Abstract
Membranes are sites of intense signaling activity within the cell, serving as dynamic scaffolds for the recruitment of signaling molecules and their substrates. The specific and reversible localization of these signaling molecules to membranes is critical for the appropriate activation of downstream signaling pathways. Phospholipid-binding domains, including C1, C2, PH, and PX domains, play critical roles in the membrane targeting of protein kinases. Recent structural studies have identified a new membrane association domain, the Kinase Associated 1 (KA1) domain, which targets a number of yeast and mammalian protein kinases to membranes containing acidic phospholipids. Despite an abundance of localization studies on lipid-binding proteins and structural studies of the isolated lipid-binding domains, the question of how membrane binding is coupled to the activation of the kinase catalytic domain has been virtually untouched. Recently, structural studies on protein kinase C (PKC) have provided some of the first structural insights into the allosteric regulation of protein kinases by lipid second messengers.
Introduction
Lipid turnover within membranes directs both signal transduction and membrane trafficking in cells. The initial targeting event is dependent on the binding of lipids to protein domains. 54 of the 518 human protein kinases contain one or more known lipid-binding modules, highlighting the importance of lipids in regulating the action of protein kinases. The mechanisms for lipid-stimulated subcellular translocation, which typically entail the binding of the newly generated lipid to a particular structural domain of the kinase, are well understood in many cases. However, the mechanism of enzymatic activation of protein kinases is equally important, yet it has been more challenging to address experimentally and answers have come more slowly. The allosteric regulation of protein kinases by other proteins, soluble small molecules, and phosphorylation has been elucidated structurally for a growing number of examples [1,2]. Only now, in contrast, are examples of allosteric regulation of protein kinases by lipids coming to be understood at the structural level. In this review, we examine how membrane-embedded lipids both target and activate protein kinases, with a strong emphasis on the latter.
Protein kinases that translocate to membranes via lipid-binding domains
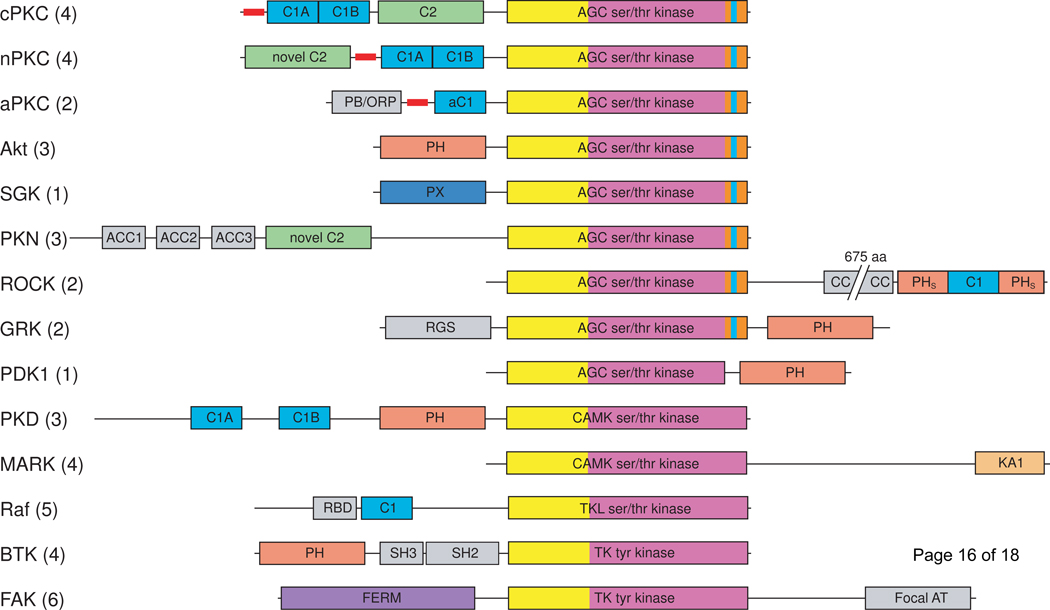
At least six discrete membrane interacting domains occur in mammalian protein kinases. These include the C1, C2, FERM, PH, and PX domains [3] with the recent addition of the KA1 domain [4]. Examples can be found of protein kinases lacking canonical lipid-binding domains that are nevertheless activated by lipids [5]. This review will focus, however, on those protein kinases that contain discrete, conserved lipid binding domains. The majority of serine/threonine kinases containing lipid-binding modules belong to the AGC (protein kinases A, G, and C) branch of the kinome [6,7]. AGC kinases have in common a C-terminal extension that wraps around the N-lobe of the catalytic domain, regulates the structure of the N-lobe, and so regulates activity. As described below, the C-terminal extension is critically important in coupling activation and lipid binding. Members of the Ca2+/calmodulin [6,8] and tyrosine kinase-like (TKL) kinase families [9] also contain membrane interaction domains. Two subfamilies of tyrosine kinases, BTK and FAK, contain lipid-binding domains (Figure 1).
