Abstract
The unintended and often promiscous inhibition of the cardiac human Ether-à-go-go related gene (hERG) potassium channel is a common cause for either delay or removal of therapeutic compounds from development and withdrawal of marketed drugs. The clinical manifestion is prolongation of the duration between QRS complex and T-wave measured by surface electrocardiogram (ECG)—hence Long QT Syndrome. There are several useful online resources documenting hERG inhibition by known drugs and bioactives. However, their utilities remain somewhat limited because they are biased toward well-studied compounds and their number of data points tends to be much smaller than many commercial compound libraries. The hERGCentral (www.hergcentral.org) is mainly based on experimental data obtained from a primary screen by electrophysiology against more than 300,000 structurally diverse compounds. The system is aimed to display and combine three resources: primary electrophysiological data, literature, as well as online reports and chemical library collections. Currently, hERGCentral has annotated datasets of more than 300,000 compounds including structures and chemophysiological properties of compounds, raw traces, and biophysical properties. The system enables a variety of query formats, including searches for hERG effects according to either chemical structure or properties, and alternatively according to the specific biophysical properties of current changes caused by a compound. Therefore, the hERGCentral, as a unique and evolving resource, will facilitate investigation of chemically induced hERG inhibition and therefore drug development.
Introduction
Cardiac toxicity of developing compounds or marketed drugs is often caused by small molecule-induced abnormalities in cardiophysiology of different ionic currents, noticeably associated with reduction of repolarizing potassium currents, commonly known as IKr and IKs. Nowadays, it is a routine practice to examine developing compounds, regardless of their intended targets, for their effects on human Ether-à-go-go related gene (hERG) potassium channel, the most common target of promiscous inhibition by small molecules (e.g., see reviews by Sanguinetti et al.1,2). Various studies have compared different assays to assess compound effects. It is generally agreed that data collected by electrophysiological recording, relative to other biochemical or fluorescence assays, are more reliable predictors for QT prolongation. Several online resources are constructed to document hERG effects by small molecules, primarily known drugs or bioactives (see Table 1). Detailed structure-and-function studies have led to better understanding of compound–hERG interactions. However, it is well appreciated that bioactives and drugs, as a whole, are selective and a relatively small sample size. Therefore, there is no evidence that these well-studied bioactive compounds could serve as a general representation of drug-like compound libraries that often consist of 50,000 to a few million diverse structures.
Table 1.
Comparison of Human Ether-à-go-go Related Gene Online Resources
| |
hERG assay data |
|
|
Cross database links |
Structure search |
||||
|---|---|---|---|---|---|---|---|---|---|
| hERG resources | Compounds | Properties | Source | Other resources or main features | Activity exploration features | Link-out | Link-in | Structure inputs | Drawer based on |
| hERGCentrala | 318,941 | Activities, raw traces | Assay results | hERG literature, compound profiling | Histogram, dynamic range selection by dragging, ranking | PubChem records | Search by PubChem IDs | MOL2, load external structure by PubChem IDs | Pure Javascript |
| ChEMBL (Target 165)b | 5,650 | Activities | Assay results | Target data, compound profiling | Data table, histogram, ranking | EMBL-EBI database, etc. | IUPHAR, ChEMBL, etc. | SMILES, Keywords | N/A |
| PubChem (AID 376)c | 1,953 | Activities | Assay results | Compound profiling, SAR | Data table, histogram, ranking | Entrez cross- database, etc. | Entrez cross- database, etc. | MOL2, SMILES | Pure Javascript |
| QSAR Datasetsd | 572 | Activities | Literature | Downloadable data files | Data table | Literature sources | N/A | SMILES | Java applet |
