Abstract
Avirulent strains of a bacterial pathogen could be useful tools for investigating immunological responses to infection and potentially effective vaccines. We have therefore constructed an auxotrophic TIGR4 Δpab strain of Streptococcus pneumoniae by deleting the pabB gene Sp_0665. The TIGR4 Δpab strain grew well in complete medium but was unable to grow in serum unless it was supplemented with para-aminobenzoic acid (PABA). The TIGR4 Δpab strain was markedly attenuated in virulence in mouse models of S. pneumoniae nasopharyngeal colonization, pneumonia, and sepsis. Supplementing mouse drinking water with PABA largely restored the virulence of TIGR4 Δpab. An additional Δpab strain constructed in the D39 capsular serotype 2 background was also avirulent in a sepsis model. Systemic inoculation of mice with TIGR4 Δpab induced antibody responses to S. pneumoniae protein antigens, including PpmA, PsaA, pneumolysin, and CbpD, but not capsular polysaccharide. Flow cytometry demonstrated that IgG in sera from TIGR4 Δpab-vaccinated mice bound to the surface of TIGR4 and D39 bacteria but not to a capsular serotype 3 strain, strain 0100993. Mice vaccinated with the TIGR4 Δpab or D39 Δpab strain by intraperitoneal inoculation were protected from developing septicemia when challenged with the homologous S. pneumoniae strain. Vaccination with the TIGR4 Δpab strain provided only weak or no protection against heterologous challenge with the D39 or 0100993 strain but did strongly protect against a TIGR4 capsular-switch strain expressing a serotype 2 capsule. The failure of cross-protection after systemic vaccination with Δpab bacteria suggests that parenteral administration of a live attenuated vaccine is not an attractive approach for preventing S. pneumoniae infection.
INTRODUCTION
Streptococcus pneumoniae is a common cause of pneumonia, meningitis, and septicemia both in adults and in children. It is estimated to cause over 800,000 deaths in children under 5 years of age worldwide, 11% of all mortality in this age group (29). Although precise data are lacking, S. pneumoniae is also an important cause of adult morbidity and mortality. Prevention of S. pneumoniae infections is therefore an important public health priority, which the recent introduction of the conjugated capsular polysaccharide antigen vaccine has partially addressed (20). This vaccine provides effective prevention against colonization and invasive infection caused by strains of S. pneumoniae expressing capsular serotypes included in the vaccine. However, at present this vaccine has generally been used in children and not adults (14), and because it protects only against selected S. pneumoniae strains, its routine use has had a profound effect on S. pneumoniae ecology, with an increasing incidence of previously uncommon strains as colonizers and as causes of invasive disease (25). The conjugated vaccine is also very expensive and is complex in design, making its introduction to the low-income countries in which the burden of S. pneumoniae infections is the highest more difficult (22, 32). Routine vaccination of adults uses a nonconjugated capsular antigen vaccine which, although it has broader coverage against 23 of the 91 capsular serotypes, has weak efficacy against pneumonia, the commonest severe manifestation of S. pneumoniae infection (12).
As a consequence of these difficulties with the existing conjugated and nonconjugated S. pneumoniae vaccines, other approaches have been suggested for an S. pneumoniae vaccine. These include vaccines using protein antigens, killed whole-cell S. pneumoniae, or attenuated live S. pneumoniae cells (22, 32). Given the ability of bacteria to acquire new genetic traits or undergo mutation which could revert attenuated bacteria to virulence, there are important questions about safety that make live attenuated vaccines against bacterial infections a relatively unattractive option. Furthermore, there are important practical difficulties about preservation and transport of a live attenuated vaccine compared to non-live attenuated vaccines. As a consequence, only a limited number of live bacterial vaccines have been used in practice, including the Mycobacterium bovis BCG vaccine for prevention of tuberculosis (43) and more recently developed vaccines for prevention of typhoid (11). However, the wide range of antigens and pathogen-associated molecular patterns (PAMPs) present in a live attenuated vaccine (26) suggests that the immune responses that they induce are likely to be very powerful and may also more closely mimic those obtained after natural infection than immunity to a subcomponent or dead bacterial vaccine. This may provide a mechanism for inducing protective immunity under circumstances where other types of vaccines may not or for creating highly protective sera that could be used for passive immunization during outbreaks of S. pneumoniae infection or to assist with the treatment of antibiotic-resistant strains. In addition, the wide range of different patient populations that are susceptible to serious S. pneumoniae infections suggests that there is a need for a range of different preventative strategies. Hence, a live vaccine could be potentially useful against S. pneumoniae, and there are published data on possible live attenuated vaccine candidates. Roche et al. used live unencapsulated attenuated S. pneumoniae strains to vaccinate mice via a colonization model and showed effective systemic and mucosal protection (32), and Richards et al. demonstrated that prior nasopharyngeal colonization with a pneumolysin-negative S. pneumoniae mutant resulted in significant serotype cross-protection against invasive pneumococcal disease (31).