Figure 1.
Domain composition of the major protein kinase families containing lipid-binding domains. Lipid-binding domains are: C1 domain (light blue), C2 domain (green), PH domain (salmon), PX domain (dark blue), KA1 domain (pale orange), FERM domain (purple). The kinase domains are shown in yellow (N-lobe) and magenta (C-lobe); the AGC kinase family C-terminal tail is orange with the NFD motif highlighted in cyan. Pseudosubstrate segments are represented as thick red lines. Additional domains are shown in gray: Bem1 (PB/ORP), antiparallel coiled coil (ACC), coiled coil (CC), regulator of G-protein signaling (RGS), Ras binding domain (RBD), Src homology 2 (SH2). cPKC, conventional protein kinase C; nPKC, novel protein kinase C; aPKC, atypical protein kinase C; SGK, serum/glucocorticoid regulated kinase; PKN, protein kinase N; ROCK, Rho-activated kinase; GRK, Gprotein coupled receptor kinase; PDK1, phosphoinositide-dependent kinase 1; PKD, protein kinase D; MARK, microtubule affinity regulating kinase; focal AT, focal adhesion targeting. Numbers in parentheses indicate the number of mammalian kinases in each family.
Mechanism of lipid activation of a conventional PKC
The protein kinase C (PKC) isozymes have been by far the best-studied paradigm of an enzyme family that is both relocalized and enzymatically activated by lipid signals [6,10]. Hydrolysis of phosphatidylinositol-(4,5)-bisphosphate (PIP2) by phospholipase C (PLC) generates the classic lipid second messenger, diacylglycerol (DAG), and inositol-1,4,5-trisphosphate (IP3). IP3 stimulates the release of calcium from intracellular stores. Conventional PKCs, which contain a calcium- and phospholipid- binding C2 domain, are recruited to the membrane, where Ca2+ ions bridge the C2 domain to phosphatidylserine (PS). Once at the membrane, PKC is activated by binding to DAG via its C1 domains [10–14]. Biochemical [15] and imaging [16] studies on PKC have illustrated how multiple lipid binding modules can cooperate to drive stable membrane localization. In a general sense, binding of DAG provides the energy for displacement of the autoinhibitory pseudosubstrate from the catalytic cleft, therefore activating the kinase [17]. What has remained to be understood have been the molecular details whereby DAG binding to the C1 domain triggers activation.
The C1-DAG-membrane interaction is at the heart of PKC activation by DAG. C1 domains contain a rim of hydrophobic residues surrounding the DAG binding cleft. These residues mediate insertion into the lipid bilayer. Stable insertion in the membrane requires occupancy of the DAG binding cleft, while high-affinity binding to DAG occurs only in the context of membrane penetration and bulk hydrophobic interactions with the rim. The hydrophobic rim of the C1 domain is critical for the allosteric regulation of the proteins that contain it. C1 domains can form interdomain interactions in the inactive state via hydrophobic interactions with the rim. For example, in the C1-containing RacGAP protein β2-chimaerin, the hydrophobic rim is almost completely buried in interdomain contacts that stabilize the closed, inactive state of the enzyme [18].
The crystal structure of full-length PKCβII in a partially activated state showed that activation of the kinase by DAG goes through two stages [19]. Highresolution data is still unavailable for the first stage in activation. The general picture is that the membrane and DAG compete with an intramolecular site on the N-lobe of the kinase for binding to the C1A domain. The C1A domain is very near the pseudosubstrate region. In the current view, membrane binding by the C1A hydrophobic rim is sterically incompatible with the presence of the pseudosubstrate in the active site. Thus DAG and membrane binding to the C1A forces the pseudosubstrate out of the active site, allowing access to substrates. A crystallographic structure of the completely autoinhibited conformation of a PKC with the pseudosubstrate and C1A domain in place is still an important missing piece of the puzzle.
