Abstract
Vascular endothelial growth factor (VEGF) plays a central role in breast cancer development and progression, but the mechanisms that control its expression are poorly understood. Breast cancer tissue microarrays revealed an inverse correlation between the Forkhead transcription factor FOXO3a and VEGF expression. Using the lapatinib-sensitive breast cancer cell lines BT474 and SKBR3 as model systems, we tested the possibility that VEGF expression is negatively regulated by FOXO3a. Lapatinib treatment of BT474 or SKBR3 cells resulted in nuclear translocation and activation of FOXO3a, followed by a reduction in VEGF expression. Transient transfection and inducible expression experiments showed that FOXO3a represses the proximal VEGF promoter whereas another forkhead member, FOXM1, induces VEGF expression. Chromatin immunoprecipitation and oligonucleotide pull-down assays demonstrated that both FOXO3a and FOXM1 bind a consensus Forkhead response element (FHRE) in the VEGF promoter. Upon lapatinib stimulation, activated FOXO3a displaces FOXM1 bound to the FHRE before recruiting histone deacetylase 2 (HDAC2) to the promoter, leading to decreased histones H3 and H4 acetylation, and concomitant transcriptional inhibition of VEGF. These results show that FOXO3a-dependent repression of target genes in breast cancer cells, such as VEGF, involves competitive displacement of DNA-bound FOXM1 and active recruitment of transcriptional repressor complexes.
Keywords: VEGF, FOXO3a, FOXM1, breast cancer, transcription
Introduction
The vascular endothelial growth factor (VEGF) family of growth factors, consisting of 6 members, VEGF-A (commonly called VEGF), VEGF-B,-C,-D,-E and the placental growth factor (PIGF), plays a crucial role in tissue development and maintenance through regulating the processes of vasculogenesis, angiogenesis and lymphangiogenesis (Lohela et al 2009). These VEGF ligands bind to 3 distinct primary receptors and 2 co-receptors to trigger downstream intracellular signalling. Of the primary receptors, VEGFR-1 (Flt-1) and VEGFR-2 (KDR/Flk-1) are associated predominantly with angiogenesis, and VEGFR-3 (Flt-4) to lymphangiogenesis. VEGFR-2 is expressed ubiquitously on almost all endothelial cell types, whereas the expression of VEGFR-1 and -3 is restricted to particular vascular supporting tissues. The neuropilin-1 and -2 receptors are co-receptors that can enhance the binding affinity of the various VEGF-ligands to the primary receptors. Upon ligand-binding, the VEGF receptors activate downstream signalling cascades, including the phosphatidylinositol 3-kinase(PI3K)-Akt(PKB), the p38-MAPK, and the Raf pathways, which in turn control the endothelial cell survival, proliferation and migration (Lentzsch et al 2004, Pytel et al 2009). VEGF and its receptors are frequently overexpressed in human tumours, especially in breast, non-small cell lung, colorectal, and prostate cancers (Ferrer et al 1998, Heist et al 2008, Jain et al 2009, Schneider and Sledge 2007, Yamaguchi et al 2007). VEGF mediates angiogenesis, a process that plays a central role in the growth, progression, and metastasis of solid tumours (Kitadai 2010, Makrilia et al 2009). In consequence, VEGF and associated signalling pathways have been the targets for many novel anti-cancer targeted therapeutics (Margolin 2002). For instance, bevacizumab, an anti-VEGF antibody, has been shown to enhance response rates and prolonged progression-free survival in metastatic breast cancer. Similarly, inhibition of the VEGF signalling by receptor tyrosine kinase inhibitors (RTKIs), including sunitinib, decrease proliferation of numerous cancer cells in vitro (Ikezoe et al 2006a, Ikezoe et al 2006b). Besides being a therapeutic target, VEGF is also a rational prognostic marker in many cancers (Margolin 2002). For example, VEGF expression in gastric cancer has been shown to be an independent negative prognostic marker (Ferrer et al 1998, Heist et al 2008, Jain et al 2009, Schneider and Sledge 2007, Yamaguchi et al 2007).
The PI3K-Akt cell proliferation and survival signalling pathway plays a key role in tumorigenesis of many cancers as well as in development of anti-cancer chemotherapy resistance. The Forkhead box class O (FOXO) transcription factors are crucial downstream effectors of the PI3K-Akt signalling pathway and are implicated in a wide variety of cellular functions including cell proliferation, apoptosis, differentiation and resistance to oxidative stress and DNA damage (Arden 2008, Burgering 2008, Calnan and Brunet 2008, Fu and Tindall 2008, Gomes et al 2008, Ho et al 2008, Huang and Tindall 2007, Lam et al 2006, Maiese et al 2008, Myatt and Lam 2007, Reedquist et al 2006). As such, deregulation of FOXO proteins is associated with tumorigenesis and cancer progression. In addition, emerging evidence has also demonstrated that FOXO proteins, in particular the FOXO3a, has a central role in mediating the cytostatic and cytotoxic effects of chemotherapy (Fernandez de Mattos et al 2004, Fernandez de Mattos et al 2008, Gomes et al 2008, Ho et al 2008, Hui et al 2008a, Hui et al 2008b, McGovern et al 2009, Myatt and Lam 2007, Sunters et al 2003, Sunters et al 2006). The mammalian FOXO family of transcription factors comprises of 4 members, FOXO1, FOXO3a, FOXO4 and FOXO6, and they are direct substrates of Akt (Myatt and Lam 2007). FOXO proteins interact with a core consensus DNA sequence GTAAA(C/T)A to modulate target gene expression. Phosphorylation of FOXOs by Akt results in their nuclear exclusion and inactivation.
Lapatinib (GW572016) is a small molecule dual tyrosine kinase inhibitor (TKI) for HER2 and EGFR that acts through competitive inhibition of ATP-binding to the receptor tyrosine kinase domain (Ciardiello 2005, Nelson and Dolder 2006, Wakeling 2002). Lapatinib has been shown to cause growth delay and cell death in breast cancer cell lines and human tumour xenografts expressing high levels of EGFR and/or HER2. Recent phase II/III clinical studies also demonstrated that lapatinib was well tolerated and provided anti-tumour activity in patients with breast as well as with other types of cancer when used as a monotherapy or in combination with other anti-cancer treatments (Ciardiello 2005, Montemurro et al 2007). Most recent studies showed lapatinib displays antiangiogenic effect in a lung cancer model (Diaz et al 2010) and that combination treatment of lapatinib with paclitaxel, but not lapatinib alone, effectively inhibits angiogenesis in head and neck squamous cell carcinoma (HNSCC) cells (Kondo et al 2010). However, whilst enhanced HER2/EGFR expression may have been shown to function primarily through two pathways the ERK1/2 MAP kinase and PI3K-Akt signalling cascades (Montemurro et al 2007, Yarden and Sliwkowski 2001, Zhang et al 2007), a complete understanding of the mechanism by which HER2/EGFR promotes tumorigenesis remains lacking. Latest work demonstrates that FOXO3a plays an essential role in mediating the cytostatic and cytotoxic function of lapatinib as well as the EGFR specific TKI gefitinib (Hegde et al 2007, Krol et al 2007, McGovern et al 2009).
A recent cDNA microarray study revealed that FOXO3a can potentially repress VEGF expression in a colon carcinoma cell line (Delpuech et al 2007). In the present study, we validated this notion in breast cancer patient samples and then went on to investigate the molecular mechanism by which FOXO represses VEGF expression.
Results
Inverse correlation between FOXO3a and VEGF expression in breast cancer
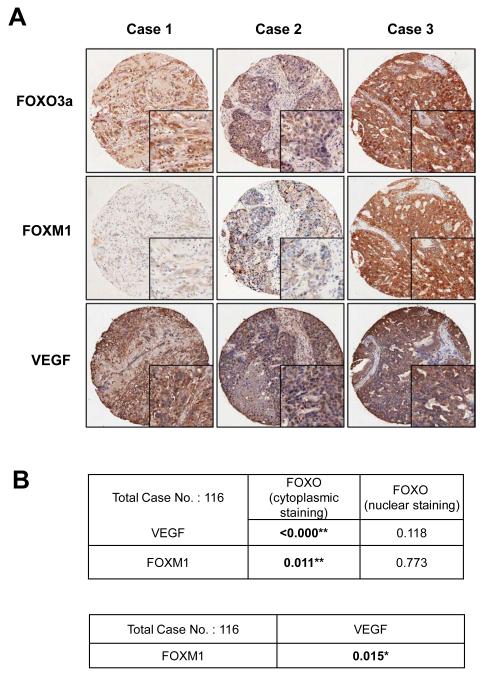
The expression patterns of FOXO3a, FOXM1 and VEGF were examined in a panel of breast cancer samples by immunohistochemistry. Representative patterns of staining are shown in Figure 1A. FOXO3a immunoreactivity was predominantly cytoplasmic in most tumour samples and correlated positively with VEGF (P = <0.001, Chi-square test) and FOXM1 (P = 0.011, Chi-square test) staining irrespective of histological type, suggesting that the activated nuclear FOXO3a inhibits FOXM1 and VEGF expression in vivo in most breast cancer samples (Fig. 1B and S1-5). Notably, there was also an inverse association between nuclear FOXO3a and VEGF expression but it was not statistically significant. Moreover, FOXM1 expression also significantly correlated with the expression of VEGF (P=0.015, Chi-square test), suggesting FOXM1 promotes VEGF expression in breast cancer cells (Fig. 1B and S6).
