate-limiting step of Rtt109 and Rtt109-Vps75 has been identified as the transfer of the acetyl group [20,21]. Further analysis of Rtt109 K290 auto-acetylation has also revealed it to be essential for Rtt109 activity in vitro and in vivo [23,25], and that it acts to increase the binding affinity of acetyl-CoA [23]. Consistent with K290 being near the putative substrate-binding pocket (Figure 1b) [26], K290Ac occurs intra-molecularly and is not influenced by the presence of Vps75 [23]. The latter implies that Vps75 is involved in histone H3 presentation to Rtt109, and reinforces that the potential for Rtt109 activation is a direct read-out of cellular acetyl-CoA concentration. Acetyl-CoA concentration is an indicator of the cells overall metabolic state. The catalytic mechanism of Rtt109-Asf1 has not been as closely examined, potentially because of the low affinity between Rtt109 and Asf1.
Rtt109-Vps75 Structures
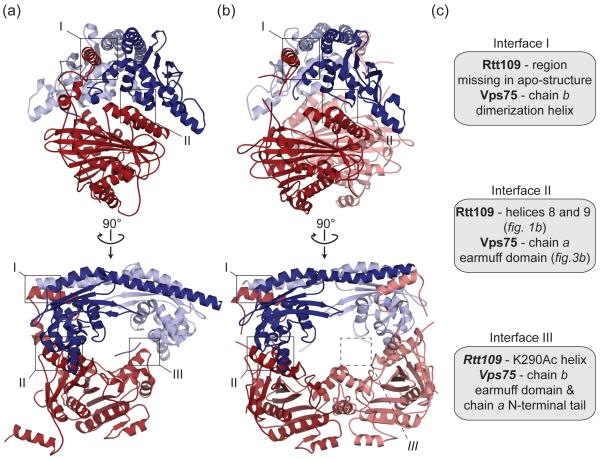
Insight into Rtt109 has also emerged from recent crystallographic analysis with Vps75 [20,22,27]. Three independent groups have reported a total of five Rtt109-Vps75 structures, with only two of the structures being from the same crystal form (Table 1). Notably, the data obtained by Kolonko et al. is of poor resolution, limiting structural detail and preventing refinement [20]. Collectively the remaining structures reveal three potential interfaces between Rtt109 and Vps75 (interface I, II, III) (Figure 2). These interfaces account for the high affinity between Rtt109 and Vps75, with interface I and II together burying 3725 Å2 SAS [22]. These interfaces are both hydrophobic and electrostatic in nature, and occur in context of the constitutive Vps75 homo-dimer [17,19,28]. The Vps75 homo-dimer is formed through an anti-parallel arrangement of the Vps75 dimerization helix, located upstream of the so-called earmuff domain (Figure 3a). The Vps75 homo-dimer is either present in the asymmetric unit (Figure 2a) [20,27], or generated through a crystallographic 2-fold symmetry axis (Figure 2b) [22]. Vps75 dimerization is essential for Rtt109 binding [22], with interface I and II involving different Vps75 chains (Figure 2c). The relevance of these interfaces in solution has been verified through enzymatic assays with Rtt109 mutants [22].
Table 1.
Crystal Structures of Rtt109-Vps75
| PDB Code |
Purificatino Procedure |
Tagged Protein |
Res. (Å) |
Space Group |
Vps75: Rtt109 |
Other | Interface1 | Ref |
|---|---|---|---|---|---|---|---|---|
| n/a | 1. Ni-NTA AC 2. Cation EC 3. SEC |
Rtt109 | 4.3 | P21 | 2:1 | Low resolution prevented refinement |
II2,3 | [20] |
| 3Q66 | 1. GST AC 2. SEC |
Rtt109 | 2.7 | P6122 | 2:1 | Protein buffer had acetyl-CoA; no acetyl-CoA in PDBs |
I, II, III | Su et al. [27] |
| 3Q68 | 2.7 | P212121 | 2:1 | I, II, III | ||||
| 3Q33 | 1. Ni-NTA AC 2. Anion EC 3. SEC |
Vps75 | 2.8 | P212121 | 2:2 | Crystal soaked with CoA and an Ac-peptide |
I, II3 | Tang et al. [22] |
| 3Q35 | 3.3 | - |
See Figure 2.
nterface I not observed as limited resolution prevented building beyond the Rtt109 alone and Vps75 alone molecular replacement search models [20].
Interface III not observed as a truncated Vps75 construct was used [22].