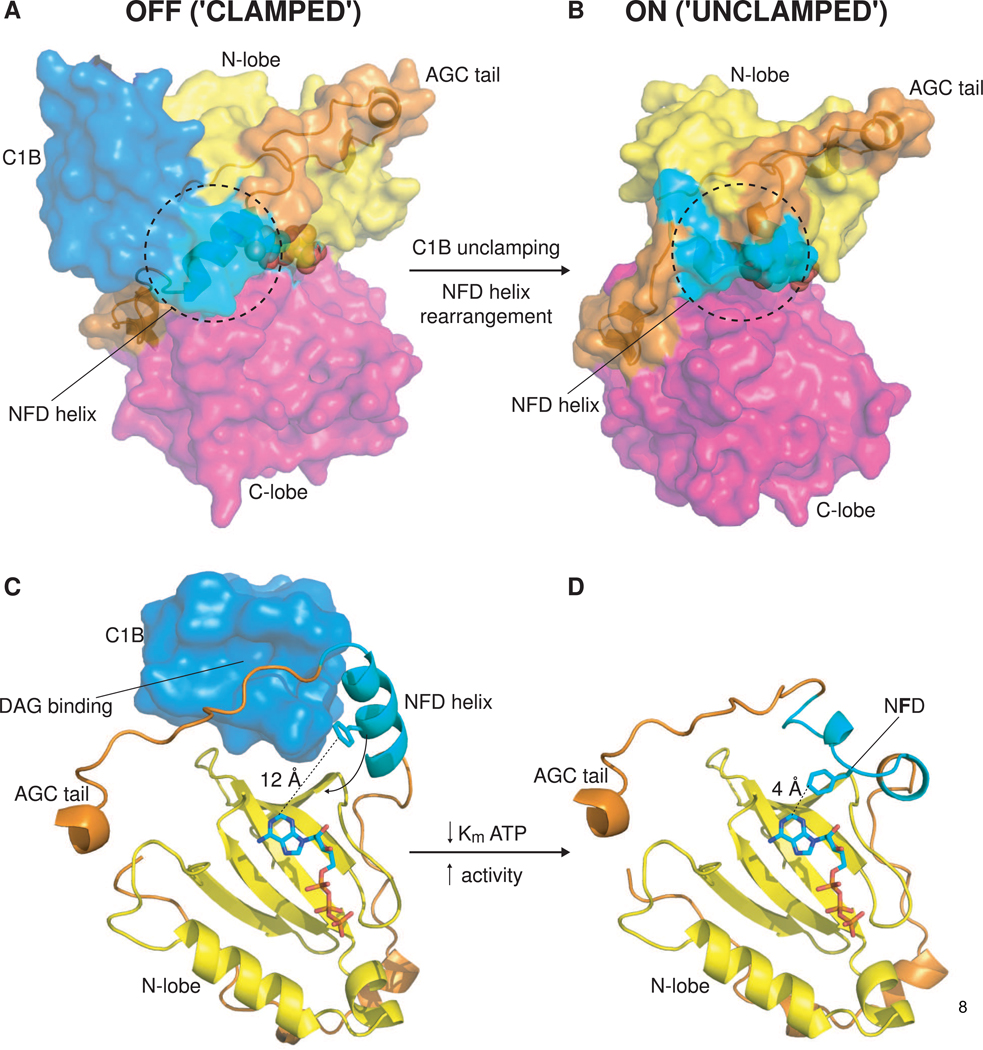
Pseudosubstrate release is not the end of the activation story. The C1B domain and the catalytic domain form an extensive interface involving its hydrophobic rim, a state that can persist even when the C1A is displaced. Residues in the tail segment of the PKC catalytic domain sequester the DAG binding cleft of the C1B domain in the absence of lipid binding (Figure 2A). DAG binding triggers displacement of the tail segment from the C1B cleft, and the subsequent conformational change in the C-terminal segment of the kinase domain results in the insertion of the conserved NFD motif Phe side-chain into the nucleotide-binding site (Figure 2B). The Phe side chain interacts with the adenine moiety of ATP, stabilizing its binding and contributing a factor of ~50 to catalytic efficiency. Thus full activation of the conventional PKC isozymes (and probably the novel isozymes) that contain two C1 domains requires engagement of both of the C1 domains with the membrane, each contributing to activation in distinct ways (Figure 2C–D).