Figure 1. Representative expression patterns of FOXO3a, FOXM1 and VEGF in tissue microarray.
One hundred and thirty-three cases of breast cancer diagnosed between the years 1992 to 2001 with clinical follow up data available were retrieved from the records of the Department of Pathology, Queen Mary Hospital of Hong Kong. The patients’ ages at diagnosis ranged from 30 to 90 years old, with a mean of 53 years. Histological sections of all cases were reviewed by the pathologist, the representative paraffin tumour blocks chosen as donor block for each case and the selected areas marked for construction of tissue microarray blocks. Tissue sections were deparaffinised, rehydrated and stained with a previously described primary polyclonal FOXO3a specific antibody (Nordigarden et al 2009, Rosivatz et al 2006) (diluted at 1:1400), the VEGF antibody (dilution 1:250) and FOXM1 (C-20, dilution 1:450, Santa Cruz). A total of 116 could be assessed and scored for FOXO3a, FOXM1 and VEGF expression using a scanscope (Scanscope Aperio Technologies, Inc, Vista, Calif) connected to a personal computer as described in figure S1. The expression pattern and subcellular localization were correlated with histological type, histological grade, clinical stage, estrogen and progestrogen receptor status, HER2 oncoprotein overexpression, lymph node metastasis and survival time (Fig. S2 and S3). Tumour tissue samples obtained from breast cancer patients that had been formalin-fixed and paraffin-embedded were immunohistochemically stained with FOXO3a, FOXM1 and VEGF antibodies using the streptavidin-biotin-peroxidase technique. Scoring was performed as described in Fig. S1. A) Three representative tumour cases showing corresponding FOXO3a FOXM1 and VEGF staining patterns (magnification: x 100; insets x400). The three cases 1, 2 and 3 represent low, medium and high FOXO3a cytoplasmic staining. Cases 1 and 2 also show nuclear FOXO3a staining and low FOXM1 and VEGF staining, while case 3 shows predominantly strong cytoplasmic FOXO3a staining, and strong FOXO3a and VEGF staining. B) Correlation analysis of FOXO3a, FOXM1 and VEGF staining in 116 breast carcinoma cases. The correlation between predominant nuclear/cytoplasmic FOXO3a expression with FOXM1 and VEGF expression and FOXM1 with VEGF expression was studied using Chi-Square Tests and was considered significant * at p≤0.05 and very significant ** at p≤0.01.
FOXO3a activation correlates with down-regulation of FOXM1 and VEGF expression
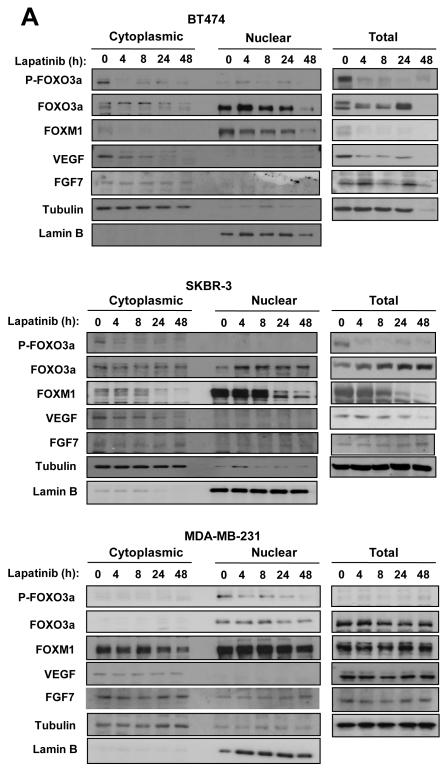
FOXM1 has recently been suggested to regulate VEGF expression (Zhang et al 2008) and to be regulated by FOXO3a (Francis et al 2009). To determine if FOXO3a and FOXM1 also modulates VEGF transcription, we first monitored the expression of VEGF, FOXM1, and FOXO3a upon lapatinib treatment of responsive and resistant breast cancer cell lines. Western blot analysis showed that lapatinib treatment of sensitive BT474 and SKBR3 cells caused a decline in phosphorylation but an increase in nuclear FOXO3a levels, indicating activation of this transcription factor (Fig. 2A). FOXO3a activation upon lapatinib treatment was accompanied by a decrease in VEGF and FOXM1 levels. The result also showed that another growth factor FGF7 was not down-regulated by lapatinib, suggesting that the repression of VEGF expression by lapatinib and FOXO3a is specific. Notably, all factors were down-regulated in BT474 cells after 48 h, probably reflecting global protein degradation and cell death. In contrast, there were no appreciable changes in P-FOXO3a, nuclear FOXO3a, FOXM1, or VEGF levels upon treatment of lapatinib-resistant MDA-MB-231 breast cancer cells.
Figure 2. Expression of FOXO3a, FOXM1 and VEGF in response to lapatinib treatment in breast cancer cell lines.
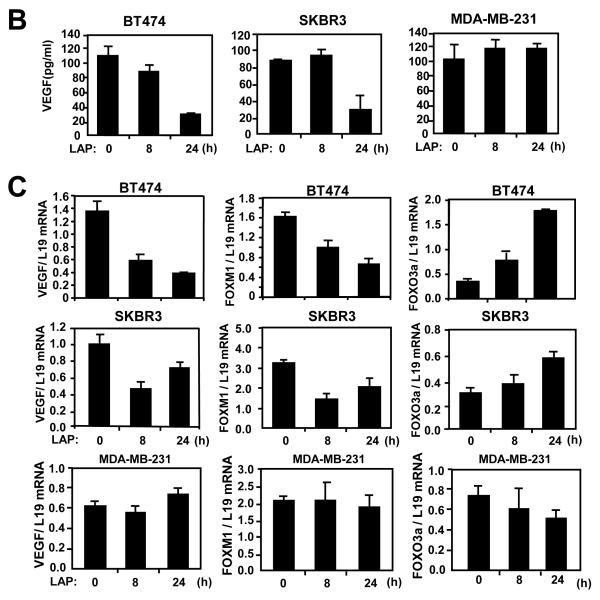
The lapatinib sensitive BT474 and SKBR-3 and resistant MDA-MB-231 cells were cultured in 10% FCS medium for 24 h before treatment with lapatinib. A) At times indicated, cells were collected and analysed for P-FOXO3a, total FOXO3a, FOXM1, VEGF, FGF7, LaminB and tubulin expression by western blotting of nuclear/cytoplamic (left panel) and total (right panel) lysates. B) VEGF concentrations in supernatants of the lapatinib-treated breast cancer cells were measured by a quantitative sandwich enzyme immunoassay according to the manufacturer’s protocol (Quantikine ELISA, R&D Systems, Abingdon, UK). The optical density was measured at 450nm using a Sunrise-Tecan plate reader (TECAN Ltd, Reading, UK) and VEGF concentrations normalised using standard curves. C) In parallel, VEGF mRNA levels of these lapatinib-treated breast cancer cells were also analysed by qRT-PCR and normalized to L19 RNA expression. Total RNA (2 μg) isolated using the RNeasy Mini kit (Qiagen, Crawley, UK) was reverse transcribed using the Superscript III reverse transcriptase and random primers (Invitrogen, Paisley, UK), and the resulting first strand cDNA was used as template in the real-time PCR. All experiments were performed in triplicate. The following gene-specific primer pairs were designed using the ABI Primer Express software: FOXM1-sense: 5′-TGCAGCTAGGGATGTGAATCTTC-3′ and FOXM1-antisense: 5′-GGAGCCCAGTCCATCAGAACT-3′; ERα-sense: 5′-CAGATGGTCAGTGCCTTGTTGG-3′ and ERα-antisense: 5′-CCAAGAGCAAGTTAGGAGCAAACAG-3′; L19-sense 5′-GCGGAAGGGTACAGCCAAT-3′ and L19-antisense 5′-GCAGCCGGCGCAAA-3′. Specificity of each primer was determined using NCBI BLAST module. Real time PCR was performed with ABI PRISM 7700 Sequence Detection System using SYBR Green Mastermix (Applied Biosystems, Brackley, UK). The RT-qPCR results shown are representative of 3 independent experiments.