Abbreviations: PDB – Protein Databank, AC - Affinity chromatography, EC - Exchange Chromatography, SEC – Size Exclusion Chromatography, Res. – Resolution, Ref - Reference
Figure 2. Interfaces observed in Vps75-Rtt109 at a 2:1 [Su et al.] (a) and 2:2 [Tang et al.] (b) stoichiometry.
(a), (b) Cartoon representation of the Rtt109 (red or pink) and Vps75 (blue or pale blue) complex [PDB 3Q66 in (a) and 3Q33 in (b)]. Bottom views are 90° rotations of top views and key interfaces are boxed. The asymmetric unit is shown in (a), while the asymmetric unit and a Rtt109-Vps75 pair related by a crystallographic 2-fold symmetry axis are shown in (b). Interface III is not found in the 2:2 complex (broken box). (c) Description of the Rtt109 and Vps75 regions involved in interfaces I, II and III.
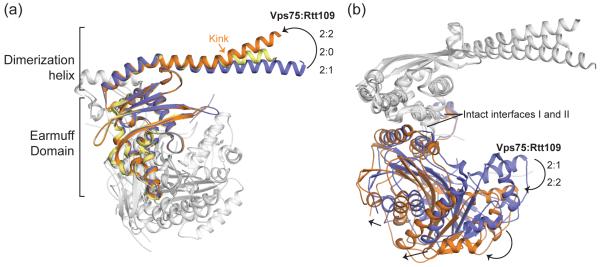
Figure 3. Conformational differences between Vps75-Rtt109 at a 2:1 and 2:2 stoichiometry.
Cartoon superposition (via a single Vps75 chain) of Vps75 alone (PDB 3DM7 chain a – yellow) and Rtt109-Vps75 (PDB 3Q66 chains a and c – blue and 3Q33 – orange). (a) Compared to free Vps75, the Vps75 dimerization helix is kinked in the 2:2 complex and arcs downwards in the 2:1 complex. Rtt109 chains are shown in white. (b) Rtt109 is shifted ‘outward’ in the 2:2 complex compared to the 2:1 complex. Arrows indicate example secondary structure shifts, and Vps75 chains are shown in white. Interface I and II remain intact.
A notable feature of interface I is the involvement of Rtt109 residues 130-175. Limited proteolysis indicates that these residues undergo a disorder-to-order transition upon Vps75 binding [22]. These residues were disordered or removed from the Rtt109 alone structures (Figure 1b). This is consistent with the instability of Rtt109 observed in a Vps75 deletion strain [6,14]. Further comparison of free- and Vps75-bound Rtt109, however, fails to identify differences in the putative active or substrate-binding site (Figure 1b). Thus, while Vps75 is important for Rtt109 stability, it does not appear to induce an ‘active’ Rtt109 conformation.
The major difference between the structures is the stoichiometry of the Rtt109-Vps75 complex. Structures by Tang et al. have one Rtt109 per Vps75 monomer (Figure 2b) [22], while those by Su et al. have one Rtt109 per Vps75 dimer (Figure 2a) [20,27]. This difference arises as the Tang et al. structures do not contain interface III such that the Rtt109 is tilted outward compared to the Su et al. structures (Figures 2b and 3b). Interface III is missing as Tang et al. used truncated Vps75, and/or because it might not be physiologically relevant (see next paragraph). The Tang et al. structures also contain a kink in the dimerization helix of Vps75 such that the region involved in interface I is tilted upwards (Figure 3a). This contrasts the Su et al. structures, where this region arcs downward. The outward shift of Rtt109 and kink in Vps75 facilitate binding of a second Rtt109 in the Tang et al. structures. Importantly, this different stoichiometry is not a result of mixing different Rtt109-Vps75 ratios as in all cases the proteins were co-expressed and co-purified (Table 1). It may however, be a result of different purification protocols. Interestingly, unlike the Tang et al. structures, the Su et al. structures don’t contain acetyl-coA (Table 1). Resolving the issue of stoichiometry is important as it influences the size of the cavity between Vps75 and Rtt109. Based on the orientation of Rtt109, sequence conservation and mutagenesis, this cavity is most likely the histone-binding site [20,22,27]. The size of the cavity will determine if Rtt109-Vps75 binds H3-H4 dimer and/or tetramer and if acetylation occurs on a single or both H3 chains. In vitro Rtt109-Vps75 can acetylate a H3-H4 dimer [20] but, like Vps75 alone, it does not split a H3-H4 tetramer [15,27].