Figure 2.
Allosteric activation of PKC by DAG binding to the C1B domain. (A) ‘Clamped’ conformation of PKCβII. Surface representation of PKCβII, illustrating the interface between the lipid-binding C1B domain (light blue) and the kinase domain (yellow/magenta). A novel helix containing the conserved NFD motif found in the Cterminal tail of AGC kinases packs against the C1B domain, while residues preceding the novel helix occupy the DAG binding site of the C1B domain. ATP is represented with spheres. (B) ‘Unclamped’ conformation of PKCι. Surface representation of PKCι, illustrating the conformational change that occurs in the NFD helix upon DAG/membrane binding by the C1B domain. (C) View of the DAG-binding cleft of the C1B domain and the nucleotide-binding site (sandwiched between the N-lobe (yellow) and C-lobe (not shown) of the kinase domain). The lipid-binding site is inaccessible in this conformation and the critical phenylalanine of the NFD motif is sequestered 12 Å away from the nucleotide. (D) View of the active conformation of PKCι in which the NFD helix has undergone a conformational rearrangement in the absence of the C1 domain. The central phenylalanine of the NFD motif is brought into close (4 Å) contact with the adenine ring of ATP. Contacts between the phenylalanine side chain and the adenine moiety lower the Km for ATP, thereby increasing the catalytic rate, Kcat, of the enzyme.
Other C1 domain-containing kinases
Raf and KSR
Kinase suppressor of Ras (KSR) is a ceramide-activated protein kinase [20] that contains a C1 domain homologous to the C1 domain of PKCζ. Ceramide activates KSR1 to autophosphorylate and transactivate RAF1. RAF1, like KSR1, contains a ceramide-binding C1 domain, which is necessary for its activation. The C1 domain of KSR1 mediates its translocation to ceramide-containing membranes [21]. While recent work has made progress towards understanding the allosteric activation of the RAF1 kinase domain through a side-to-side homodimer or heterodimer with KSR1 [22], less is known about the molecular mechanism whereby ceramide activates RAF1/KSR1. The C1 domain-containing regulatory region of RAF1 inhibits kinase activity via an intramolecular interaction with the kinase domain [23]. Mutations in the C1 domain activate RAF1 by relieving an autoinhibitory interaction with the kinase domain. It is tempting to imagine that RAF1/KSR1 may be regulated by ceramide binding in a manner analogous to that by which PKCβII is activated by DAG. There must be many differences in detail, however, since RAF1 and KSR1 contain only one C1 domain, and are lacking an obvious counterpart to the NFD motif of the PKCs. B-RAF is the most frequently mutated oncogene in the kinase superfamily and more than 160 kinases are associated with human diseases; all nine clinically approved, intracellularly active, kinase inhibitors target the ATP-binding site, so pharmacological studies would benefit from more full-length structures in inactive conformations.
Rho kinases
Rho kinases contain a split PH domain into which a C1 domain is inserted [24]. Rho kinases associate with the membrane via this PH-C1 module in a manner that is dependent on the presence of both. The PH-C1 unit binds to 3’-phosphoinositides, though the contributions of the individual PH and C1 domains to lipid binding remain to be dissected. The split PH-C1 domain of the Rho kinases, like the C1B domain of PKCβII, couples specific lipid binding to kinase activity. The C-terminal regulatory region suppresses kinase activity by sequestering the N-terminal kinase domain in an intramolecular interaction. Membrane binding, activation by RhoA [25], or in vivo caspase-mediated cleavage of the C-terminal domains [26] relieves the inhibitory intramolecular interactions, thereby activating the kinase. This mechanism seems to parallel the C1A-dependent activation step of PKC. It will be interesting to see if more subtle mechanisms, analogous perhaps to the C1B-dependent unclamping of the AGC extension, are operative amongst the Rho kinases.