To develop a safer potential vaccine, a strain containing two or more mutations profoundly affecting virulence would be beneficial, and there is a need to identify mutations that can effectively attenuate the virulence of S. pneumoniae, preferably without markedly altering the surface properties of the bacteria to preserve immune stimulation and potential antigenic targets. An auxotrophic mutation affecting in vivo growth but dispensable for growth under at least some laboratory culture conditions would fulfill these criteria, and the folate biosynthetic pathway is an attractive target to achieve this (3). Folate is synthesized by microorganisms from chorismate via the intermediate para-aminobenzoic acid (PABA), and disruption or deletion of the gene encoding the enzyme converting chorismate to PABA, PABA synthetase, in Gram-negative bacteria or the fungal pathogen Aspergillus fumigatus creates PABA auxotrophs. These PABA auxotrophs are totally attenuated in virulence due to the low levels of PABA present in mammalian systems (5), and a Salmonella enterica serovar Typhi PABA auxotroph has been used as a live attenuated vaccine (36). A signature-tagged mutagenesis screen for S. pneumoniae virulence determinants identified that a mutant with disruption of the pabB gene was attenuated for virulence in mouse models of colonization, sepsis, and pneumonia (9). These data suggest that deletion of pabB would result in a live attenuated strain of S. pneumoniae with conditional virulence that could be a useful tool for investigating disease pathogenesis and potentially contribute to a live attenuated vaccine.
We have therefore created an S. pneumoniae strain containing a deletion of the gene encoding PABA synthetase (pabB, Sp_0665 in the strain TIGR4 genome) and characterized its virulence and the potential efficacy of this mutant strain at inducing protective immune responses using murine models of S. pneumoniae infection.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The S. pneumoniae strains used in this work are listed in Table 1. The mutant strain was constructed with TIGR4 capsular serotype 4 and D39 capsular serotype 2 clinical isolates. Additional experiments used the capsular serotype 3 clinical isolate 0100993 (19) and TIGR4 strains expressing capsular serotypes 2 and 3 (kind gifts from M. Lipsitch) (37, 40). S. pneumoniae strains were cultured at 37°C in 5% CO2 on Columbia agar (Oxoid) supplemented with 5% horse blood (TCS Biosciences) or in Todd-Hewitt broth supplemented with 0.5% yeast extract (THY). The antibiotic kanamycin (500 μg ml−1) was added to blood agar plates where appropriate. Growth of strains was compared in broth culture by measuring the optical density at 580 nm (OD580) at hourly intervals. Working stocks of bacterial cultures in THY (OD580, 0.3 to 0.4) were stored at −80°C with 10% glycerol.
Table 1.