A possible explanation for the different stoichiometries is that both exist, depending on the relative concentrations of Rtt109 and Vps75. This is supported by native gels with Vps75 shifting at 2:1 (Vps75:Rtt109), and super-shifting at 2:2 [17]; or native gels where free Vps75 is only detected in greater than 2:2 mixtures [20]. A 2:2 complex has also been observed by analytical gel filtration [28]. Such an explanation implies that the protocol employed by Su et al. enriched for Vps75. This is feasible as the proteins were expressed from separate plasmids, possibly in different copy number, as well as because Vps75 is a constitutive homo-dimer and Rtt109 is notorious for aggregation and degradation [21]. As such, compared to Vps75, Rtt109 could easily have been depleted during purification. A 2:2 complex is also incompatible with interface III. Interface III may be an artifact as it is small, involves few sequence-specific contacts and occurs near the Vps75 nuclear localization sequence that is bound by proteins which import Rtt109-Vps75 [14]. It is difficult to visualize how Rtt109 and import proteins could simultaneously bind Vps75. The Tang et al. structure clearly reveals that a 2:2 complex can form, albeit in the absence of the Vps75 N-terminal tail. While the Vps75 N-terminal tail is not involved in the Vps75 interaction with H3-H4, it is required for maximal Rtt109-Vps75 activity in vitro [17,22]. A 2:2 complex with equal affinity sites however, conflicts with difficult to interpret analytical ultracentrifugation data [17,28] and the near maximal Rtt109 activity observed at 2:1 [20]. The question is; does Rtt109-Vps75 have maximal activity in the cell? To truly resolve the Rtt109-Vps75 stoichiometry conundrum, more experiments are required. In particular, the stoichiometry of the complex and the influence of the Vps75 N-terminal tail can easily be quantified using relatively straightforward in vitro fluorescence-based techniques.
Future Directions
Regardless of the outstanding issues relating to the Rtt109-Vps75 structures, they imply that Vps75 stimulates Rtt109 activity through positioning the histone H3 substrate. As such, there is a clear demand for a more molecular view of Vps75-histone complexes. Initial work with Vps75 homolog Set, clearly paves the way [29]. It will be interesting to see if Vps75 physically blocks K56. It will also be intriguing to determine why Vps75 paralogue Nap1 can bind to but not activate Rtt109 [17], and to determine how Rtt109 enhances histone deposition by Vps75 [19]. Perhaps the most interesting questions however are those relating to cross-talk or histone hand-offs between Asf1 and Vps75, as well as the fate of the histones acetylated by Rtt109. On a broader level, the identification of other HAT-histone chaperone pairs in both yeast and higher eukaryotes will undoubtedly be informative. Studies have indicated interactions between Vps75 orthologs and p300/CBP [30,31]. Continued characterization seems to uncover more non-canonical functions of the histone chaperone family. Understanding these functions, as well as the regulation and activity of HATS, is of paramount importance if we are to completely decipher the dynamics of chromatin in the cell.
Highlights.
Histone chaperones Vps75 and Asf1 stimulate Rtt109 acetyltransferase towards histone H3 approximately 100-fold.
Rtt109-Asf1 acetylates H3 K56, while Rtt109-Vps75 acetylates H3 K9 and K27.
Auto-acetylation of Rtt109 at K290 is essential and is not influenced by Vps75.
Based on structural work, the stoichiometry of Vps75-Rtt109 is uncertain; it may be 2:1 or 2:2
Vps75 imports Rtt109 into the nucleus, stabilizes Rtt109, and positions H3 for acetylation by Rtt109.
Acknowledgments
S.D. and K.L are supported by the Howard Hughes Medical Institute. K.L. is also supported by the NIH (GM067777 and GM088409). We thank members of the Luger laboratory for helpful discussion.