| Tox-Portal.nete | 263 | IC50 | Literature | Toxicity prediction | Data table | N/A | N/A | N/A | |
| hERGAPDbasef | 165 | Activities | Literature | hERG literature | Data table | PubMed (PMID) | N/A | MOL2,SMILES, chemical name | N/A |
| QTDrugsg | 123 | Drug annotation | Advisory board | QT drugs list | Data table, ranking | PubMed | N/A | N/A | N/A |
| Robert R. Fenichelh | 106 | IC50, IKr | Literature | Literature | Data table, ranking | Literature sources | N/A | N/A | N/A |
| CDT (KCNH2)i | 100 | Interactions | Literature | Toxicogenomics database | N/A | PharmGKB, UniProt, etc. | IUPHAR, ChEMBL, etc. | N/A | N/A |
| SHCCj | N/A | N/A | Literature | hERG literature, mutations table, etc. | N/A | N/A | N/A | N/A | N/A |
| Ionchannels.orgk | N/A | N/A | Literature | hERG literature, citations | N/A | Literature sources | N/A | N/A | N/A |
| OMIM (152427)l | N/A | N/A | Literature | Genome profiling | N/A | Ensembl, UniProt, etc. | IUPHAR, ChEMBL, etc | N/A | N/A |
| PharmGKB Gene (PA212)m | N/A | N/A | Literature | Genome profiling | N/A | Ensembl, UniProt, etc. | UniProt, HGNC, etc. | N/A | N/A |
| Protein Ontology (PRO:000000791)n | N/A | N/A | Literature | Protein ontology report | N/A | UniProt, HGNC, etc. | IUPHAR, ChEMBL, etc. | N/A | N/A |
| IUPHAR Receptor (Kv 11.1)o | N/A | N/A | Literature | Ion channels characterization | N/A | ChEMBL, Ensembl, etc. | UniProt, HGNC, etc. | N/A | N/A |
| UniProtKB (Q12809)p | N/A | N/A | Literature | Protein knowledge base | N/A | PharmGKB, Ensembl, etc. | IUPHAR, ChEMBL, etc. | N/A | N/A |
www.qsarworld.com/qsar-datasets.php; nine datasets, compound number based on distinct compound names (compound identity in different datasets may vary; repeat count is likely).
www.grt.kyushu-u.ac.jp/hergapdbase/; compound number based on distinct compound names.
www.azcert.org/medical-pros/drug-lists/list-03.cfm?sort=Generic_name; compound number based on distinct drug names.
www.fenichel.net/pages/Professional/subpages/QT/Tables/pbyclass.htm; compound number based on distinct drug names.
hERG, human Ether-à-go-go related gene; ID, identity; N/A, not applicable; SAR, structure-activity relationship.
hERGCentral (www.hergcentral.org) is an online resource center for researchers who work on hERG potassium channels and/or develop therapeutic compounds without cardiac side effects. The key component is the combination of annotated primary experimental data of more than 300,000 diverse compounds and bioactives. The resource has incoporated effective search functions for both general inhibition profile and detailed biophysical changes for general pharmacologists and ion channel investigators. Therefore, hERGCentral is unique and will facilitate research and drug discovery.
hERGCentral Overview
Despite its important role in cardiotoxicity profiling, there are a relatively small number of online hERG resources (Table 1). Fenichel's database collected hERG assay bioactivity data from scientific literature. Arizona Center for Research and Education on Therapeutics (Arizona CERT) and the University of Lausanne provide complete and up-to-date lists of drugs that prolong the QT-interval. Recently, hERGAPDbase has advanced this effort by collecting and displaying two types of electrophysiological assay data from the scientific literature. In addition to allowing users to browse data according to compound names, it provides a Web-based interface for structure-based similarity search. A summary of these databases is outlined in Table 1.
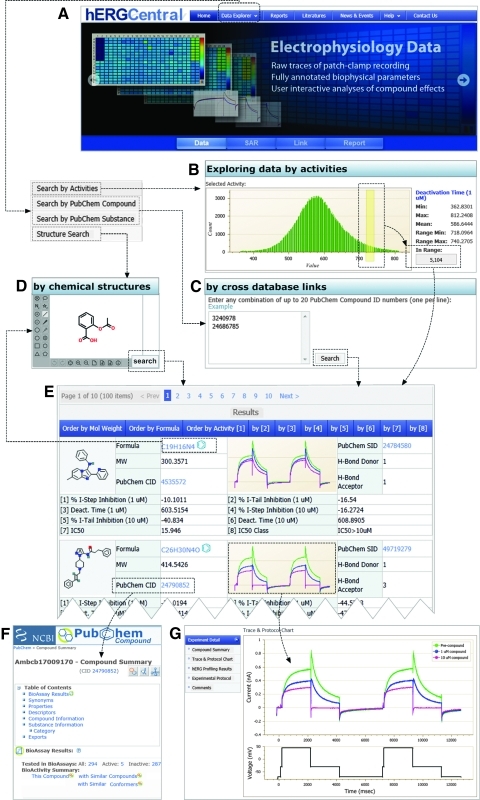
Unlike the resources mentioned above, hERGCentral is based on experimental data obtained from a primary screen by electrophysiology against more than 300,000 diverse structural compounds. Both structures/chemophysioproperties of tested compounds and their specific effects on biophysical properties of hERG channels have been annotated. Such a large database should be helpful to the scientific community to better understand compound–hERG channel interactions. To explore properly, effective methods are necessary for users to visualize the data. In the current version of deployment, users may explore the data in a number of ways as outlined in Figure 1.