Strains and primers used in this study
| Name | Description or sequence (source or reference) |
|---|---|
| Strains | |
| TIGR4 | S. pneumoniae serotype 4 clinical isolate |
| D39 | S. pneumoniae serotype 2 clinical isolate |
| D39DΔ | Unencapsulated D39 due to deletion of cpsD (27) |
| 0100993 | S. pneumoniae serotype 3 clinical isolate |
| 0100993 Δcps | Unencapsulated 0100993 due to replacement of the cps locus with kanamycin resistance cassette (unpublished strain); Kmra |
| T4-4+4 | TIGR4 capsular-switch strain reexpressing serotype 4 (40) |
| T4-4+2 | TIGR4 capsular-switch strain expressing serotype 2 (40) |
| T4-4+3 | TIGR4 capsular-switch strain expressing serotype 3 (40) |
| TIGR4 Δpab | TIGR4 with in-frame deletion of Sp_0665; Kmr (this study) |
| D39 Δpab | D39 with in-frame deletion of Sp_0665; Kmr (this study) |
| Primers | |
| Sp0664F | GGACACCGTGAAGGTACTGATG |
| KmSp0664R | ATCTGCCTCCTCATCCTCTTTTATTTAAAGATTGAAGTTTTAA |
| KmSp0666F | CGCTAGATAGGGGTCCCGAGCATGAAATTAATATTTTTGCATGG |
| Sp0667R | GGTTCTCCCTCGTGAATATCCG |
| KmF | AAGAGGATGAGGAGGCAGAT |
| KmR | GCTCGGGACCCCTATCTA |
| Sp0665RTF | CTTGAACAGGCCAGTTGGTT |
| Sp0665RTR | ACAAGGTCCACATCCTCTCG |
| Sp0666RTF(3) | TCAGCACCTAGCTCAAGAGACA |
| Sp0666RTR(3) | CTCCTCAGAAACTCCGACCA |
| Sp0667RTF | GAGGGAGAACCTGGTTATTCTG |
| Sp0667RTR | TCAGCTTTCAGGTAATAGCTTCC |
| Sp0668RTF | TGTTGGTGGTGGTATTGTCG |
| Sp0668RTR | TGACAATCCCTGTTGCTGAA |
| Sp0669RTF(3) | TTGGAAACCAATCCTTGGAA |
| Sp0669RTR(3) | TGCAAAGCCACATACTGCAT |
Kmr, kanamycin resistance.
Construction of the Sp_0665 deletion mutant.
For the in-frame deletion of Sp_0665, a construct was created with the following 3 fragments. Two fragments consisting of 989 bp of flanking DNA 5′ to the Sp_0665 ATG (primers Sp0664F and KmSp0664R) and 818 bp of flanking DNA 3′ to the Sp_0665 open reading frame (ORF; starting from the ATG of the overlapping Sp_0666 ORF; primers KmSp0666F and Sp0667R) with 3′ and 5′ linkers complementary to the 5′ and 3′ portions of the kan cassette (aphIII gene), respectively, were amplified by PCR from TIGR4 genomic DNA. A third fragment consisting of the kan cassette was amplified from nonencapsulated S. pneumoniae TIGR4 mutant strain genomic DNA (30) (a kind gift from Tim Mitchell, University of Glasgow) with the primers KmF and KmR and fused with the flanking fragments of the Sp_0665 gene by overlap extension PCR (16). Primers used for the overlap extension PCR are shown in Table 1. The construct was transformed into S. pneumoniae by homologous recombination and allelic replacement using competence-stimulating peptide (CSP-2; a kind gift from Don Morrison, University of Illinois) and standard protocols (10).
DNA, RNA extraction, and RT-PCR.
Genomic DNA and total RNA were isolated from S. pneumoniae strains using a Wizard genomic DNA isolation kit and SV total RNA isolation system (Promega), respectively, following the manufacturer's instructions, except that cells were incubated with 0.1% deoxycholic acid (Sigma) at 37°C for 10 min before extraction. RNasin (0.5%; Promega) was added to extracted RNA to prevent it from degradation. cDNA was derived and amplified from RNA using an Access reverse transcription-PCR (RT-PCR) system (Promega) and target-specific primers. Primers used for the transcriptional analysis of the Sp_0665-0669 operon are described in Table 1. The National Center for Biotechnology Information website (http://blast.ncbi.nlm.nih.gov/Blast) was used for DNA and protein BLAST searches.
Flow cytometry assays for C3b/iC3b deposition and neutrophil phagocytosis.
To assess the effect of pabB deletion on the complement deposition on S. pneumoniae and on the interaction with phagocytes, flow cytometry assays were performed according to previously described methods (13). For C3b/iC3b deposition, 2 × 106 CFU of bacterial pellets, human serum (1:4 dilution in PBS), and fluorescein isothiocyanate-conjugated polyclonal goat anti-human C3 antibody (1:300; ICN Cappel, Aurora, OH) were used. The proportion of bacteria positive for C3b/iC3b and mean fluorescence intensity (MFI) were obtained using a FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA), collecting data from at least 20,000 bacteria. For the opsonophagocytosis assay, the proportion of freshly extracted human neutrophils associated with 5,6-carboxyfluorescein-succinimidyl ester (FAM-SE; Molecular Probes, Eugene, OR)-labeled fluorescent bacteria (1 × 106 CFU) was measured by flow cytometry after opsonization with 1/8 and 1/4 dilutions of commercially available baby rabbit serum (S7764; Sigma) and at a multiplicity of infection of 10.