FOXM1 mRNA levels of these cells were also analysed by RT-qPCR, and normalized with L19 RNA expression.
To confirm that lapatinib represses VEGF expression, secreted levels of VEGF were determined by ELISA in the three cell lines (Fig. 2B). Whereas secreted VEGF levels remained unchanged upon lapatinib treatment of MDA-MB-231 cells, the levels declined markedly after 24 h treatment of the sensitive BT474 and SKBR3 cells. As a control, we also measured the secreted levels of FGF7 by ELISA (Fig. S7). The results showed that the concentrations of the irrelevant control growth factor FGF7 did not alter significantly after lapatinib treatment in BT474, SKBR3 and MDA-MB-231 cells, suggesting that the repression of VEGF by FOXO3a and lapatinib is specific. We then tested if lapatinib regulated VEGF, FOXM1 or FOXO3a expression at the transcriptional level. RT-qPCR analysis confirmed that lapatinib inhibited VEGF and FOXM1 mRNA expression in the sensitive SKBR3 but not the resistant MDA-MB-231 cells (Fig. 2C). Notably, FOXO3a transcript levels were also up-regulated in SKBR3 cells. Together these results demonstrate that lapatinib treatment of sensitive breast cancer cells induces and activates FOXO3a but inhibits FOXM1 and VEGF expression.
FOXO3a represses VEGF and FOXM1 expression
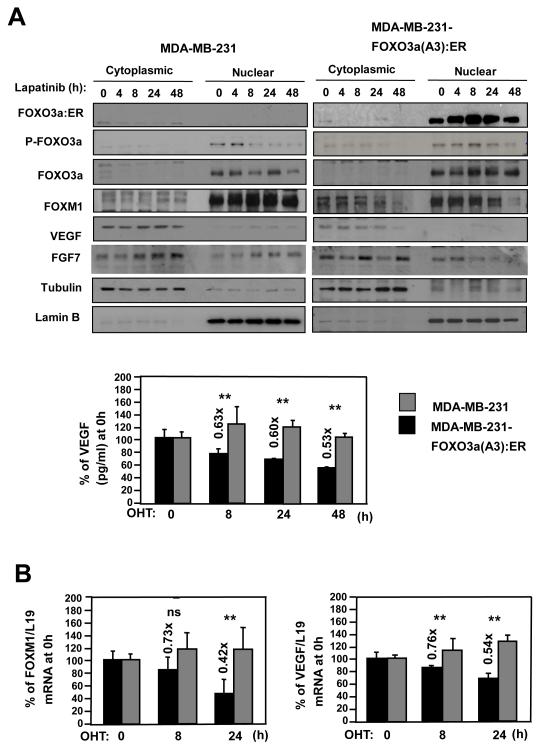
To study the mechanism underlying the reciprocal relationship between FOXO3a activation and VEGF and FOXM1 inhibition, we used an estrogen receptor α (ER)-negative MDA-MB-231 cell line expressing a fusion protein containing a constitutively active FOXO3a(A3) and ligand-binding domain of ER. In MDA-MB-231-FOXO3a(A3):ER cells, FOXO3a can be conditionally activated by 4-hydroxytamoxifen (4-OHT). As shown in Figure 3A, 4-OHT not only induced nuclear accumulation of activated FOXO3a but also inhibited expression of both VEGF and FOXM1. This down-regulation of VEGF and FOXM1 upon 4-OHT treatment was dependent upon FOXO3a activation, as no response was observed upon treatment of control MDA-MB-231 cells. As anticipated, induction of FOXO3a activity also decreased secreted VEGF levels, apparent at 8 h of 4-OHT stimulation, whereas this response was absent in control MDA-MB-231 cells (Fig. 3A). Consistently, breast cancer cells migrated at slower rates in scratch wound healing assays when cultured in supernatants derived from FOXO3a-induced MDA-MB-231 cells (Fig. S8). Further, 4-OHT also down-regulated VEGF and FOXM1 mRNA levels in MDA-MB-231-ER:FOXO3a(A3) cells, relative to control cells (Fig. 3B), inferring that FOXM1 and VEGF expression is negatively regulated by FOXO3a at a transcriptional level.
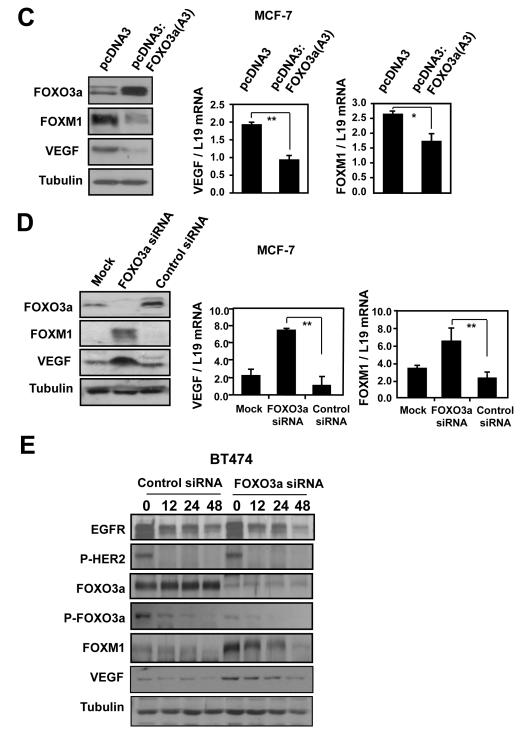
Figure 3. FOXO3a represses the expression of FOXM1 and VEGF in the breast carcinoma cells MDA-MB-231 and MCF-7.
A) MDA-MB-231-FOXO3a(A3):ER and MDA-MB-231 cells were treated with 200 nmol/L 4-OHT for the indicated times. Nuclear and cytoplasmic extracts were prepared at the times indicated, separated on polyacrylamide gels, and subjected to immunoblotting with specific antibodies. The expression levels of FOXO3a(A3):ER, FOXO3a, P-FOXO3a, FOXM1, VEGF, FGF7, tubulin and Lamin B1 were analyzed by Western blotting and the VEGF concentrations in supernatants of the 4-OHT-treated MDA-MB-231 cells cells were measured by the quantitative sandwich enzyme immunoassay as described in Fig. 2B. B) Total RNA was extracted from these cells and analyzed for FOXM1 and VEGF mRNA expression using RT-qPCR as described in the text and normalized to the level of L19 RNA. C) MCF-7 cells transiently transfected with the constitutively active FOXO3a(A3) or control vector were analysed for VEGF and FOXM1 expression by western blot and RT-qPCR analysis. D) MCF-7 cells transiently transfected with FOXO3a or control siRNA, or mock transfected were analysed by western blot using specific antibodies FOXO3a, FOXM1, VEGF and tubulin as indicated and by RT-qPCR for VEGF and FOXM1 mRNA expression. All data shown represent the averages of data from three experiments, and the error bars show the standard deviations. E) BT474 cells transiently transfected with control or FOXO3a siRNA, were treated with lapatinib for 0, 16, 24 and 48 h. Protein lysates were prepared at the times indicated and analyzed by western blot using specific antibodies P-HER2, HER-2, P-FOXO3a, FOXO3a, FOXM1, VEGF and tubulin as indicated.
To corroborate these observations, MCF-7 breast carcinoma cells were transiently transfected with the constitutively active FOXO3a(A3) or control empty expression vectors, and VEGF and FOXM1 expression monitored. Western blot and RT-qPCR analyses demonstrated that the FOXO3a(A3) mutant inhibited FOXM1 and VEGF expression, at protein and mRNA levels, respectively (Fig. 3C). Conversely, transiently transfection of MCF-7 cells with a FOXO3a targeting siRNA pool or non-targeting control siRNA increased VEGF and FOXM1 expression (Fig. 3D). To demonstrate further that FOXO3a has a role in the down-regulation of FOXM1 and VEGF by lapatinib treatment, we transfected the BT474 breast carcinoma cells with either a FOXO3a-specific or a nonspecific control siRNA pool and studied the expression of VEGF and FOXM1 after lapatinib treatment (Fig. 3E). Western blot analysis showed that the FOXO3a-specific siRNA, but not control siRNA, effectively knocked down the expression of endogenous FOXO3a in the BT474 cells. As observed previously, Lapatinib treatment led to a decrease in P-HER2 in both control and FOXO3a siRNA cells. However, silencing of FOXO3a elevated the basal expression levels of FOXM1 and VEGF, and alleviated the down-regulation of FOXM1 and VEGF by lapatinib. Notably, the expression levels of FOXM1 and VEGF did eventually decline at 48 h after lapatinib, which could be due to the functional compensation by other FOXO isoforms or the fact that FOXM1 and/or VEGF are also repressed by lapatinib through other transcription factors or at the post-transcriptional level. Together these data further confirmed that FOXO3a negatively regulates VEGF and FOXM1 expression, through a mechanism likely to involve transcriptional inhibition.