PH domain-containing serine/threonine kinases
Protein kinase B (PKB/Akt) is an AGC kinase with a N-terminal PIP3-binding PH domain. The PI 3-kinase pathway, in which PKB is a critical effector, is the most commonly dysregulated pathway in human cancers. The PH domain of PKB is involved in both membrane targeting and allosteric activation of the kinase. Very recently a crystal structure of full-length PKB was determined, in the presence of the inhibitor compound VIII [27]. The structure is in the “PH-in” conformation [28]. The PH domain binds across the catalytic cleft, contacting both the N- and C-lobes of the kinase and directly blocking substrate access to the active site. Direct interactions between the PH and kinase domains are extensive. These interactions are further stabilized by bridging interactions with compound VIII, which is wedged between the domains and seems to act as a sort of molecular glue. The exposed side chain of Trp80 of the PH domain plays a role loosely analogous to the hydrophobic rim of the C1 domains of the PKCs, forming contacts with both the inhibitor and hydrophobic residues of the N-lobe of the kinase. The PH domain seems to be playing a role closer to that of the pseudosubstrate-C1A portion of PKC, as opposed to the C1B-NFD motif nexus. Indeed, the AGC extension is disordered in this structure. The main open question concerning this structure and PKB activation by lipids is to what extent compound VIII has trapped a normal inactive conformation as opposed to inducing a conformation that is off of the normal activation pathway.
The AGC kinase G-protein coupled receptor kinase 2 (GRK2) contains a single PH domain that targets it to membranes. The structure of GRK2 in complex with Gβγ shows that the PH domain forms extensive intramolecular interfaces with both the regulator of G-protein signaling (RGS) domain and with the Gβγ subunit [29]. An extension of the C-terminal tail of the kinase domain acts as an essential scaffold for the RGS and PH domains. Mutation of the interface between the PH and RGS domains leads to a deficiency in phospholipid-mediated activation of GRK2, suggesting that allosteric communication occurs between the PH, RGS, and kinase domains. While it is known that Gβγ binding to the PH domain (which serves as a model for Gβγ binding to other PH domain-containing effector proteins such as PLCβ) enhances phospholipid binding, more focused analysis of the mechanism by which phospholipid binding is coupled to kinase activation would be helpful.
PH domain-containing tyrosine kinases
Tec family tyrosine kinases contain a N-terminal PH domain essential for signaling. Engagement of the B-cell receptor (BCR) by antigen activates phosphatidylinositol-3-kinase (PI3K), leading to the accumulation of phosphatidylinositol-3,4,5-trisphosphate (PIP3) in the inner leaflet of the plasma membrane. PIP3 binding by the PH domain mediates the recruitment of Bruton’s tyrosine kinase (Btk) to the membrane, where it controls critical B-cell proliferation and survival pathways [30]. Mutation of Arg 29, a key residue in PIP3 binding, results in X-linked agammaglobulinaemia (XLA). Charge-switching mutations in Glu 41 and Asp 43 result in a gain-of-function in Btk, leading to increased membrane targeting and robust transforming activity in vitro, though mutations at these sites have not been associated with disease. It has been proposed that additional electrostatic interactions with anionic phospholipids may explain this effect, though similar charge-switching mutations in the vicinity of Glu 41 and Asp 43 do not differ markedly from wild-type Btk. In the absence of a structure of a full-length Tec tyrosine kinase, such as Btk, it is not possible to conclusively say whether such mutations affect membrane binding directly, or whether they allosterically activate the kinase by destabilizing an intramolecular interface between the PH domain and the rest of the protein. Structures of the PH domain suggest that it may mediate dimerization of Btk at the membrane, though Btk is monomeric in solution. The lipid-specific recruitment of Btk to the membrane might be allosterically coupled to Btk activation through PH domain-mediated dimerization.
PX domain-containing kinases
Serum and glucocorticoid kinase 3 (SGK3) is one of three known PX domain-containing kinases. Like the PH domain of Akt, the PX domain of SGK3 is N-terminal to the kinase domain, and both Akt and SGK3 contain the canonical AGC kinase C-terminal tail. The PX domain of SGK3 binds preferentially to phosphatidylinositol-3-phosphate (PI3P) and targets it to endosomes [31]. A functional PX domain is required for PDK1-dependent activation loop phosphorylation and activation of SGK3. Replacement of the hydrophobic motif of SGK3 with that of PKC-related kinase 2 (PRK2) generates a constitutively active enzyme; in contrast to wild type SGK3, activation loop phosphorylation of this chimeric protein is independent of its PX-domain. PX domain-mediated dimerization of SGK3 may stabilize the interaction between SGK3 and the membrane in a multivalent manner, analogous to PH domain-mediated dimerization of Btk at the membrane [32]. Minimally, PI3P binding by the PX domain is required for activation loop phosphorylation of SGK3, though whether there is an additional allosteric effect from displacement of the PX domain from intramolecular contacts with the kinase domain is not known.