ELISA.
Antibody levels in mouse immune sera were measured by standard whole-cell enzyme-linked immunosorbent assays (ELISAs) or by an anti-capsular polysaccharide/cell wall polysaccharide protocol as described previously (41). ELISA plates (Nunc Maxisorp) were coated with S. pneumoniae bacterial suspension at 50 μl/well for whole-cell ELISAs or with S. pneumoniae capsular polysaccharide antigen at 100 μl/well (10 μg/ml). For the anti-capsular polysaccharide ELISA, serum samples were preincubated in buffer containing both cell wall polysaccharide and 22F polysaccharide for 30 min at room temperature prior to addition to the ELISA plates to increase the specificity of captured and measured antibody. The plates were developed using monoclonal goat anti-mouse IgG heavy-chain alkaline phosphatase conjugate (Sigma), and antibody levels were assessed by measuring ODs at different serum dilutions.
IgG binding to live S. pneumoniae TIGR4 and immunoblots.
Flow cytometry assays of IgG deposition on the surface of S. pneumoniae were performed using a previously described protocol of Jomaa et al. (15). Bacterial pellets containing 5 × 106 CFU, pooled mouse serum (1:5 dilution in phosphate-buffered saline [PBS]), and a 1:50 dilution of phycoerythrin-conjugated goat anti-mouse IgG (Jackson ImmunoResearch) were used for the assay. Results are presented as the percentage of bacteria positive for IgG binding. For the immunoblots, whole-cell lysates (16) of S. pneumoniae were separated on SDS-polyacrylamide gels using 4 to 12% bis-Tris gels (Invitrogen, United Kingdom), blotted onto nitrocellulose membranes, and probed with pooled mouse sera (1:100 dilution) using conventional techniques.
Luminex bead assay.
Antibody responses to multiple protein antigens were measured using a multiplex flow cytometry Luminex assay based on S. pneumoniae proteins conjugated to xMAP beads (35). Recombinant TIGR4-, D39-, or serotype 23 strain-derived proteins were conjugated to xMAP beads (Luminex) as described previously (35). Combined beads (3,000 per antigen) were incubated with 10% or 1% serum in PBS–1% bovine serum albumin and then with goat anti-mouse IgG-phycoerythrin (Jackson ImmunoResearch). IgG binding was subsequently assessed using a Bioplex instrument (Bio-Rad Labs) and Bio-Plex Manager software. Data are presented as log10 MFIs of IgG binding to each bead type, after subtraction of the results for blank beads as well as the MFI for that antigen for beads incubated in control serum obtained from PBS-treated mice.
Animal models of infection.
All animal experiments conformed to institutional and governmental guidelines and regulations. Outbred CD1 female white mice (Charles Rivers Breeders) weighing approximately 25 g were used for animal infection experiments. Aliquots of −80°C stocks of bacteria grown in THY (i.e., grown in the presence of PABA) were used for challenge and vaccination experiments. Mice were anesthetized by inhalation of halothane (Zeneca) and inoculated intranasally (i.n.) with 5 × 106 CFU in an inoculum volume of 50 μl (pneumonia model and vaccination via the respiratory tract) or by intraperitoneal (i.p.) inoculation of 5 × 104, 5 × 105, or 5 × 106 CFU in an inoculum volume of 100 μl (sepsis model and systemic vaccination). To compare the course of disease between the Δpab and wild-type strains, groups of mice inoculated with S. pneumoniae strains were closely observed over the next 14 days. Mice were killed by humane methods when they exhibited the following signs of disease: hunched posture, poor mobility, weight loss, coughing, and tachypnea. Mixed-infection experiments were used to calculate competitive indexes (CIs; the ratio of mutant strain to wild-type strain recovered from the mice divided by the ratio of mutant strain to wild-type strain in the inoculum), using 7.5 × 103 CFU each of the wild-type and mutant strains in the sepsis model and groups of 4 or 6 mice (16). Mice were killed after 24 h, and spleens were recovered and homogenized in 0.5 ml of 0.9% saline, before plating dilutions on plain and kanamycin-containing blood agar plates for calculation of the CI. A CI of less than 1 indicates that the mutant strain is attenuated in virulence compared to the wild-type strain, and the lower that the CI is, the more attenuated the mutant strain is.
Statistical analysis.