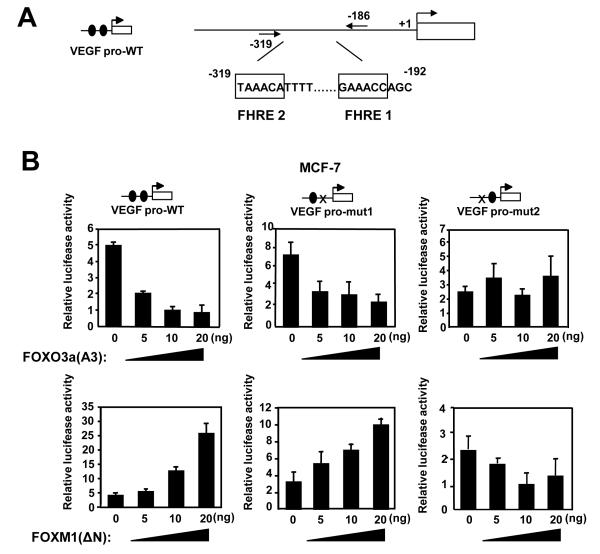
FOXO3a and FOXM1 modulate VEGF promoter activity
We postulated that FOXO3a could suppress VEGF transcription, either by modulating promoter activity or, indirectly, by inhibiting FOXM1 expression. To differentiate between these scenarios, a 1741 bp region of the putative VEGF promoter, representing positions −1,926 to −186 relative to the predominant 5′-transcription start site, was cloned upstream of a luciferase reporter (Fig. 4A). Co-transfection studies showed that expression of the FOXO3a(A3) mutant represses the activity from the putative VEGF promoter whereas exogenous expression of FOXM1 transactivated the reporter construct in a dose-dependent manner (Fig. 4B). Sequence analysis identified 2 consensus forkhead transcription response elements (FHREs) in the proximal promoter region. Mutation of the distal (−319) but not the proximal (−178) FHRE abrogated the ability of FOXO3a(A3) and FOXM1 to inhibit and activate, respectively, this promoter-reporter construct. Thus, a single response element, designated FHRE2, appears to mediate the effects of both transcription factors on the VEGF promoter.
Figure 4. FOXO3a represses and FOXM1 induces the transcriptional activity of the human VEGF gene through a FHRE consensus site proximal to the transcription start site.
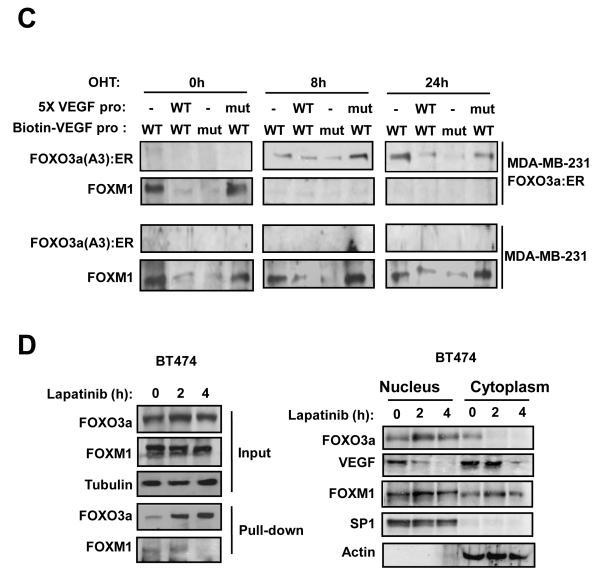
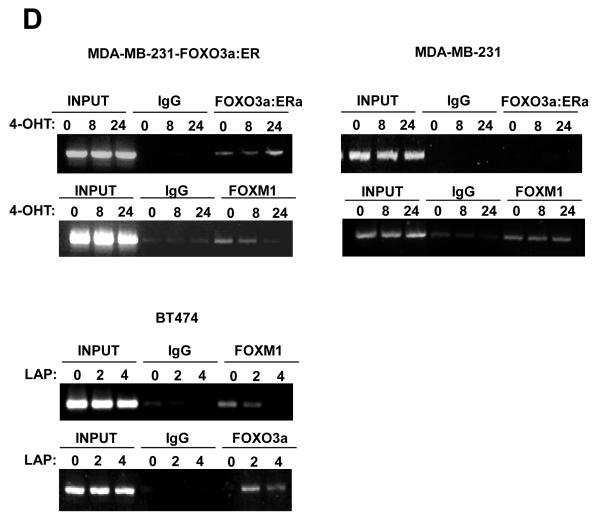
A) Effect of expression of FOXO3a and FOXM1 on VEGF promoter activity. Schematic representation of the VEGF-luciferase reporter construct, showing the consensus FHRE sequences. A 1741 bp VEGF promoter construct (positions −1,926 to −186 relative to the predominant 5′-transcription start site) was cloned into the XhoI and HindIII sites of the pGL3 basic vector (Promega, Southampton, United Kingdom). Putative forkhead site mutagenesis was performed using a Stratagene QuikChange site-directed mutagenesis kit with the oligonucleotides: Site1 (−178) (5′-ATCCCTCTTCTTTTTTCTTGGGCATTTTTTTTTAAAACTGTATTGT-3′), and Site2 (−319) (5′- TTGCTCTACTTCCCCGGGTCACTGTGGATTTTGGGGGCCAGCAGA-3′). B) MCF-7 cells were transiently transfected with 20 ng of either the wild-type, (VEGF pro-WT), mutant FHRE1 (VEGF pro-mut1), or mutant FHRE2 (VEGF pro-mut2) VEGF promoter/reporter and 0, 5, 10 or 20 ng of either the constitutively active FOXO3a(A3) or FOXM1(ΔN) expression vector. Cells were harvested 24 h after transfection and assayed for luciferase activity. All relative luciferase activity values are corrected for cotransfected Renilla activity. All data shown represent the averages of data from three independent experiments, and the error bars show the standard deviations. C) MDA-MB-231-FOXO3a(A3):ER and MDA-MB-231 cells were treated with 200 nmol/L 4-OHT for the indicated times. Nuclear extracts prepared were incubated with biotinylated wild-type or mutant FHRE2 oligonucleotides in the presence or absence of 5x molar excess of non-biotinylated wild-type or mutant FHRE2 oligonucleotides. Proteins binding to the biotinylated oligonucleotides were pulled-down using streptavidine agarose beads and analysed by western blot using specific antibodies as indicated. D) The nuclear and cytoplasmic extracts prepared from BT474 cells treated with lapatinib for 0, 2 and 4 h were western blotted for proteins indicated (right panel). The nuclear extracts from the lapatinib-treated cells were also examined by pull-down assays using biotinylated wild-type or mutant FHRE2 oligonucleotides as described above. E) Chromatin immunoprecipitation (ChIP) analysis of the human VEGF promoter. MDA-MB-231-FOXO3a(A3):ER, MDA-MB-231 and BT474 cells described above were used for ChIP assays using IgG, anti-FOXO3a and anti-FOXM1 antibodies as indicated. After crosslink reversal, the co-immunoprecipitated DNA was amplified by PCR using primers amplifying the VEGF FHRE2 containing region (−351/−186) and resolved in 2% agarose gel. Representative data from three independent experiments are shown.
FOXO3a and FOXM1 compete for binding to FHRE2
To provide more insight into the mechanism by which FOXO3a and FOXM1 regulates the VEGF promoter, we performed oligonucleotide pull-down assay with nuclear lysates from unstimulated MDA-MB-231-FOXO3a(A3):ER and MDA-MB-231 cells or cells treated with 4-OHT for 8 and 24 h. Western blot analysis of the pulled-down complexes showed that both FOXO3a and FOXM1 bind to the wild-type FHRE2 of VEGF, but not the mutated FHRE2 site (Fig. 4C). The binding of FOXO3a and FOXM1 to the FHRE2 could be competed off by excess amounts of the wild-type but not mutated FHRE2 oligonucleotides, indicating that both transcription factors bind directly to this response element (Fig. 4C). The results also revealed that FOXM1 is constitutively bound to FHRE2 in untreated MDA-MB-231-FOXO3a(A3):ER and the MDA-MB-231 cells. However, FOXM1 was replaced by the FOXO3a(A3):ER in response to 4-OHT stimulation of MDA-MB-231-FOXO3a(A3):ER but not of MDA-MB-231 cells, suggesting that activated FOXO3a down-regulates VEGF expression by competitive displacing FOXM1 bound to FHRE2.