FERM domain-containing tyrosine kinases
Binding of acidic phospholipids to the FERM domain is required for the activation of focal adhesion kinase (FAK). FAK is maintained in an autoinhibited state by steric occlusion of the catalytic cleft with the FERM domain [33]. Mutation of this interface results in a constitutively active kinase. Recently, it has been demonstrated that the FERM domain mediates the interaction of FAK with PIP2-containing membranes [34]. PIP2 binding is a feature of the proteins ezrin, radixin, and moesin, but the mode of binding differs to that of FAK. In radixin, the PIP2 headgroup is bound in a basic pocket between the F1 and F3 subdomains of the FERM domain, whereas FAK lacks this basic pocket. Despite similarity of the F3 subdomain to the PH domain fold, binding to phospholipids is not possible due to the lack of the basic pocket. Instead, the interaction of FAK with PIP2 is mediated by a basic patch on the F2 subdomain. This patch is near the interface with the catalytic domain as observed in the crystal structure of full length FAK [33]. Lipid binding causes a conformational change of FAK both in vitro and in vivo [34]. The F2 basic patch is required for activation of FAK following growth factor stimulation and PIP2 depletion results in decreased FAK activity. These observations support the hypothesis that lipid binding allosterically activates FAK by relieving autoinhibitory intramolecular interactions.
Conclusions
Lipids have long been known to stimulate both the translocation and enzyme activity of certain protein kinases, of which the PKCs are the historical paradigm. These properties of PKCs led to the discovery of the C1 and C2 domains and thence to the concept of protein regulation by lipid binding domains. The translocation properties of lipid binding domains proved easier to study than their allosteric regulatory properties. Thus, by the early 2000s we had an extensive knowledge of lipid binding domain-mediated protein translocation, but far less information on enzyme activation. The labor-intensive nature of crystallizing full-length lipidactivated kinases has been a factor in this gap. Once crystallized, relating the structures to allosteric properties can still be a challenge in cases where the enzymological groundwork is incomplete. The recent structural studies of PKC benefited greatly from building on the groundwork of classical biochemical studies in the 1980s and early 1990s [10,35,36]. The crystal structure of PKCβII is the first structural glimpse at how a membrane-embedded second messenger, DAG, can activate a kinase. More work is needed to gain a complete picture of this complex system. Given the availability of full-length structures of PKB and FAK, these kinases also offer promise to yield more mechanistic insight in the relatively near term.
Highlights.
10% of the human kinome contains lipid-binding domains
Kinases with novel lipid-binding domains are still being discovered
Recent kinase structures have provided new insights into their activation by lipids
A novel interface between the C1B and kinase domains controls PKCbII activation
Acknowledgements
This research was supported by the Intramural Program of the NIH, NIDDK (J.H.H.). Thomas A. Leonard was supported by a EMBO Long Term Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Thomas A. Leonard, Email: leonardt@niddk.nih.gov.
James H. Hurley, Email: hurley@helix.nih.gov.
References
- 1.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 2.Pellicena P, Kuriyan J. Protein-protein interactions in the allosteric regulation of protein kinases. Curr Opin Struct Biol. 2006;16:702–709. doi: 10.1016/j.sbi.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 3.Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 4. Moravcevic K, Mendrola JM, Schmitz KR, Wang YH, Slochower D, Janmey PA, Lemmon MA. Kinase associated-1 domains drive MARK/PAR1 kinases to membrane targets by binding acidic phospholipids. Cell. 2010;143:966–977. doi: 10.1016/j.cell.2010.11.028. •• This work identifies a new lipid-binding domain found in protein kinases and highlights the importance of coincidence detection in signaling at the membrane.