All the in vitro growth curves were performed in triplicate and are represented as means and standard deviations (SDs). Results of growth curves and binding assays were analyzed using two-tailed t tests and analysis of variance (ANOVA) with post hoc tests. The numbers of target organ CFU were compared between strains using the Mann-Whitney U test, and the survival of infected mice was compared using the log rank test.
RESULTS
Construction of Δpab S. pneumoniae strain.
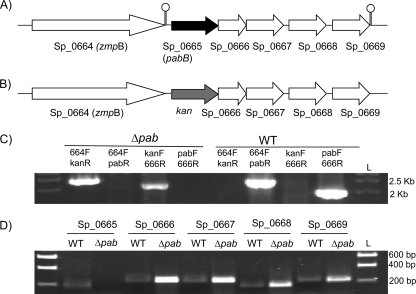
Searches of the S. pneumoniae TIGR4 genome identified a single gene, Sp_0665, which encodes a product with 38% identity and 57% similarity to the Escherichia coli PabB synthase (Table 2). Sp_0665 was the first gene in a putative 5-gene operon that also contained genes encoding a putative pyrimidine utilization protein (Sp_0666), pneumococcal surface protein (Sp_0667), glucokinase (Sp_0668), and thymidylate synthase (Sp_0668) (Fig. 1). To construct a PABA auxotrophic mutant strain, the TIGR4 strain was transformed with a construct made by overlap extension PCR in which Sp_0665 was replaced with the kanamycin resistance cassette (kan) in frame. The identities of the resulting TIGR4 Δpab mutant strains were confirmed by PCR (Fig. 1C) and sequencing. RT-PCR demonstrated that the mutation was nonpolar with continued transcription of the remaining genes in this putative operon (Fig. 1D). The stability of the mutant strain was confirmed by growing it in THY without kanamycin for two consecutive growth cycles and then plating it onto blood agar plates with and without kanamycin. All bacteria remained kanamycin resistant.
Table 2.
BLAST alignments of Sp_0665 amino acid sequence to other organisms
| Organism | Gene no. or name | Gene description | Size (no. of amino acids) | % identity/% similaritya |
|---|---|---|---|---|
| S. pyogenes | pabB | Putative PABA synthase | 585 | 56/73 (573) |
| Streptococcus equi | SEQ2030 | Chorismate binding enzyme | 578 | 54/73 (574) |
| Listeria monocytogenes | LmonH_020100001050 | PABA synthase | 568 | 41/57 (566) |
| Lactococcus lactis | pabB | PABA synthase Component I | 628 | 32/48 (484) |
| Bacillus subtilis | pabB | PABA synthase | 470 | 39/60 (257) |
| Staphylococcus aureus | SAB0663 | PABA synthase component I | 386 | 48/64 (258) |
| E. coli | ECP_1755 | PABA synthase component I | 453 | 38/57 (275) |
Numbers in parentheses indicate the length (number) of amino acids compared.
Fig. 1.
Construction of Δpab strains. (A) Schematic of the Sp_0665-0669 locus with the TIGR4 genome gene number and the assigned gene names in parentheses, if available. Arrows indicate transcriptional direction, lollipop structures indicate transcriptional termination, and the putative pabB gene is shaded black. (B) Structure of the Sp_0665-0669 locus in the Δpab mutant strain derived from the wild-type TIGR4 strain, showing replacement of Sp_0665 with an in-frame copy of kan, which is shaded gray. (C) Ethidium bromide-stained agarose gels showing PCR analysis of the TIGR4 Δpab strain confirming replacement of Sp_0665 with kan. The primer pairs (Table 1) used for each reaction are given above each lane. Lane L, DNA ladder size marker, with sizes listed on the right. (D) Ethidium bromide-stained agarose gels of products of RT-PCRs using internal primers for each gene within the putative Sp_0665-0669 operon (primers are listed in Table 1), confirming the nonpolar deletion of Sp_0665 in the Δpab strain. Reaction mixtures not containing reverse transcriptase gave no products (data not shown). Lane L, DNA ladder size marker, with sizes listed on the right. WT, wild type.
In vitro characterization of Δpab S. pneumoniae strain.