The FHRE pull-down experiment was then repeated in the BT474 cells following lapatinib treatment in the presence of molar excess of mutated FHRE oligonucleotides (Fig. 4C). Parallel Western blot analysis of nuclear and cytoplasmic lysates showed that lapatinib induces nuclear accumulation of FOXO3a after 2 to 4 hours, concomitant with the down-regulation of VEGF expression but without discernible change in FOXM1 levels at these time-points (Fig. 4D). The pull-down results, however, indicated that the lapatinib-activated FOXO3a displaces FOXM1 from the FHRE2 of the VEGF promoter at these time-points. Thus, although prolonged activation of FOXO3a will down-regulate FOXM1 levels, inhibition of VEGF expression is an early event and mediated, at least in part, by displacing FOXM1 and binding to FHRE2. Consistent with this, we have also obtained data from FHRE pull-down and chromatin immunoprecipitation (ChIP) assays, suggesting that FOXO3a can displace FOXM1 binding to the FHRE2 of the VEGF promoter (Fig. S9). Conversely, FOXM1 was unable to compete FOXO3a off the VEGF promoter. The finding that FOXO3a can displace FOXM1 from the VEGF FHRE2 and not vice versa is further supported by a recent structural study of the FOXM1 DNA-recognition domain demonstrating that FOXM1 has a lower DNA-binding affinity to the consensus ‘TAAACA’ recognition sequence compared with other forkhead proteins (Littler et al 2010).
FOXO3a is recruited to the proximal region of the VEGF promoter in vivo
We next performed chromatin ChIP assays to determine the in vivo occupancy of the VEGF promoter in the BT474 cells in response to lapatinib treatment. The anti-FOXO3a antibody, but not the control antibody (IgG), precipitated the proximal region, encompassing FHRE2, of the VEGF promoter in BT474 cells (Fig 4E). The amount of precipitated DNA increased significantly following 2 h of lapatinib treatment, reflecting enhanced occupancy of FOXO3a to this region of the VEGF promoter in vivo, consistent with the DNA pull-down results. In contrast, the binding of FOXM1 decreased at 2 h following lapatinib treatment. Notably, the binding of both the FOXO3a and FOXM1 to the VEGF promoter decreased substantially by 4 h, probably suggesting decreased accessibility to the proximal region of the VEGF promoter. This observation pointed to the possibility that FOXO3a play a role in recruiting chromatin remodelling enzymes, such as histone deacetylases (HDACs), to repress the VEGF transcription.
FOXO3a recruits HDAC2 to the VEGF promoter
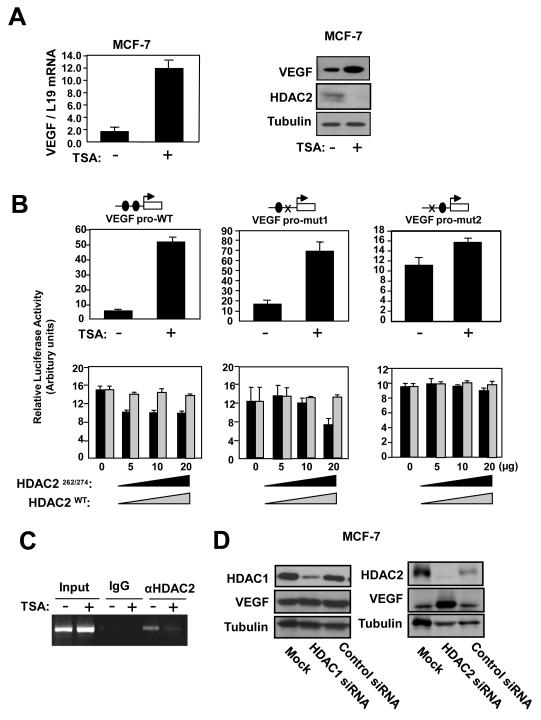
To test the hypothesis that FOXO3a recruits HDACs to repress VEGF transcription, we first treated MCF-7 cells with the HDAC inhibitor TSA and monitored VEGF expression. RT-qPCR and Western blot analyses demonstrated that TSA strongly enhances VEGF mRNA and protein levels (Fig. 5A). TSA also triggered a marked induction in VEGF promoter activity, which was abolished upon mutation of the FHRE2, but not FHRE1, site. Conversely, overexpression of the dominantly active HDAC2C262A/C274A mutant, but not the wild-type HDAC2, repressed VEGF promoter activity in a dose-dependent manner (Fig. 5B). This ability of HDAC2 to repress VEGF promoter activity was again dependent on a functional FHRE2. The inability of wild-type HDAC2 to repress VEGF promoter activity could be due to the high levels of endogenous HDAC2 in MCF-7 cells. ChIP assays further demonstrated that TSA induced a decrease in HDAC2 binding to the proximal VEGF promoter (Fig. 5C). Finally, HDAC2 knockdown using siRNA significantly up-regulated VEGF expression whereas silencing of HDAC1 silencing had little or no effect on VEGF expression. Combined, the data provide compelling evidence that HDAC2 mediates transcriptional inhibition of the VEGF promoter in breast cancer cells.
Figure 5. Effects of HDAC2 overexpression and depletion on the expression of VEGF in MCF-7 cells.
A) MCF-7 cells treated with vehicle or 100 nM trichostatin A (TSA; Sigma, UK) for 24 h were harvested for RT-qPCR and western blot analysis for VEGF expression. B) MCF-7 cells transiently transfected with 20 ng of either the wild-type, (VEGF pro-WT), mutant FHRE1 (VEGF pro-mut1), or mutant FHRE2 (VEGF pro-mut2) VEGF promoter/reporter were either untreated or treated with 100 nM TSA, or co-tranfected with 0, 5, 10 or 20 ng of either the wild-type or constitutively active HDAC2 expression vector. The transfected cells were then harvested for luciferase assays after 24 h. C) MCF-7 cells untreated or treated with TSA for 24 h were analysed for HDAC2 binding on the VEGF promoter by ChIP assays as described. D) MCF-7 cells were transiently transfected with control and smart pool siRNA against either HDAC1 or HDAC2 and analysed by western blotting for protein expression as indicated.
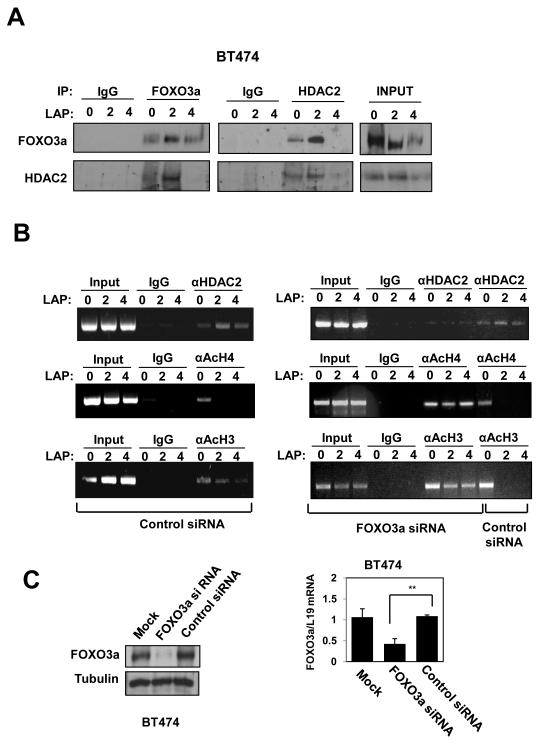
To examine if FOXO3a recruits HDAC2 to the VEGF promoter, we performed immunoprecipitation and ChIP experiments on BT474 cells treated with lapatinib. HDAC2 and FOXO3a co-immunoprecipitated and this interaction was enhanced upon lapatinib treatment, probably reflecting nuclear translocation of FOXO3a (Fig. 6A). ChIP assays showed increased recruitment of HDAC2 to the proximal VEGF promoter after 2 h of lapatinib treatment (Fig. 6B). Histone H3 and H4 acetylation are epigenetic marks associated with activated promoters (Bernstein et al 2005, Davie and Candido 1978). HDAC2 recruitment coincided with a decrease in bound acetylated histones H3 and H4, indicating active chromatin remodelling and compaction of the proximal VEGF promoter. Further, siRNA-mediated FOXO3a knockdown (Fig. 6C and 6D) in BT474 cells abolished the recruitment of HDAC2 to the proximal VEGF promoter upon lapatinib treatment as well as the concomitant decrease in acetylated histones H3 and H4. These findings demonstrate that FOXO3a activation in breast cancer cells results in displacement of DNA-bound FOXM1, binding to FHRE2, recruitment of HDAC2, and transcriptional repression of VEGF.
Figure 6. OXO3a recruits HDAC2 to the VEGF promoter in response to lapatinib in BT474 cells.
