Abstract
Summary: Sequencing pooled DNA samples (Pool-Seq) is the most cost-effective approach for the genome-wide comparison of population samples. Here, we introduce PoPoolation2, the first software tool specifically designed for the comparison of populations with Pool-Seq data. PoPoolation2 implements a range of commonly used measures of differentiation (FST, Fisher's exact test and Cochran-Mantel-Haenszel test) that can be applied on different scales (windows, genes, exons, SNPs). The result may be visualized with the widely used Integrated Genomics Viewer.
Availability and Implementation: PoPoolation2 is implemented in Perl and R. It is freely available on http://code.google.com/p/popoolation2/
Contact: christian.schloetterer@vetmeduni.ac.at
Supplementary Information: Manual: http://code.google.com/p/popoolation2/wiki/Manual Test data and tutorial: http://code.google.com/p/popoolation2/wiki/Tutorial Validation: http://code.google.com/p/popoolation2/wiki/Validation
1 INTRODUCTION
Next-generation sequencing of pooled DNA samples (Pool-Seq) allows the comparison of population samples on a genomic scale, thus facilitating the transition from single marker studies to population genomics. Due to its cost-effectiveness (Futschik and Schlötterer, 2010), Pool-Seq can be used for a range of applications. The most intuitive application is the comparison of natural populations to perform standard population genetic analyses on a genomic scale (e.g. Begun et al., 2007). The comparison of natural Arabidopsis lyrata populations from different habitats allowed the characterization of genes involved in heavy metal tolerance (Turner et al., 2010). Also in experimental evolution studies, Pool-Seq has been used to identify genomic regions that show high differentiation between different selective treatments (Burke et al., 2010; Parts et al., 2011; Turner et al., 2011). Finally, Pool-Seq offers an enormous potential for selective genotyping (Darvasi and Soller, 1994; Hillel et al., 1990; Lander and Botstein, 1989).
While several tools for analyzing Pool-Seq data of single populations are already available (Bansal, 2010; Kofler et al., 2011; Pandey et al., 2011), to our knowledge no standalone software tool is available for the comparison of Pool-Seq data for multiple populations. PoPoolation2 is a software tool dedicated to the comparison of allele frequencies between populations.
2 IMPLEMENTATION
As input PoPoolation2 requires a ‘pileup’ file for every population (sample) of interest or alternatively a single multi ‘pileup’ file (mpileup) may be used. These files can be obtained by mapping the reads of a Pool-Seq experiment to a reference genome and subsequently converting the mapping results into the ‘pileup/mpileup’ format with samtools (Li et al., 2009) (For Manual see http://code.google.com/p/popoolation2/wiki/Manual; Test data and tutorial http://code.google.com/p/popoolation2/wiki/Tutorial). PoPoolation2 requires Pool-Seq data from at least two populations, but may be used with an unlimited number of populations.
To assess allele frequency differences between population samples PoPoolation2 implements a wide variety of statistics.
As the most intuitive measure of population differentiation, the allele frequency differences are reported.
The fixation index (FST) can be calculated to measure differentiation between populations. FST values may either be calculated with the classical approach (Hartl and Clark, 2007) or with an approach adapted to digital data (Karlsson et al., 2007)
The statistical significance of allele frequency differences is determined with Fisher's exact test (Fisher, 1922).
Since in experimental evolution experiments and selective genotyping studies often biological replicates are available, we implemented the Cochran–Mantel–Haenszel (CMH) test (Landis et al., 1978) to test for the statistical significance between groups.
When data from more than two populations are available, PoPoolation2 automatically computes all pairwise comparisons for these tests (except for the CMH test).
All these analyses can be performed on different levels. We have implemented a sliding window analysis, which permits a genome-wide scan for differentiation using a specified window size. For the analysis of single SNPs, a window size of 1 may be used. Finally, with a user-provided GTF file the analysis of genes, coding sequence, introns, etc. is possible. To visualize the population differentiation across the genome, PoPoolation2 converts the results into file formats that are compatible with the Integrative Genomics Viewer (Robinson et al., 2011).
Finally, PoPoolation2 also implements the functionality to randomly subsample the data to achieve a uniform coverage. The subsampling is based on a user-defined quality threshold. For analyzing the data with standard software, such as Mega5 (Tamura et al., 2011) and Arlequin (Excoffier and Lischer, 2010), PoPoolation2 allows exporting the data as artificial chromosomes as ‘multi-fasta’ files and as ‘GenePop’ files (Raymond and Rousset, 1995).