- 5.Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science. 2001;294:1942–1945. doi: 10.1126/science.1066015. [DOI] [PubMed] [Google Scholar]
- 6.Newton AC. Lipid activation of protein kinases. J Lipid Res. 2009;50 Suppl:S266–S271. doi: 10.1194/jlr.R800064-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol. 2010;11:9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 8.Marx A, Nugoor C, Panneerselvam S, Mandelkow E. Structure and function of polarity-inducing kinase family MARK/Par-1 within the branch of AMPK/Snf1-related kinases. FASEB J. 2010;24:1637–1648. doi: 10.1096/fj.09-148064. [DOI] [PubMed] [Google Scholar]
- 9.Udell CM, Rajakulendran T, Sicheri F, Therrien M. Mechanistic principles of RAF kinase signaling. Cell Mol Life Sci. 2011;68:553–565. doi: 10.1007/s00018-010-0520-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishizuka Y. Intracellular signaling by hydrolysis of phospholipids and activation of protein kinase C. Science. 1992;258:607–614. doi: 10.1126/science.1411571. [DOI] [PubMed] [Google Scholar]
- 11.Ono Y, Fujii T, Igarashi K, Kuno T, Tanaka C, Kikkawa U, Nishizuka Y. Phorbol ester binding to protein kinase C requires a cysteine-rich zinc-finger-like sequence. Proc Natl Acad Sci U S A. 1989;86:4868–4871. doi: 10.1073/pnas.86.13.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hurley JH, Newton AC, Parker PJ, Blumberg PM, Nishizuka Y. Taxonomy and function of C1 protein kinase C homology domains. Protein Sci. 1997;6:477–480. doi: 10.1002/pro.5560060228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newton AC. Diacylglycerol's affair with protein kinase C turns 25. Trends Pharmacol Sci. 2004;25:175–177. doi: 10.1016/j.tips.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 14.Colon-Gonzalez F, Kazanietz MG. C1 domains exposed: from diacylglycerol binding to protein-protein interactions. Biochim Biophys Acta. 2006;1761:827–837. doi: 10.1016/j.bbalip.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 15.Newton AC, Johnson JE. Protein kinase C: a paradigm for regulation of protein function by two membrane-targeting modules. Biochim Biophys Acta. 1998;1376:155–172. doi: 10.1016/s0304-4157(98)00003-3. [DOI] [PubMed] [Google Scholar]
- 16.Oancea E, Meyer T. Protein kinase C as a molecular machine for decoding calcium and diacylglycerol signals. Cell. 1998;95:307–318. doi: 10.1016/s0092-8674(00)81763-8. [DOI] [PubMed] [Google Scholar]
- 17.Newton AC. Protein kinase C: poised to signal. Am J Physiol Endocrinol Metab. 2010;298:E395–E402. doi: 10.1152/ajpendo.00477.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Canagarajah B, Leskow FC, Ho JY, Mischak H, Saidi LF, Kazanietz MG, Hurley JH. Structural mechanism for lipid activation of the Rac-specific GAP, beta2-chimaerin. Cell. 2004;119:407–418. doi: 10.1016/j.cell.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 19. Leonard TA, Rozycki B, Saidi LF, Hummer G, Hurley JH. Crystal structure and allosteric activation of protein kinase C betaII. Cell. 2011;144:55–66. doi: 10.1016/j.cell.2010.12.013. •• The structure of full-length protein kinase C provides the first structural insight into the allosteric activation of a protein kinase by a lipid. The mechanism proceeds through two steps that involve the sequential displacement of the DAG-binding domains from contacts with the kinase domain.
- 20.Zhang Y, Yao B, Delikat S, Bayoumy S, Lin XH, Basu S, McGinley M, Chan-Hui PY, Lichenstein H, Kolesnick R. Kinase suppressor of Ras is ceramide-activated protein kinase. Cell. 1997;89:63–72. doi: 10.1016/s0092-8674(00)80183-x. [DOI] [PubMed] [Google Scholar]
- 21.Yin X, Zafrullah M, Lee H, Haimovitz-Friedman A, Fuks Z, Kolesnick R. A ceramide-binding C1 domain mediates kinase suppressor of ras membrane translocation. Cell Physiol Biochem. 2009;24:219–230. doi: 10.1159/000233248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajakulendran T, Sahmi M, Lefrancois M, Sicheri F, Therrien M. A dimerization-dependent mechanism drives RAF catalytic activation. Nature. 2009;461:542–545. doi: 10.1038/nature08314. [DOI] [PubMed] [Google Scholar]