A) Cell extracts prepared from BT474 cells 0, 2 and 4 h after treatment with lapatinib were immunoprecipitated (IP) with antibodies against FOXO3a and HDAC2 or an IgG control antibody. The precipitated complexes and the inputs were examined for FOXO3a and HDAC2 expression. B) BT474 cells were transiently transfected with smart pool siRNA against FOXO3a or control siRNA pool. Twenty-four h afterwards, the transfected BT474 cells were treated with lapatinib for 0, 2, or 4 h and then analysed for HDAC2, acetylated histone H3 and H4 binding on the VEGF promoter by ChIP assays as described. C) Western blot and RT-qPCR analyses were performed as described to demonstrate effective and specific FOXO3a knock-down.
Discussion
Signals mediated through VEGFs and their receptors have been shown to be essential for breast cancer carcinogenesis, cell migration (metastasis) and angiogenesis (Lohela et al 2009). Yet, the molecular mechanisms regulating VEGF expression in cancer cells are only partially understood. A previous cDNA microarray study using a colon carcinoma cell line DLD-1 has suggested that FOXO3a can potentially repress VEGF expression (Delpuech et al 2007). Our present analysis of breast cancer patient samples revealed that FOXO3a nuclear localisation is significantly but inversely associated with VEGF expression, suggesting FOXO3a negatively regulates VEGF expression in vivo in breast cancer.
Using the lapatinib sensitive breast cancer cell lines BT474 and SKBR3 as models for FOXO3a activation, the hypothesis that FOXO3a regulates VEGF expression was examined and the underlying mechanisms involved explored in the present study. lapatinib treatment resulted in inactivation of the phosphatidylinositol-3-kinase (PI-3K) pathway, nuclear translocation and activation of FOXO3a (Hegde et al 2007) and ultimately reduction in VEGF expression at protein, mRNA and gene promoter levels. Transient transfection and inducible FOXO3a expression experiments showed that FOXO3a represses while FOXM1 activates VEGF expression through a proximal FHRE site of the VEGF promoter, as mutation of this FHRE abrogated the regulation by FOXO3a and FOXM1. ChIP and oligonucleotide pull-down assays further demonstrated that both FOXO3a and FOXM1 bind directly to the FHRE of the VEGF promoter and that activated FOXO3a can displace FOXM1 from the FHRE, suggesting that FOXO3a can repress VEGF expression through competing off the transcriptional activator FOXM1. Consistently, FOXO3a accumulated and replaced FOXM1 at the FHRE as early as 2 h after lapatinib treatment; however, it was also noted that neither FOXO3a nor FOXM1 bound to the FHRE by 4 h. The lack of occupancy of the proximal VEGF promoter region by FOXO3a and FOXM1 at 4 h suggested that FOXO3a accumulation might lead to exclusion of transcription factors through chromatin remodelling. Histone acetylation decondenses the chromatin, making nucleosomal DNA more accessible to transcription factors, whereas inhibition of histone deacetylase activity by HDACs leads to condensation of the chromatin and exclusion of transcription factors. Consistently, we found that upon activation, FOXO3a recruits HDAC2 to the proximal region of the VEGF promoter, as revealed by ChIP analysis. Current evidence also proposes that transcriptionally active DNA is located in nucleosomes with acetylated histones H3 and H4. Our ChIP assays showed that in response to lapatinib treatment in BT474 cells, there was an increase in FOXO3a and HDAC2 binding, concomitant with a decrease in acetylated histones H3 and H4 levels. We performed co-immunoprecipitation experiments to examine the amount of HDAC2 binding to FOXO3a in response to lapatinib in BT474 cells. The results showed that the amount of HDAC2 interacting with FOXO3a increased substantially at 2 h but declined by 4 h following lapatinib treatment. The increase in FOXO3a binding to HDAC2 in response to lapatinib is likely to be due to the relocation of FOXO3a to the nucleus, while the declined in FOXO3a binding to HDAC2 was probably a result of the disassociation in binding between the two proteins as well as a decline in HDAC2 levels, as revealed by immunoprecipitaion and western blot analyses, respectively.
Overexpression of FOXM1 has been implicated with metastasis and angiogenesis in a number of malignancies, including glioma, gastric and pancreatic cancer. Consistent with our findings, a recent study has also demonstrated that FOXM1 transcriptionally regulates VEGF expression in glioma cells (Zhang et al 2008). It is notable that the FOXM1 responsive sites identified previously locate over 500 bp 5′-upstream of the FOXO/FOXM1 binding site defined in this study and neither of these sites appears to be a consensus FOXO-binding element. Importantly, deletion of site 2 in the present VEGF promoter abolishes responsiveness to FOXO3a, FOXM1 and HDAC, suggesting this FHRE is targeted by FOXO3a and FOXM1. In the present study we further demonstrated that FOXM1 functions downstream of FOXO3a, and its activity and expression are negatively regulated by FOXO3a. Nevertheless, FOXM1 is not the sole effector of FOXO3a function. FOXO3a can also negatively regulate gene expression through FOXM1 independent mechanisms, such as by means of HDAC recruitment. The ability of FOXO proteins to repress VEGF expression has been documented in Foxo1 null cells where VEGF is overexpressed and angiogenesis deregulated (Furuyama et al 2004, Park et al 2009). This notion is now supported further by our finding that expression of nuclear FOXO3a expression significantly inversely correlates with VEGF expression in breast cancer patient samples. Consistently, constitutively active FOXO mutants have been shown to inhibit HUVEC cell migration, and capillary tube formation (Davis et al 2009, Lee et al 2008).
In summary, together the present results suggest that FOXO3a can potentially repress VEGF expression, through at least two mechanisms. First, activated FOXO3a can compete off the transcription activator FOXM1 from binding to the FHRE of the VEGF gene promoter. Second, FOXO3a can recruit HDACs to the VEGF promoter to induce chromatin condensation and transcription factor exclusion. Furthermore, FOXO3a has also been shown previously to be able to repress FOXM1 expression at transcriptional levels (Francis et al 2009). Consequently, FOXO3a can repress VEGF expression indirectly via regulating FOXM1 expression. The mechanisms by which FOXO3a represses VEGF expression may represent common means whereby FOXO3a negatively regulates target gene expression. Thus, the present study also provides novel understanding on the mechanisms by which FOXO transcription factors repress target gene expression. Furthermore, the findings from this study also suggest that therapeutic strategies targeting FOXO3a or FOXM1 can be used as an alternative or in parallel with anti-VEGF targeted agents as well as conventional chemotherapy in rational and effective treatment of tumours (Fernandez de Mattos et al 2008, Gomes et al 2008, Srivastava et al 2010).
Materials and Methods
Cell Culture
The human breast carcinoma cell lines BT474, SKBR3, MCF-7, and MDA-MB-231 originated from the American Type Culture Collection and were acquired from the Cell Culture Service, Cancer Research UK (London, UK), where they were tested and authenticated. Cell lines used in the present study were in culture for less than 6 months. All cells used were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 2 mM glutamine, and 100 units/ml penicillin/streptomycin at 37 °C. Lapatinib was obtained from GlaxoSmithKline, dissolved in DMSO and used at a final concentration of 0.1 μM.
Plasmids
For the generation of human VEGF promoter constructs, a 1741 bp VEGF promoter construct was generated using PCR primers 5′-ATCTCGAGGAGGCTATGCCAGCTGTAGG-3′ and 5′-GCAAGCTTTCTGCTGGTTTCCAAAATCC-3′ from genomic DNA and cloned into the pGL3 basic vector (Promega, Southampton, United Kingdom). Putative forkhead site mutagenesis was performed using a Stratagene QuikChange site-directed mutagenesis kit. The HDAC2WT and HDAC2C262A/C274A expression plasmids were kind gifts from Dr. Antonella Riccio (University College London, UK) (Nott et al 2008) and the FOXO3a(A3) or FOXM1(ΔN) expression vectors have previously been described (Essafi et al 2005, Krol et al 2007, Kwok et al 2008).
Luciferase reporter assay, Antibodies, Real-time quantitative PCR (RT-qPCR) and Gene Silencing with Small Interfering RNAs (siRNAs)
Western Blotting
Western blotting was performed on whole cell extracts as described previously (Krol et al 2007).
Preparation of nuclear and cytoplasmic extracts
Nuclear and cytoplasmic extracts were prepared as previously described (Essafi et al 2009).
Pull-down assays using biotin-labelled oligonucleotides
Pull-down assays were performed as described previously (Labied et al 2006) using the 100nmol of biotin-labelled double stranded oligonucleotides: Wild-type (5′-GTTTTATCCCTCTTCTTTTTTCTTAAACATTTTTTAAA-3′) Mutant: (5′-GTTTTATCCCTGTTCTTTTTTCTTGGGCATTTTTTAAA-3′). The pulled down complexes were then analysed by Western blotting.