Abbreviations
- Ac
Acetyl
- Asf1
Anti-silencing Function 1
- CBP
CREB-binding protein
- Gcn5
General control nonderepressible 5
- HAT
Histone acetyl-transferase
- KAT
Lysine acetyl-transferase
- Nap1
Nucleosome assembly protein 1
- Rtt109
Regulator of Ty1 Transposition 109
- Tip60
Tat interactive protein 60
- Vps75
Vacuolar Protein Sorting 75
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 2.Berndsen CE, Denu JM. Catalysis and substrate selection by histone/protein lysine acetyltransferases. Curr Opin Struct Biol. 2008;18:682–689. doi: 10.1016/j.sbi.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schneider J, Bajwa P, Johnson FC, Bhaumik SR, Shilatifard A. Rtt109 is required for proper H3K56 acetylation: a chromatin mark associated with the elongating RNA polymerase II. J Biol Chem. 2006;281:37270–37274. doi: 10.1074/jbc.C600265200. [DOI] [PubMed] [Google Scholar]
- 4.Driscoll R, Hudson A, Jackson SP. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science. 2007;315:649–652. doi: 10.1126/science.1135862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, Zhang Z. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science. 2007;315:653–655. doi: 10.1126/science.1133234. [DOI] [PubMed] [Google Scholar]
- 6.Fillingham J, Recht J, Silva AC, Suter B, Emili A, Stagljar I, Krogan NJ, Allis CD, Keogh MC, Greenblatt JF. Chaperone control of the activity and specificity of the histone H3 acetyltransferase Rtt109. Mol Cell Biol. 2008;28:4342–4353. doi: 10.1128/MCB.00182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burgess RJ, Zhou H, Han J, Zhang Z. A role for Gcn5 in replication-coupled nucleosome assembly. Mol Cell. 2010;37:469–480. doi: 10.1016/j.molcel.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuo MH, Brownell JE, Sobel RE, Ranalli TA, Cook RG, Edmondson DG, Roth SY, Allis CD. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 9.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 10.Tsubota T, Berndsen CE, Erkmann JA, Smith CL, Yang L, Freitas MA, Denu JM, Kaufman PD. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol Cell. 2007;25:703–712. doi: 10.1016/j.molcel.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- 12.Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–117. doi: 10.1038/nature07861. •• This study shows that in humans and flies, histone H3 K56 is acetylated by Rtt109 structural homolog, p300/CBP. It is shown that in vivo such acetylation relies on Asf1 and is associated with DNA damage. These results reinforce the evolutionary link between fungal-specific Rtt109 and metazoan-specific p300/CBP.
- 13.Wang L, Tang Y, Cole PA, Marmorstein R. Structure and chemistry of the p300/CBP and Rtt109 histone acetyltransferases: implications for histone acetyltransferase evolution and function. Curr Opin Struct Biol. 2008;18:741–747. doi: 10.1016/j.sbi.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keck KM, Pemberton LF. Interaction with the Histone Chaperone Vps75 Promotes Nuclear Localization and HAT Activity of Rtt109 In Vivo. Traffic. 2011 doi: 10.1111/j.1600-0854.2011.01202.x. • The presented experiments identify the Vps75 nuclear localization sequence and reveal a role for Vps75 in Rtt109 nuclear import. The authors provide evidence for Rtt109-Vps75 acetyltransferase activity in the cytoplasm, consistent with nascent histone H3 being a substrate. They also show that Vps75 stabilizes Rtt109 in vivo.
- 15.Bowman A, Ward R, Wiechens N, Singh V, El-Mkami H, Norman DG, Owen-Hughes T. The histone chaperones Nap1 and Vps75 bind histones H3 and H4 in a tetrameric conformation. Mol Cell. 2011;41:398–408. doi: 10.1016/j.molcel.2011.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park YJ, Sudhoff KB, Andrews AJ, Stargell LA, Luger K. Histone chaperone specificity in Rtt109 activation. Nat Struct Mol Biol. 2008;15:957–964. doi: 10.1038/nsmb.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donham DC, 2nd, Scorgie JK, Churchill ME. The activity of the histone chaperone yeast Asf1 in the assembly and disassembly of histone H3/H4-DNA complexes. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berndsen CE, Tsubota T, Lindner SE, Lee S, Holton JM, Kaufman PD, Keck JL, Denu JM. Molecular functions of the histone acetyltransferase chaperone complex Rtt109-Vps75. Nat Struct Mol Biol. 2008;15:948–956. doi: 10.1038/nsmb.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolonko EM, Albaugh BN, Lindner SE, Chen Y, Satyshur KA, Arnold KM, Kaufman PD, Keck JL, Denu JM. Catalytic activation of histone acetyltransferase Rtt109 by a histone chaperone. Proc Natl Acad Sci U S A. 2010;107:20275–20280. doi: 10.1073/pnas.1009860107. • In this report, the authors compare the activity of Rtt109-Vps75 to that of Rtt109 alone, and present structural data of Vps75-Rtt109. The limited resolution of the structural data, however, prevents refinement or building beyond the molecular replacement search models. The authors argue for a 2:1 stoichiometry of Vps75-Rtt109.