Fig. 1.
Overall outline of hERGCentral. (A) Home page of hERGCentral, on the top dropdown menu for three ways of data exploration can be accessed. (B) Exploring data by activities: large scale of hERG data can be explored interactively by various activities such as tail current inhibition. (C) Exploring data by cross database links: external database IDs (here, PubChem compounds and substances) can be used to browse data on hERGCentral. (D) Exploring data by chemical structures: molecular structures can be easily entered through the Web interface, which allows users to explore results with similar structures. (E) In either way of exploration, the results are presented in a tabular view, where various properties, activities, as well as thumbnails of raw current trace data are listed. By clicking any property or activity on the heading of this table, users can sort the results accordingly. (F) Live links to external public databases, for example, PubChem compound summary, are maintained. (G) Detailed results such as electrophysiological recording traces at a higher resolution, as well as voltage protocols can be accessed by clicking the current trace thumbnails. CID, compound ID; hERG, human Ether-à-go-go related gene; SID, substance ID; ID, identity.
Exploring Data by Activity
Representing a large dataset via a histogram gives an overview of the data distribution where outliers, sometimes the most interesting compounds, could easily stand out. Such a format also significantly reduces the number of data points, thereby reducing the traffic between the Web server and end users. As such, hERGCentral provides a general method of data exploration by activity. Activities of interests, for example, deactivation time and tail current inhibition, are measured from electrophysiological recording of compound libraries. Each of these parameters is presented in a histogram, as shown in Figure 2. The histograms are designed to be interactive, where users can easily select a desirable range of interest. Moreover, range indices for activities, that is, mapping from activity range to corresponding data entries, are constructed to facilitate efficient data exploration. Once a range is selected, the number of compounds whose activities fall into the range will be immediately displayed next to the histogram. Based on the number of compounds, users can revise the desired range or request information on individual compounds by a simple click and the data will be listed in a tabular view, as shown in Figure 1E. The activity search provides the user with an intuitive way to identify hERG blockers either by a single parameter or by applying multiple different parameters. These parameters include biophysical activity (e.g., tail current amplitude) and structural features of a chemical (e.g., molecular weight and number of hydrogen bond donors). An example is included in the Appendix.
Fig. 2.
(Top) The process of generating activity histograms and range indices from raw recording traces: raw current trace data obtained from electrophysiological recording of a large compound library were processed, and activity measurements of interest, for example, tail current inhibition, were calculated. Discretization and index construction are applied to generate histograms and range indices for efficient data exploration. (Bottom) Web interface for exploring data by activities: end users can first select an activity of interest (Step 1); the corresponding activity histogram presents an interactive distribution of the activity, which allows users to select a range of interest via mouse (Step 2); the Web interface then immediately displays the number of compounds whose activities fall into the selected range. Users can list these results in a tabular view by clicking the indicated button (Step 3).