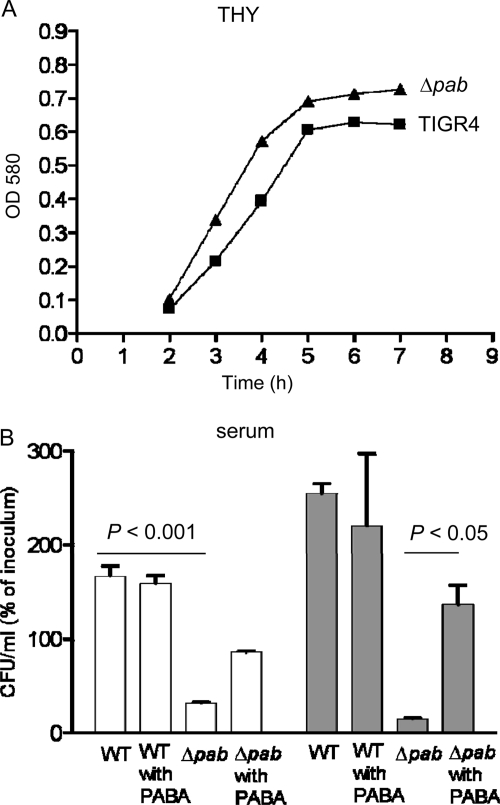
The growth of the TIGR4 Δpab strain in the complete medium THY was similar to the growth of the parental TIGR4 strain (Fig. 2 A). However, in human serum, in which there are only negligible concentrations of PABA, the TIGR4 Δpab strain had markedly impaired growth compared to TIGR4. Over 6 h, the numbers of TIGR4 Δpab CFU fell to 14.5% (SD, 2.86%) of the inoculating dose, whereas the numbers of TIGR4 CFU increased by 254% (SD, 22.5%). Supplementation of the serum with 2 mg/ml PABA partially reversed the survival defect of the TIGR4 Δpab strain, with the numbers of CFU increasing by 136% (SD, 14.3%) after 6 h. Hence, the TIGR4 Δpab strain had markedly attenuated growth under physiological conditions representative of those found during infection, and this is related to PABA auxotrophy. To assess whether interactions with important components of the host immune response to S. pneumoniae had been affected in the TIGR4 Δpab strain, the susceptibility of this strain to opsonization with complement components C3b/iC3b and to neutrophil phagocytosis was assessed using established flow cytometry assays (13). The TIGR4 Δpab strain had similar levels of C3b/iC3b deposition on its surface as the TIGR4 strain (fluorescence intensities [FIs], 4,373 [SD, 148] and 5,566 [SD, 222], respectively). Furthermore, there were no biologically significant differences in the degree of neutrophil association (a measure of phagocytosis) between the TIGR4 Δpab strain (FI, 603 [SD, 22]) and the TIGR4 strain (FI, 559 [SD, 24]).
Fig. 2.
In vitro characterization of the TIGR4 Δpab strain. (A) Growth of wild-type TIGR4 and TIGR4 Δpab strains in THY broth measured by OD580. (B) Numbers of bacterial CFU for the wild-type and the Δpab strains after incubation in human serum with and without supplementation of PABA (2 mg/ml). Results are expressed as a percentage of the inoculating CFU number, and error bars represent SDs. Clear columns, results after 3 h; gray columns, results after 6 h. For the results for the TIGR4 Δpab strain without PABA supplementation versus the wild-type strain, P was <0.001, and for the results for the TIGR4 Δpab strain without PABA supplementation versus the TIGR4 Δpab strain with PABA supplementation, P was <0.05 (ANOVA with post hoc tests).
Virulence of Δpab S. pneumoniae strains.