Tissue Microarray and Immnohistochemistry
See Fig. 1 legend
Chromatin Immunoprecipitation (ChIP) assay
ChIP assay was performed as described (Essafi et al 2005) using FOXO3a(A3):ER MDA-MB-231 cells treated with or without 200nM 4-OHT for 24 hours before harvesting. DNA fragments were purified using the QIAquick Spin Kit (Qiagen, UK). For PCR, one-twenty-fifth of the extracted DNA was used and amplified in 25 PCR cycles using specific primers. PCRs were then performed on the purified DNA using the following primers: (−351)-5′-TCCGGGTTTTATCCCTCTTC-3′; 5′-TCTGCTGGTTTCCAAAATCC-3′ (−186).
Statistical analysis
To test for the relationship between VEGF, FOXM1 and nuclear/cytoplasmic FOXO3a, statistical analysis was performed using Pearson’s correlation test and was considered significant at p≤0.05 and very significant at ≤0.01 All statistical analysis was performed with SPSS v.16 (SPSS inc, Chicago, IL, USA).
Supplementary Material
Acknowledgements
The authors declare that there is no conflict of interest arising from the submission of this manuscript. Grant support: Cancer Research UK (M. Petkovic, K.K. Ho and E.W-F. Lam), Breast Cancer Campaign (E.W-F. Lam), Portuguese Science and Technology Foundation (FCT) (A. R. Gomes).
References
- Arden KC. FOXO animal models reveal a variety of diverse roles for FOXO transcription factors. Oncogene. 2008;27:2345–2350. doi: 10.1038/onc.2008.27. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Burgering BM. A brief introduction to FOXOlogy. Oncogene. 2008;27:2258–2262. doi: 10.1038/onc.2008.29. [DOI] [PubMed] [Google Scholar]
- Calnan DR, Brunet A. The FoxO code. Oncogene. 2008;27:2276–2288. doi: 10.1038/onc.2008.21. [DOI] [PubMed] [Google Scholar]
- Ciardiello F. Epidermal growth factor receptor inhibitors in cancer treatment. Future Oncol. 2005;1:221–234. doi: 10.1517/14796694.1.2.221. [DOI] [PubMed] [Google Scholar]
- Davie JR, Candido EP. Acetylated histone H4 is preferentially associated with template-active chromatin. Proc Natl Acad Sci U S A. 1978;75:3574–3577. doi: 10.1073/pnas.75.8.3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R, Singh KP, Kurzrock R, Shankar S. Sulforaphane inhibits angiogenesis through activation of FOXO transcription factors. Oncol Rep. 2009;22:1473–1478. doi: 10.3892/or_00000589. [DOI] [PubMed] [Google Scholar]
- Delpuech O, Griffiths B, East P, Essafi A, Lam EW, Burgering B, et al. Induction of Mxi1-SR alpha by FOXO3a contributes to repression of Myc-dependent gene expression. Mol Cell Biol. 2007;27:4917–4930. doi: 10.1128/MCB.01789-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz R, Nguewa PA, Parrondo R, Perez-Stable C, Manrique I, Redrado M, et al. Antitumor and antiangiogenic effect of the dual EGFR and HER-2 tyrosine kinase inhibitor lapatinib in a lung cancer model. BMC Cancer. 2010;10:188. doi: 10.1186/1471-2407-10-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essafi A, Fernandez de Mattos S, Hassen YA, Soeiro I, Mufti GJ, Thomas NS, et al. Direct transcriptional regulation of Bim by FoxO3a mediates STI571-induced apoptosis in Bcr-Abl-expressing cells. Oncogene. 2005;24:2317–2329. doi: 10.1038/sj.onc.1208421. [DOI] [PubMed] [Google Scholar]
- Essafi A, Gomes AR, Pomeranz KM, Zwolinska AK, Varshochi R, McGovern UB, et al. Studying the subcellular localization and DNA-binding activity of FoxO transcription factors, downstream effectors of PI3K/Akt. Methods Mol Biol. 2009;462:201–211. doi: 10.1007/978-1-60327-115-8_13. [DOI] [PubMed] [Google Scholar]
- Fernandez de Mattos S, Essafi A, Soeiro I, Pietersen AM, Birkenkamp KU, Edwards CS, et al. FoxO3a and BCR-ABL regulate cyclin D2 transcription through a STAT5/BCL6-dependent mechanism. Mol Cell Biol. 2004;24:10058–10071. doi: 10.1128/MCB.24.22.10058-10071.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez de Mattos S, Villalonga P, Clardy J, Lam EW. FOXO3a mediates the cytotoxic effects of cisplatin in colon cancer cells. Mol Cancer Ther. 2008;7:3237–3246. doi: 10.1158/1535-7163.MCT-08-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer FA, Miller LJ, Andrawis RI, Kurtzman SH, Albertsen PC, Laudone VP, et al. Angiogenesis and prostate cancer: in vivo and in vitro expression of angiogenesis factors by prostate cancer cells. Urology. 1998;51:161–167. doi: 10.1016/s0090-4295(97)00491-3. [DOI] [PubMed] [Google Scholar]
- Francis RE, Myatt SS, Krol J, Hartman J, Peck B, McGovern UB, et al. FoxM1 is a downstream target and marker of HER2 overexpression in breast cancer. Int J Oncol. 2009;35:57–68. doi: 10.3892/ijo_00000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Tindall DJ. FOXOs, cancer and regulation of apoptosis. Oncogene. 2008;27:2312–2319. doi: 10.1038/onc.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama T, Kitayama K, Shimoda Y, Ogawa M, Sone K, Yoshida-Araki K, et al. Abnormal angiogenesis in Foxo1 (Fkhr)-deficient mice. J Biol Chem. 2004;279:34741–34749. doi: 10.1074/jbc.M314214200. [DOI] [PubMed] [Google Scholar]
- Gomes AR, Brosens JJ, Lam EW. Resist or die: FOXO transcription factors determine the cellular response to chemotherapy. Cell Cycle. 2008;7:3133–3136. doi: 10.4161/cc.7.20.6920. [DOI] [PubMed] [Google Scholar]
- Hegde PS, Rusnak D, Bertiaux M, Alligood K, Strum J, Gagnon R, et al. Delineation of molecular mechanisms of sensitivity to lapatinib in breast cancer cell lines using global gene expression profiles. Mol Cancer Ther. 2007;6:1629–1640. doi: 10.1158/1535-7163.MCT-05-0399. [DOI] [PubMed] [Google Scholar]
- Heist RS, Zhai R, Liu G, Zhou W, Lin X, Su L, et al. VEGF polymorphisms and survival in early-stage non-small-cell lung cancer. J Clin Oncol. 2008;26:856–862. doi: 10.1200/JCO.2007.13.5947. [DOI] [PubMed] [Google Scholar]
- Ho KK, Myatt SS, Lam EW. Many forks in the path: cycling with FoxO. Oncogene. 2008;27:2300–2311. doi: 10.1038/onc.2008.23. [DOI] [PubMed] [Google Scholar]
- Huang H, Tindall DJ. Dynamic FoxO transcription factors. J Cell Sci. 2007;120:2479–2487. doi: 10.1242/jcs.001222. [DOI] [PubMed] [Google Scholar]
- Hui RC, Francis RE, Guest SK, Costa JR, Gomes AR, Myatt SS, et al. Doxorubicin activates FOXO3a to induce the expression of multidrug resistance gene ABCB1 (MDR1) in K562 leukemic cells. Mol Cancer Ther. 2008a;7:670–678. doi: 10.1158/1535-7163.MCT-07-0397. [DOI] [PubMed] [Google Scholar]
- Hui RC, Gomes AR, Constantinidou D, Costa JR, Karadedou CT, Fernandez de Mattos S, et al. The forkhead transcription factor FOXO3a increases phosphoinositide-3 kinase/Akt activity in drug-resistant leukemic cells through induction of PIK3CA expression. Mol Cell Biol. 2008b;28:5886–5898. doi: 10.1128/MCB.01265-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikezoe T, Nishioka C, Tasaka T, Yang Y, Komatsu N, Togitani K, et al. The antitumor effects of sunitinib (formerly SU11248) against a variety of human hematologic malignancies: enhancement of growth inhibition via inhibition of mammalian target of rapamycin signaling. Mol Cancer Ther. 2006a;5:2522–2530. doi: 10.1158/1535-7163.MCT-06-0071. [DOI] [PubMed] [Google Scholar]