Exploring Data by Cross Database Links
Although hERGCentral is a very specialized bioassay database platform, it is also designed to interact with other public resources to improve its utility and user experience. Ideally, the interactions should be implemented in both ways: link-out and link-in. With link-outs of hERG data to other public databases such as PubChem and Entrez, it makes it easier for users to investigate the effects of a given compound in other assays. The second is link-in, that is, mapping other database records (usually IDs) to those on hERGCentral. Technically, the link-ins are much harder to implement than the link-outs. This is mainly because the number of records in widely used databases is very large and continuously increasing. For example, PubChem, a dynamically growing primary database, as of January 2011 consists of 31 million compound records and 75 million substance records. hERGCentral has implemented both link-in and link-out with PubChem. For the link-outs, live links to PubChem compound and substance summary are maintained, as shown in Figure 1F. For the link-ins, hERGCentral adopts an efficient method to map external public database IDs to hERG assay results on hERGCentral. Particularly, more than 300,000 tested compounds on hERGCentral are associated to a relatively small ID set, denoted by H={1,2, …,hmax}, so that core information can fit into random access memory for fast accessing. The PubChem ID set, denoted by P={1,2, …, pmax}, is mapped to H via an integer array with indices representing P and values representing H. The resultant link-in is therefore of a constant time complexity, that is, O(1) instead of O(N). Such a design offers the benefit of constant search time when accessing each single element within the integer array regardless of its size.3
The overall procedure to explore data on hERGCentral by external public database IDs is shown in Figure 3.
Fig. 3.
Flow diagram to explore data on hERGCentral using external public database IDs. Here, the widely used PubChem CIDs (A) and SIDs (B) are accepted by hERGCentral Web interfaces, which allow multiple ID submission for a given query. For those IDs with matched results found on hERGCentral, results will be returned directly; for those with no matched results found, hERGCentral still retrieves compound profile, including molecular formula, etc., from PubChem. If users click the “ ” button next to the formula, hERGCentral will fetch the structure information from PubChem database. With the fetched structure information, the structure search will run on hERGCentral; thus, results of similar chemical structures can still be returned to users.
” button next to the formula, hERGCentral will fetch the structure information from PubChem database. With the fetched structure information, the structure search will run on hERGCentral; thus, results of similar chemical structures can still be returned to users.
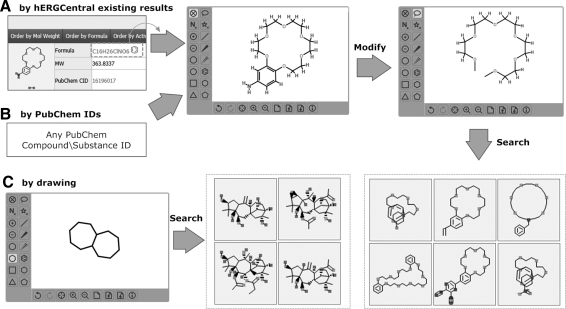
Exploring Data by Chemical Structures
Another valuable way of exploring data on hERGCentral is by chemical structure search. Although hERGCentral is meant primarily to store hERG assay data, chemical structure data are also stored on hERGCentral for those compounds that have associated hERG assay data. This affords setting up efficient query/retrieval functions for users. One of the most relevant elements of structure search is the structure input. hERGCentral Web interface integrates a Javascript-based molecular drawing tool that supports all desktop browsers (the drawing tool software is publicly available at http://metamolecular.com/chemwriter/). Chemical structures can be entered in three ways, as shown in Figure 4. An example to look up a known compound by name is included (see Appendix). One interesting way is to allow the user to modify existing structures. After the server loads molecular structure data from either hERGCentral or external database like PubChem, users can modify the structure, for example, removing/adding chemical bonds or altering the atoms, and then perform the search. The Figure 4A outlines a case where the crown ether-like structure is loaded and searched by modifying the structure of a PubChem compound (CID 16196017). Specifically, the side component (benzene) is removed. The remaining crown ether structure is then used as query to search the compound library. The returned structures are shown in Figure 4.
Fig. 4.
Flow diagram to explore data by chemical structures. A convenient structure drawer is provided via the Web interface. Chemical structures can be entered in three ways. (A) Structure information for each existing tested compound on hERGCentral is available to be loaded into the drawer by clicking the “ ” button next to the formula. (B) A PubChem compound/substance ID can be used to load the corresponding structure. Users may modify the structure in the drawing tool (top right), and perform the search. (C) For simple (sub) structures, users may enter them by drawing tools.
” button next to the formula. (B) A PubChem compound/substance ID can be used to load the corresponding structure. Users may modify the structure in the drawing tool (top right), and perform the search. (C) For simple (sub) structures, users may enter them by drawing tools.