- 21.Albaugh BN, Kolonko EM, Denu JM. Kinetic mechanism of the Rtt109-Vps75 histone acetyltransferase-chaperone complex. Biochemistry. 2010;49:6375–6385. doi: 10.1021/bi100381y. • This paper contains a thorough analysis of Rtt109-Vps75 catalytic mechanism. The data confirms a sequential mode of action where Rtt109-Vps75 binds acetyl-CoA and histone H3 substrate in any order.
- 22.Tang Y, Holbert MA, Delgoshaie N, Wurtele H, Guillemette B, Meeth K, Yuan H, Drogaris P, Lee EH, Durette C, et al. Structure of the Rtt109-AcCoA/Vps75 complex and implications for chaperone-mediated histone acetylation. Structure. 2011;19:221–231. doi: 10.1016/j.str.2010.12.012. •• This article describes a thorough investigation of Vps75-Rtt109 structure and function. It includes a Vps75-Rtt109 crystal structure with a 2:2 stoichiometry, and is the only Vps75-Rtt109 structure that contains density for acetyl-CoA. The interfaces between Vps75 and Rtt109 observed in the crystal structure are confirmed with biochemical and in vivo assays. The data reveal that Asf1 and Vps75 activate Rtt109 via different mechanisms; and that Rtt109-Asf1 is responsible for histone H3 K56Ac, while Rtt109-Vps75 is responsible for K9Ac and K27Ac.
- 23.Albaugh BN, Arnold KM, Lee S, Denu JM. Autoacetylation of the histone acetyltransferase RTT109. J Biol Chem. 2011 doi: 10.1074/jbc.M111.251579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin C, Yuan YA. Structural insights into histone H3 lysine 56 acetylation by Rtt109. Structure. 2008;16:1503–1510. doi: 10.1016/j.str.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 25.Stavropoulos P, Nagy V, Blobel G, Hoelz A. Molecular basis for the autoregulation of the protein acetyl transferase Rtt109. Proc Natl Acad Sci U S A. 2008;105:12236–12241. doi: 10.1073/pnas.0805813105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang Y, Holbert MA, Wurtele H, Meeth K, Rocha W, Gharib M, Jiang E, Thibault P, Verreault A, Cole PA, et al. Fungal Rtt109 histone acetyltransferase is an unexpected structural homolog of metazoan p300/CBP. Nat Struct Mol Biol. 2008;15:738–745. doi: 10.1038/nsmb.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su D, Hu Q, Zhou H, Thompson JR, Xu RM, Zhang Z, Mer G. Structure and histone binding properties of the vps75-rtt109 chaperone-lysine acetyltransferase complex. J Biol Chem. 2011;286:15625–15629. doi: 10.1074/jbc.C111.220715. • The two crystal structures of Rtt109-Vps75 described in this manuscript have different space groups and yet both contain a 2:1 stoichiometry of Vps75-Rtt109. The paper also describes hydrogen-deuterium exchange data implying that histone H3 is bound similarly by Vps75 alone and Vps75-Rtt109. This is consistent with Vps75 presenting the histone H3 substrate to Rtt109.
- 28.Tang Y, Meeth K, Jiang E, Luo C, Marmorstein R. Structure of Vps75 and implications for histone chaperone function. Proc Natl Acad Sci U S A. 2008;105:12206–12211. doi: 10.1073/pnas.0802393105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muto S, Senda M, Akai Y, Sato L, Suzuki T, Nagai R, Senda T, Horikoshi M. Relationship between the structure of SET/TAF-Ibeta/INHAT and its histone chaperone activity. Proc Natl Acad Sci U S A. 2007;104:4285–4290. doi: 10.1073/pnas.0603762104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asahara H, Tartare-Deckert S, Nakagawa T, Ikehara T, Hirose F, Hunter T, Ito T, Montminy M. Dual roles of p300 in chromatin assembly and transcriptional activation in cooperation with nucleosome assembly protein 1 in vitro. Mol Cell Biol. 2002;22:2974–2983. doi: 10.1128/MCB.22.9.2974-2983.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito T, Ikehara T, Nakagawa T, Kraus WL, Muramatsu M. p300-mediated acetylation facilitates the transfer of histone H2A-H2B dimers from nucleosomes to a histone chaperone. Genes Dev. 2000;14:1899–1907. [PMC free article] [PubMed] [Google Scholar]