- Ikezoe T, Yang Y, Nishioka C, Bandobashi K, Nakatani H, Taguchi T, et al. Effect of SU11248 on gastrointestinal stromal tumor-T1 cells: enhancement of growth inhibition via inhibition of 3-kinase/Akt/mammalian target of rapamycin signaling. Cancer Sci. 2006b;97:945–951. doi: 10.1111/j.1349-7006.2006.00263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain L, Vargo CA, Danesi R, Sissung TM, Price DK, Venzon D, et al. The role of vascular endothelial growth factor SNPs as predictive and prognostic markers for major solid tumors. Mol Cancer Ther. 2009;8:2496–2508. doi: 10.1158/1535-7163.MCT-09-0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitadai Y. Angiogenesis and lymphangiogenesis of gastric cancer. J Oncol. 2010:468725. doi: 10.1155/2010/468725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo N, Tsukuda M, Ishiguro Y, Kimura M, Fujita K, Sakakibara A, et al. Antitumor effects of lapatinib (GW572016), a dual inhibitor of EGFR and HER-2, in combination with cisplatin or paclitaxel on head and neck squamous cell carcinoma. Oncol Rep. 2010;23:957–963. doi: 10.3892/or_00000720. [DOI] [PubMed] [Google Scholar]
- Krol J, Francis RE, Albergaria A, Sunters A, Polychronis A, Coombes RC, et al. The transcription factor FOXO3a is a crucial cellular target of gefitinib (Iressa) in breast cancer cells. Mol Cancer Ther. 2007;6:3169–3179. doi: 10.1158/1535-7163.MCT-07-0507. [DOI] [PubMed] [Google Scholar]
- Kwok JM, Myatt SS, Marson CM, Coombes RC, Constantinidou D, Lam EW. Thiostrepton selectively targets breast cancer cells through inhibition of FOXM1 expression. Mol Cancer Ther. 2008;7 doi: 10.1158/1535-7163.MCT-08-0188. In Press. [DOI] [PubMed] [Google Scholar]
- Labied S, Kajihara T, Madureira PA, Fusi L, Jones MC, Higham JM, et al. Progestins regulate the expression and activity of the forkhead transcription factor FOXO1 in differentiating human endometrium. Mol Endocrinol. 2006;20:35–44. doi: 10.1210/me.2005-0275. [DOI] [PubMed] [Google Scholar]
- Lam EW, Francis RE, Petkovic M. FOXO transcription factors: key regulators of cell fate. Biochem Soc Trans. 2006;34:722–726. doi: 10.1042/BST0340722. [DOI] [PubMed] [Google Scholar]
- Lee HY, Youn SW, Kim JY, Park KW, Hwang CI, Park WY, et al. FOXO3a turns the tumor necrosis factor receptor signaling towards apoptosis through reciprocal regulation of c-Jun N-terminal kinase and NF-kappaB. Arterioscler Thromb Vasc Biol. 2008;28:112–120. doi: 10.1161/ATVBAHA.107.153304. [DOI] [PubMed] [Google Scholar]
- Lentzsch S, Chatterjee M, Gries M, Bommert K, Gollasch H, Dorken B, et al. PI3-K/AKT/FKHR and MAPK signaling cascades are redundantly stimulated by a variety of cytokines and contribute independently to proliferation and survival of multiple myeloma cells. Leukemia. 2004;18:1883–1890. doi: 10.1038/sj.leu.2403486. [DOI] [PubMed] [Google Scholar]
- Littler DR, Alvarez-Fernandez M, Stein A, Hibbert RG, Heidebrecht T, Aloy P, et al. Structure of the FoxM1 DNA-recognition domain bound to a promoter sequence. Nucleic Acids Res. 2010;38:4527–4538. doi: 10.1093/nar/gkq194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohela M, Bry M, Tammela T, Alitalo K. VEGFs and receptors involved in angiogenesis versus lymphangiogenesis. Curr Opin Cell Biol. 2009;21:154–165. doi: 10.1016/j.ceb.2008.12.012. [DOI] [PubMed] [Google Scholar]
- Maiese K, Chong ZZ, Shang YC. OutFOXOing disease and disability: the therapeutic potential of targeting FoxO proteins. Trends Mol Med. 2008;14:219–227. doi: 10.1016/j.molmed.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makrilia N, Lappa T, Xyla V, Nikolaidis I, Syrigos K. The role of angiogenesis in solid tumours: an overview. Eur J Intern Med. 2009;20:663–671. doi: 10.1016/j.ejim.2009.07.009. [DOI] [PubMed] [Google Scholar]
- Margolin K. Inhibition of vascular endothelial growth factor in the treatment of solid tumors. Curr Oncol Rep. 2002;4:20–28. doi: 10.1007/s11912-002-0044-9. [DOI] [PubMed] [Google Scholar]
- McGovern UB, Francis RE, Peck B, Guest SK, Wang J, Myatt SS, et al. Gefitinib (Iressa) represses FOXM1 expression via FOXO3a in breast cancer. Mol Cancer Ther. 2009;8:582–591. doi: 10.1158/1535-7163.MCT-08-0805. [DOI] [PubMed] [Google Scholar]
- Montemurro F, Valabrega G, Aglietta M. Lapatinib: a dual inhibitor of EGFR and HER2 tyrosine kinase activity. Expert Opin Biol Ther. 2007;7:257–268. doi: 10.1517/14712598.7.2.257. [DOI] [PubMed] [Google Scholar]
- Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7:847–859. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- Nelson MH, Dolder CR. Lapatinib: a novel dual tyrosine kinase inhibitor with activity in solid tumors. Ann Pharmacother. 2006;40:261–269. doi: 10.1345/aph.1G387. [DOI] [PubMed] [Google Scholar]
- Nott A, Watson PM, Robinson JD, Crepaldi L, Riccio A. S-Nitrosylation of histone deacetylase 2 induces chromatin remodelling in neurons. Nature. 2008;455:411–415. doi: 10.1038/nature07238. [DOI] [PubMed] [Google Scholar]
- Park SH, Sakamoto H, Tsuji-Tamura K, Furuyama T, Ogawa M. Foxo1 is essential for in vitro vascular formation from embryonic stem cells. Biochem Biophys Res Commun. 2009;390:861–866. doi: 10.1016/j.bbrc.2009.10.063. [DOI] [PubMed] [Google Scholar]
- Pytel D, Sliwinski T, Poplawski T, Ferriola D, Majsterek I. Tyrosine kinase blockers: new hope for successful cancer therapy. Anticancer Agents Med Chem. 2009;9:66–76. doi: 10.2174/187152009787047752. [DOI] [PubMed] [Google Scholar]
- Reedquist KA, Ludikhuize J, Tak PP. Phosphoinositide 3-kinase signalling and FoxO transcription factors in rheumatoid arthritis. Biochem Soc Trans. 2006;34:727–730. doi: 10.1042/BST0340727. [DOI] [PubMed] [Google Scholar]
- Schneider BP, Sledge GW., Jr. Drug insight: VEGF as a therapeutic target for breast cancer. Nat Clin Pract Oncol. 2007;4:181–189. doi: 10.1038/ncponc0740. [DOI] [PubMed] [Google Scholar]
- Srivastava RK, Unterman TG, Shankar S. FOXO transcription factors and VEGF neutralizing antibody enhance antiangiogenic effects of resveratrol. Mol Cell Biochem. 2010;337:201–212. doi: 10.1007/s11010-009-0300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunters A, Fernandez de Mattos S, Stahl M, Brosens JJ, Zoumpoulidou G, Saunders CA, et al. FoxO3a transcriptional regulation of Bim controls apoptosis in paclitaxel-treated breast cancer cell lines. J Biol Chem. 2003;278:49795–49805. doi: 10.1074/jbc.M309523200. [DOI] [PubMed] [Google Scholar]
- Sunters A, Madureira PA, Pomeranz KM, Aubert M, Brosens JJ, Cook SJ, et al. Paclitaxel-induced nuclear translocation of FOXO3a in breast cancer cells is mediated by c-Jun NH2-terminal kinase and Akt. Cancer Res. 2006;66:212–220. doi: 10.1158/0008-5472.CAN-05-1997. [DOI] [PubMed] [Google Scholar]
- Wakeling AE. Epidermal growth factor receptor tyrosine kinase inhibitors. Curr Opin Pharmacol. 2002;2:382–387. doi: 10.1016/s1471-4892(02)00183-2. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Bando H, Mori T, Takahashi K, Matsumoto H, Yasutome M, et al. Overexpression of soluble vascular endothelial growth factor receptor 1 in colorectal cancer: Association with progression and prognosis. Cancer Sci. 2007;98:405–410. doi: 10.1111/j.1349-7006.2007.00402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- Zhang H, Berezov A, Wang Q, Zhang G, Drebin J, Murali R, et al. ErbB receptors: from oncogenes to targeted cancer therapies. J Clin Invest. 2007;117:2051–2058. doi: 10.1172/JCI32278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang N, Dai B, Liu M, Sawaya R, Xie K, et al. FoxM1B transcriptionally regulates vascular endothelial growth factor expression and promotes the angiogenesis and growth of glioma cells. Cancer Res. 2008;68:8733–8742. doi: 10.1158/0008-5472.CAN-08-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.