3 VALIDATION
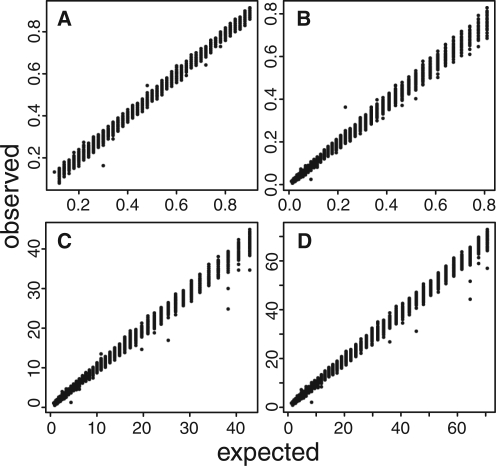
To test PoPoolation2, we placed 10 000 SNPs for two populations on chromosome 2R of Drosophila melanogaster (v5.38). For these SNPs, we simulated 75 bp reads such that the coverage was 100× and the allele frequency differences between the two populations ranged from 0.1 to 0.9. Subsequently, the simulated reads were mapped to the reference genome (D.melanogaster, chromosome 2R, v5.38) with BWA (0.5.8) (Li and Durbin, 2009) and a ‘mpileup’ file was created using samtools (0.1.13) (Li et al., 2009). Finally, we compared the expected values with the observed ones and found an almost perfect correlation between the simulated data and the estimates based on PoPoolation2 for all implemented tests (allele frequency differences: R2=0.9979, P<2.2e-16; FST: R2=0.9967, P<2.2e-16; Fisher's exact test: R2=0.9974, P<2.2e-16; CMH test: R2=0.9978, P<2.2e-16; Fig. 1). These high correlations confirm that PoPoolation2 yields highly reliable results (for details, see http://code.google.com/p/popoolation2/wiki/Validation).
Fig. 1.
Expected versus observed values for the tests implemented in PoPoolation2 using 10 000 simulated SNPs. (A) allele frequency difference; (B) FST; (C) Fisher's exact test [−log 10(P-value)]; (D) CMH test [−log 10(P-value)].
To ensure that all scripts continue to work properly, we implemented Unit-tests for the main scripts (which may be run by providing the parameter ‘–test’).
ACKNOWLEDGEMENTS
We are grateful to V. Nolte, M. Kapun and P. Orozco-ter Wengel for helpful comments and discussions. We thank all members of the ‘Institut für Populationsgenetik’ for early testing and feedback.
Funding: Austrian Science Fund (FWF): P19467-B11, P22725-B11.
Conflict of Interest: none declared.
REFERENCES
- Bansal V. A statistical method for the detection of variants from next-generation resequencing of DNA pools. Bioinformatics. 2010;26:i318–i324. doi: 10.1093/bioinformatics/btq214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun D.J., et al. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 2007;5:e310. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke M.K., et al. Genome-wide analysis of a long-term evolution experiment with Drosophila. Nature. 2010;467:587–590. doi: 10.1038/nature09352. [DOI] [PubMed] [Google Scholar]
- Darvasi A., Soller M. Selective DNA pooling for determination of linkage between a molecular marker and a quantitative trait locus. Genetics. 1994;138:1365–1373. doi: 10.1093/genetics/138.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L., Lischer H.E. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Fisher R.A. On the interpretation of χ2from contingency tables, and the calculation of P. J. R. Stat. Soc. 1922;85:87–94. [Google Scholar]
- Futschik A., Schlötterer C. The next generation of molecular markers from massively parallel sequencing of pooled DNA samples. Genetics. 2010;186:207–218. doi: 10.1534/genetics.110.114397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl D.L., Clark A.G. Principles of Population Genetics. Sunderland, Massachusetts: Sinauer Associates; 2007. [Google Scholar]
- Hillel J., et al. DNA fingerprints from blood mixes in chickens and in turkeys. Animal Biotechnol. 1990;1:201–204. [Google Scholar]
- Karlsson E.K., et al. Efficient mapping of Mendelian traits in dogs through genome-wide association. Nat. Genet. 2007;39:1321–1328. doi: 10.1038/ng.2007.10. [DOI] [PubMed] [Google Scholar]
- Kofler R., et al. PoPoolation: a toolbox for population genetic analysis of next generation sequencing data from pooled individuals. PLoS One. 2011;6:e15925. doi: 10.1371/journal.pone.0015925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E.S., Botstein D. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis J.R., et al. Average partial association in 3-way contingency-tables - review and discussion of alternative tests. Int. Stat. Rev. 1978;46:237–254. [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R.V., et al. PoPoolation DB: a user-friendly web-based database for the retrieval of natural polymorphisms in Drosophila. BMC Genet. 2011;12:27. doi: 10.1186/1471-2156-12-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parts L., et al. Revealing the genetic structure of a trait by sequencing a population under selection. Genome Res. 2011;21:1131–1138. doi: 10.1101/gr.116731.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M., Rousset F. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J. Heredity. 1995;86:248–249. [Google Scholar]
- Robinson J.T., et al. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner T.L., et al. Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils. Nat. Genet. 2010;42:260–263. doi: 10.1038/ng.515. [DOI] [PubMed] [Google Scholar]
- Turner T.L., et al. Population-based resequencing of experimentally evolved populations reveals the genetic basis of body size variation in Drosophila melanogaster. PLoS Genet. 2011;7:e1001336. doi: 10.1371/journal.pgen.1001336. [DOI] [PMC free article] [PubMed] [Google Scholar]