- 23.Cutler RE, Jr., Stephens RM, Saracino MR, Morrison DK. Autoregulation of the Raf-1 serine/threonine kinase. Proc Natl Acad Sci U S A. 1998;95:9214–9219. doi: 10.1073/pnas.95.16.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wen W, Liu W, Yan J, Zhang M. Structure basis and unconventional lipid membrane binding properties of the PH-C1 tandem of rho kinases. J Biol Chem. 2008;283:26263–26273. doi: 10.1074/jbc.M803417200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishizaki T, Maekawa M, Fujisawa K, Okawa K, Iwamatsu A, Fujita A, Watanabe N, Saito Y, Kakizuka A, Morii N, et al. The small GTP-binding protein Rho binds to and activates a 160 kDa Ser/Thr protein kinase homologous to myotonic dystrophy kinase. EMBO J. 1996;15:1885–1893. [PMC free article] [PubMed] [Google Scholar]
- 26.Sebbagh M, Renvoize C, Hamelin J, Riche N, Bertoglio J, Breard J. Caspase-3-mediated cleavage of ROCK I induces MLC phosphorylation and apoptotic membrane blebbing. Nat Cell Biol. 2001;3:346–352. doi: 10.1038/35070019. [DOI] [PubMed] [Google Scholar]
- 27. Wu WI, Voegtli WC, Sturgis HL, Dizon FP, Vigers GP, Brandhuber BJ. Crystal structure of human AKT1 with an allosteric inhibitor reveals a new mode of kinase inhibition. PLoS One. 2010;5:e12913. doi: 10.1371/journal.pone.0012913. • The structure of full-length Akt1 with an allosteric inhibitor reveals an extensive interface between the lipid-binding PH domain and the kinase domain. The inhibitor locks the kinase in an inactive conformation that cannot respond to lipid signals.
- 28.Calleja V, Alcor D, Laguerre M, Park J, Vojnovic B, Hemmings BA, Downward J, Parker PJ, Larijani B. Intramolecular and intermolecular interactions of protein kinase B define its activation in vivo. PLoS Biol. 2007;5:e95. doi: 10.1371/journal.pbio.0050095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lodowski DT, Pitcher JA, Capel WD, Lefkowitz RJ, Tesmer JJ. Keeping G proteins at bay: a complex between G protein-coupled receptor kinase 2 and Gbetagamma. Science. 2003;300:1256–1262. doi: 10.1126/science.1082348. [DOI] [PubMed] [Google Scholar]
- 30.Mohamed AJ, Yu L, Backesjo CM, Vargas L, Faryal R, Aints A, Christensson B, Berglof A, Vihinen M, Nore BF, et al. Bruton's tyrosine kinase (Btk): function, regulation, and transformation with special emphasis on the PH domain. Immunol Rev. 2009;228:58–73. doi: 10.1111/j.1600-065X.2008.00741.x. [DOI] [PubMed] [Google Scholar]
- 31.Tessier M, Woodgett JR. Role of the Phox homology domain and phosphorylation in activation of serum and glucocorticoid-regulated kinase-3. J Biol Chem. 2006;281:23978–23989. doi: 10.1074/jbc.M604333200. [DOI] [PubMed] [Google Scholar]
- 32.Xing Y, Liu D, Zhang R, Joachimiak A, Songyang Z, Xu W. Structural basis of membrane targeting by the Phox homology domain of cytokine-independent survival kinase (CISK-PX) J Biol Chem. 2004;279:30662–30669. doi: 10.1074/jbc.M404107200. [DOI] [PubMed] [Google Scholar]
- 33.Lietha D, Cai X, Ceccarelli DF, Li Y, Schaller MD, Eck MJ. Structural basis for the autoinhibition of focal adhesion kinase. Cell. 2007;129:1177–1187. doi: 10.1016/j.cell.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cai X, Lietha D, Ceccarelli DF, Karginov AV, Rajfur Z, Jacobson K, Hahn KM, Eck MJ, Schaller MD. Spatial and temporal regulation of focal adhesion kinase activity in living cells. Mol Cell Biol. 2008;28:201–214. doi: 10.1128/MCB.01324-07. • Relief of an intramolecular autoinhibitory interaction in FAK is known to be essential for its activation. This work makes a strong case for the interaction of FAK with acidic phospolipids via a conserved basic patch on its FERM domain and its consequent activation.
- 35.Newton AC. Protein kinase C: structure, function, and regulation. J Biol Chem. 1995;270:28495–28498. doi: 10.1074/jbc.270.48.28495. [DOI] [PubMed] [Google Scholar]
- 36.Mellor H, Parker PJ. The extended protein kinase C superfamily. Biochem J. 1998;332(Pt 2):281–292. doi: 10.1042/bj3320281. [DOI] [PMC free article] [PubMed] [Google Scholar]