The structure search function is developed with JChem for .NET. The default search method is based on substructure similarity. After a similarity search, the hERGCentral displays table style results listing the top 100 chemical compounds in the order of their similarity scores.
hERGCentral Implementation and Future Directions
hERGCentral was developed using a three-tier architecture. The back-end consists of a MySQL database cluster storing chemical library collections, a high-performance embeded database, Berkeley DB, for storing electrophysiological raw traces and other kinetic data, as well as a text-based search engine, Lucene, for indexing and retrieving literature and online reports. The middle tier, which handles application logic and core functionality, was developed with the C++ programming language for performance and cross-platform consideration. Specifically, the middle tier groups logic and functionalities into middleware services, including structure search service, compound profile service, and bioactivity range indexing service. The front-end Web client, which acts as a consumer of the middleware services, was implemented using ASP.NET MVC 3 architecture.
As an evolving platform, hERGCentral will continue to develop to both meet the needs and enhance the experience of end users. Major efforts are on the way to improve data and functionality enrichment. It is of benefit to allow for more data entries of hERGCentral by acquiring or annotating more experimental data that are widely scattered among literature and public databases. It is also likely that several different assays would be applied to one particular compound. These data may be available from different databases. Therefore, another improvement would be to include links to other databases or other reported screens in a tabular view so that a user can compare data from more than one source. For existing data entries, other useful properties, both chemical and biophysical, may also be added. Although hERGCentral is meant primarily to provide hERG bioassay results, pharmacological prediction indices, such as ADME (absorption, distribution, metabolism, and excretion) and toxicity profiling, are valuable to facilitate structure-activity relationship (SAR) analysis and compound profiling.
In addition to the search-by-name feature (Appendix), the functionality enrichments will include Web interface improvements. The database may also be augmented with data export options. For instance, many in silico prediction models have been developed for hERG blockers; however, the data sets utilized are usually relatively small and vary in size and data format, and therefore are not easy to compare. To address this issue, hERGCentral could provide a unified hERG data service, with which existing prediction programs can access a much larger data set and present their prediction results. It is conceivable that hERGCentral could serve as a benchmark database for hERG prediction models.
Experimental Procedure
The hERG primary screen was performed in an electrophysiological assay using the population patch clamp (PPC) mode on the Ionworks Quattro™ (MDC, Sunnyvale, CA), an automated patch clamp instrument. The Molecular Libraries Small Molecule Repository (MLSMR) library was screened, which consisted of 318,950 compounds in 384-well plate format as single compound concentration at 5 mM in dimethyl sulfoxide (DMSO) (additional detail describing the library can be obtained from http://mlsmr.glpg.com). The automated patch clamp assay on hERG channel has been reported earlier by Zou et al.4 Briefly, compound effects were tested using dual compound additions at 1 μM and subsequently at 10 μM. The Chinese hamster ovary (CHO) cells stably expressing hERG channels were freshly dislodged from flasks and dispensed into a 384-well PPC plate. Activity of hERG was then measured with the recording protocol as followings. Leak currents were linear subtracted extrapolating the current elicited by a 100-ms step to −80 mV from a holding potential of −70 mV. The screen was completed with two voltage protocols with slight voltage difference. First protocol: hERG currents were evoked by two identical voltage pulses with a 3-s interval, in which the voltage pulses consisted of a 100-ms step to −30 mV, a conditioning pre-pulse (2-s duration, +45 mV) followed by a test pulse (2-s duration, −30 mV) from a holding potential at −70 mV. Second protocol: The first voltage pulse consisted of a 100-ms step to −30 mV, a conditioning pre-pulse (2-s duration, +25 mV) followed by a test pulse (2-s duration, −30 mV) from a holding potential at −70 mV. After a 3-s interval at −70 mV, a second pulse protocol was applied consisting of a 100-ms step to −30 mV, a pre-pulse (2-s duration, +45 mV) followed by a test pulse (2-s duration, −30 mV). DMSO concentration in the assay buffer for all the screened compounds was kept at 0.02% (1 μM) and 0.2% (10 μM) (v/v) DMSO, which did not cause hERG activity change. The peak amplitudes of tail currents and steady-state currents from the second pulse before and after compound treatment were measured. Compound effects were assessed by the percentage changes in the hERG peak tail current and steady-state currents, which were calculated by dividing the difference between pre- and post-compound hERG currents by the respective pre-compound control currents in the same well. Only cells with a peak tail current amplitude >0.2 nA, a seal resistance >30 MOhms, and seal resistance drop rate <25% were included for data analysis.
