Abstract
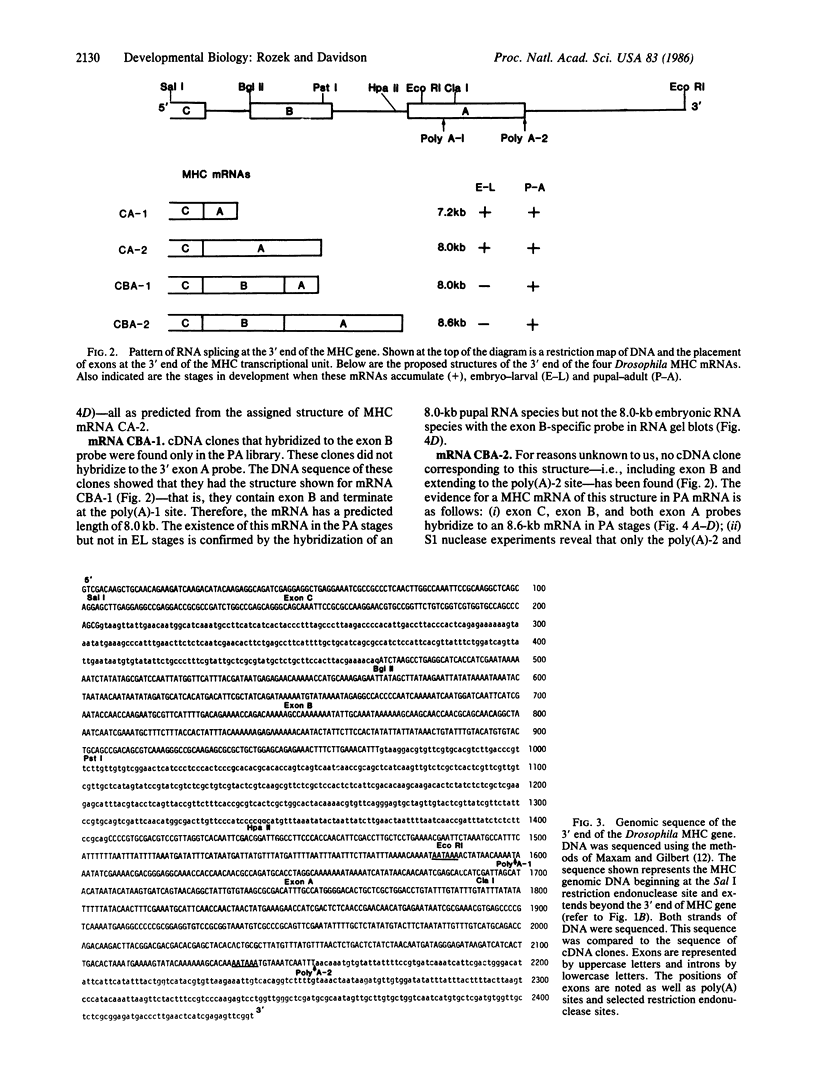
We report the sequence of genomic DNA at the 3' end of the single-copy Drosophila myosin heavy chain (MHC) gene and the structure and sequence at the 3' end of four MHC mRNAs. Two mRNAs, 7.2-kilobases (kb) and 8.0 kb in length, are expressed in all stages of development in which detectable levels of muscle-specific mRNAs accumulate. These mRNAs differ by alternate choice of two poly(A) sites within the same exon. Sequence information predicts that these two mRNAs can encode one MHC polypeptide. Two additional MHC mRNAs, 8.0 kb and 8.6 kb in length, are expressed only in late pupal and adult stages of development. These two stage-specific MHC mRNAs use the same poly(A) sites as the MHC mRNAs described above but have a different splicing pattern and thus include an additional exon. Sequence information predicts that these two stage-specific MHC mRNAs encode a second MHC polypeptide with a different COOH terminus.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amara S. G., Jonas V., Rosenfeld M. G., Ong E. S., Evans R. M. Alternative RNA processing in calcitonin gene expression generates mRNAs encoding different polypeptide products. Nature. 1982 Jul 15;298(5871):240–244. doi: 10.1038/298240a0. [DOI] [PubMed] [Google Scholar]
- Benyajati C., Spoerel N., Haymerle H., Ashburner M. The messenger RNA for alcohol dehydrogenase in Drosophila melanogaster differs in its 5' end in different developmental stages. Cell. 1983 May;33(1):125–133. doi: 10.1016/0092-8674(83)90341-0. [DOI] [PubMed] [Google Scholar]
- Bernstein S. I., Mogami K., Donady J. J., Emerson C. P., Jr Drosophila muscle myosin heavy chain encoded by a single gene in a cluster of muscle mutations. 1983 Mar 31-Apr 6Nature. 302(5907):393–397. doi: 10.1038/302393a0. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenthal S., Parker V. P., Davidson N. Developmental variations in the splicing pattern of transcripts from the Drosophila gene encoding myosin alkali light chain result in different carboxyl-terminal amino acid sequences. Proc Natl Acad Sci U S A. 1985 Jan;82(2):449–453. doi: 10.1073/pnas.82.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenthal S., Parker V. P., Mattox W. W., Davidson N. Drosophila melanogaster has only one myosin alkali light-chain gene which encodes a protein with considerable amino acid sequence homology to chicken myosin alkali light chains. Mol Cell Biol. 1984 May;4(5):956–965. doi: 10.1128/mcb.4.5.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Forbes D. J., Kirschner M. W., Caput D., Dahlberg J. E., Lund E. Differential expression of multiple U1 small nuclear RNAs in oocytes and embryos of Xenopus laevis. Cell. 1984 Oct;38(3):681–689. doi: 10.1016/0092-8674(84)90263-0. [DOI] [PubMed] [Google Scholar]
- Keller E. B., Noon W. A. Intron splicing: a conserved internal signal in introns of Drosophila pre-mRNAs. Nucleic Acids Res. 1985 Jul 11;13(13):4971–4981. doi: 10.1093/nar/13.13.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehart D. P., Kaiser D. A., Pollard T. D. Direct localization of monoclonal antibody-binding sites on Acanthamoeba myosin-II and inhibition of filament formation by antibodies that bind to specific sites on the myosin-II tail. J Cell Biol. 1984 Sep;99(3):1015–1023. doi: 10.1083/jcb.99.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner M. R., Boyle J. A., Mount S. M., Wolin S. L., Steitz J. A. Are snRNPs involved in splicing? Nature. 1980 Jan 10;283(5743):220–224. doi: 10.1038/283220a0. [DOI] [PubMed] [Google Scholar]