Abbreviations
- CID
compound ID
- DMSO
dimethyl sulfoxide
- hERG
human Ether-à-go-go related gene
- ID
identity
- PPC
population patch clamp
- SID
substance ID.
Appendix
Appendix Fig. A1.
An example of identifying hERG blockers by using the activity search. Two activity parameters are chosen: % Step Current Inhibition (1 μM), the range of (−39.7, −37.0), is selected (795 out of ∼300,000 compounds fall into this range); % Tail Current Inhibition (1 μM), the range of (−54.9, −52.0), is selected (524 compounds). Application of the two conditions together identifies 16 compounds, 4 of which are displayed. CID, compound ID; hERG, human Ether-à-go-go related gene; ID, identity.
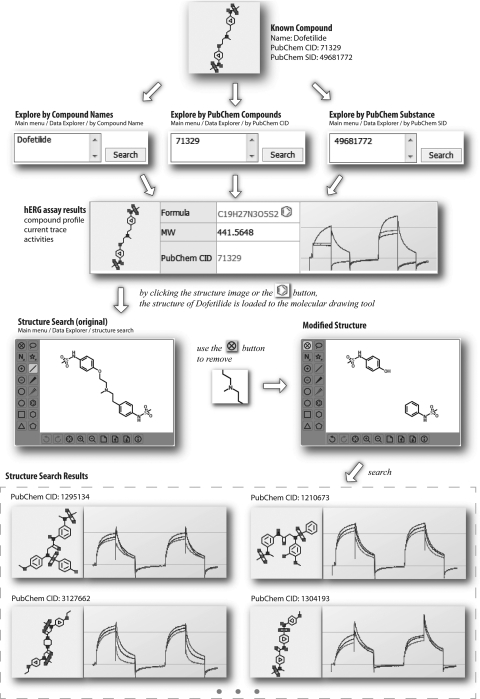
Appendix Fig. A2.
An example of demonstrating to look up a known compound (e.g., dofetilide). The hERG data of a known compound can be looked up by its name, PubChem CID, or PubChem SID. The compound structure can be loaded to the molecular drawing tool, which facilitates the structure-based search. The example indicates a search result after removal of part of dofetilide structure. The subsequent structure search returns nine compounds, four of which are displayed. SID, substance ID.
Acknowledgments
The authors would like to thank all the members in the Johns Hopkins Ion Channel Center (JHICC) for their valuable discussions and Alison Neal for editorial assistance on this article. This work was supported in part by grants from the National Institutes of Health (GM078579 and MH084691) and Maryland Stem Cell Research Foundation (2010-MSCRFE-0164-00) to M.L.
Authorship Note
Database design, implementation, and Web deployment (F.D., J.B., and S.L.); experimental data collection and annotation (H.Y. and B.Z.); project design and manuscript preparation (M.L. and F.D.).
Disclosure Statement
No competing financial interests exist.
References
- 1.Sanguinetti MC. Mitcheson JS. Predicting drug-hERG channel interactions that cause acquired long QT syndrome. Trends Pharmacol Sci. 2005;26:119–124. doi: 10.1016/j.tips.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Sanguinetti MC. Tristani-Firouzi M. hERG potassium channels and cardiac arrhythmia. Nature. 2006;440:463–469. doi: 10.1038/nature04710. [DOI] [PubMed] [Google Scholar]
- 3.Cormen TH. Leiserson CE. Rivest RL. Stein C. Introduction to Algorithms. 3rd. MIT Press; Cambridge, MA: 2009. [Google Scholar]
- 4.Zou B. Yu H. Babcock JJ, et al. Profiling diverse compounds by flux- and electrophysiology-based primary screens for inhibition of human Ether-a-go-go related gene potassium channels. Assay Drug Dev Technol. 2010;8:743–754. doi: 10.1089/adt.2010.0339. [DOI] [PMC free article] [PubMed] [Google Scholar]