- Mahdavi V., Periasamy M., Nadal-Ginard B. Molecular characterization of two myosin heavy chain genes expressed in the adult heart. Nature. 1982 Jun 24;297(5868):659–664. doi: 10.1038/297659a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McLachlan A. D., Karn J. Periodic charge distributions in the myosin rod amino acid sequence match cross-bridge spacings in muscle. Nature. 1982 Sep 16;299(5880):226–231. doi: 10.1038/299226a0. [DOI] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins J. R., Blanchard J. M., Darnell J. E., Jr Transcription units of adenovirus type 2. Termination of transcription beyond the poly(A) addition site in early regions 2 and 4. J Mol Biol. 1980 Dec 15;144(3):377–386. doi: 10.1016/0022-2836(80)90096-0. [DOI] [PubMed] [Google Scholar]
- Okayama H., Berg P. A cDNA cloning vector that permits expression of cDNA inserts in mammalian cells. Mol Cell Biol. 1983 Feb;3(2):280–289. doi: 10.1128/mcb.3.2.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okayama H., Berg P. High-efficiency cloning of full-length cDNA. Mol Cell Biol. 1982 Feb;2(2):161–170. doi: 10.1128/mcb.2.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J., Early P., Carter C., Calame K., Bond M., Hood L., Wall R. Two mRNAs with different 3' ends encode membrane-bound and secreted forms of immunoglobulin mu chain. Cell. 1980 Jun;20(2):303–312. doi: 10.1016/0092-8674(80)90616-9. [DOI] [PubMed] [Google Scholar]
- Rozek C. E., Davidson N. Drosophila has one myosin heavy-chain gene with three developmentally regulated transcripts. Cell. 1983 Jan;32(1):23–34. doi: 10.1016/0092-8674(83)90493-2. [DOI] [PubMed] [Google Scholar]
- Schachat F., Garcea R. L., Epstein H. F. Myosins exist as homodimers of heavy chains: demonstration with specific antibody purified by nematode mutant myosin affinity chromatography. Cell. 1978 Oct;15(2):405–411. doi: 10.1016/0092-8674(78)90009-0. [DOI] [PubMed] [Google Scholar]
- Setzer D. R., McGrogan M., Nunberg J. H., Schimke R. T. Size heterogeneity in the 3' end of dihydrofolate reductase messenger RNAs in mouse cells. Cell. 1980 Nov;22(2 Pt 2):361–370. doi: 10.1016/0092-8674(80)90346-3. [DOI] [PubMed] [Google Scholar]
- Sinha A. M., Umeda P. K., Kavinsky C. J., Rajamanickam C., Hsu H. J., Jakovcic S., Rabinowitz M. Molecular cloning of mRNA sequences for cardiac alpha- and beta-form myosin heavy chains: expression in ventricles of normal, hypothyroid, and thyrotoxic rabbits. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5847–5851. doi: 10.1073/pnas.79.19.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner P. Controlling roles for snurps. Nature. 1985 Jul 11;316(6024):105–106. doi: 10.1038/316105a0. [DOI] [PubMed] [Google Scholar]
- Winkelmann D. A., Lowey S., Press J. L. Monoclonal antibodies localize changes on myosin heavy chain isozymes during avian myogenesis. Cell. 1983 Aug;34(1):295–306. doi: 10.1016/0092-8674(83)90160-5. [DOI] [PubMed] [Google Scholar]