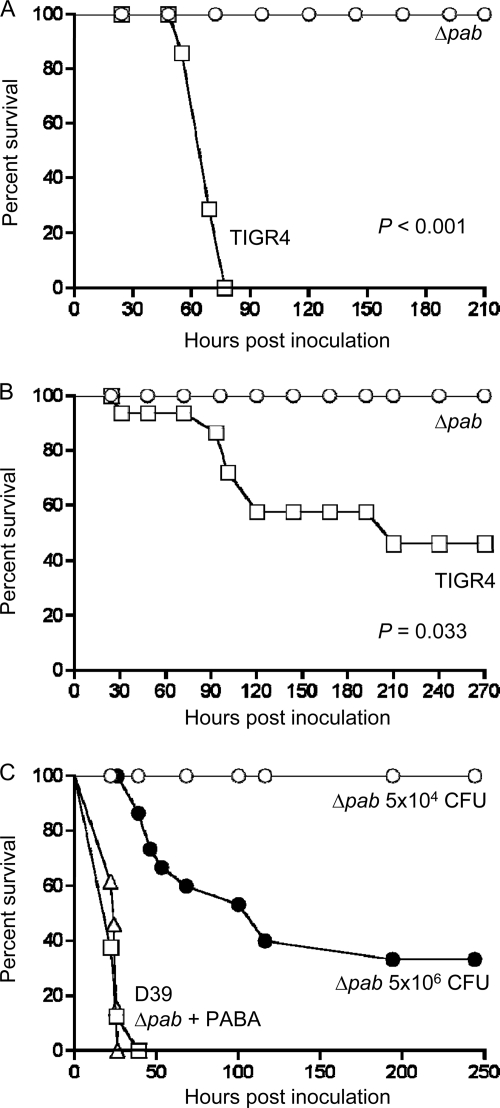
The in vitro growth data suggested that the Δpab strains will be attenuated during infection, and this was investigated using mouse models of sepsis (i.p. inoculation), pneumonia (i.n. inoculation), and nasopharyngeal colonization (small-volume i.n. inoculation under light anesthesia). In the sepsis model, all mice inoculated with the TIGR4 strain (5 × 104 CFU per mouse) developed fatal disease within 4 days, whereas none of the mice inoculated with the TIGR4 Δpab strain developed fatal disease (Fig. 3 A). Similar results were obtained with the pneumonia model (5 × 106 CFU per mouse), with over 50% of mice inoculated with TIGR4 developing fatal disease within 9 days, whereas all the mice inoculated with the TIGR4 Δpab strain survived (Fig. 3B). In the nasopharyngeal colonization model, by 2 days after inoculation, no TIGR4 Δpab bacteria were found in the nasal washes, whereas significant numbers of the wild-type strain persisted (Table 3). To assess whether this loss of virulence was related to PABA auxotrophy, additional experiments were performed in which mice infected with the TIGR4 Δpab strain had PABA added to their drinking water (1 mg/ml). After infection, mice were culled at 24 h (i.p. inoculation, 7 × 103 CFU/mouse) or 48 h (i.n. inoculation, 5 × 106 CFU/mouse), and the numbers of bacterial CFU in target organs were calculated by plating serial dilutions on plain and antibiotic-containing blood plates. No TIGR4 Δpab strain bacteria were recovered in either model from mice without PABA supplementation, whereas similar numbers of CFU of the Δpab strain were found in the spleens and lungs of mice supplemented with PABA and those of mice infected with the TIGR4 strain (Table 3).
Fig. 3.
Time course of development of fatal infection for mice inoculated with the TIGR4 Δpab or D39 Δpab strain. (A) Results for mice inoculated i.p. with 5 × 104 CFU of the TIGR4 or TIGR4 Δpab strain; (B) results for mice inoculated i.n. with 5 × 106 CFU of the TIGR4 or TIGR4 Δpab strain; (C) results for mice inoculated i.p. with 5 × 104 CFU of the D39 strain (triangles), D39 Δpab strain (open circles), or D39 Δpab strain with PABA supplementation to the water (squares) or 5 × 106 CFU of the D39 Δpab strain (black circles). P values were obtained using log rank tests, and for panel C, P was <0.001 for the D39 strain or D39 Δpab strain with PABA supplementation versus both CFU doses for the D39 Δpab strains.
Table 3.
Virulence of Δpab mutanta
| Route of inoculation | Target organ | No. of CFU of S. pneumoniae straind: |
P valuee |
|||
|---|---|---|---|---|---|---|
| TIGR4 | TIGR4 Δpabf | TIGR4 Δpab + PABA | TIGR4 vs TIGR4 Δpab | TIGR4 Δpab vs TIGR4 Δpab+ PABA | ||
| i.p. | Spleenb | 5.8 (0.2) | 0 | 4.5 (0.25) | <0.01 | <0.05 |
| i.n. | Lungc | 4.3 (0.5) | 0 | 3.1 (0.8) | <0.05 | <0.05 |
| i.n. | Spleenc | 1.5 (0.2) | 0 | 1.7 (0.5) | <0.05 | <0.05 |
| NP | Nasal washesc | 4.79 (0.36) | 0 | NDg | <0.05 | |
Virulence was assessed from the numbers of log10 CFU/ml recovered from target organs in mouse models of sepsis (i.p. inoculation), pneumonia (i.n. inoculation), or nasopharyngeal colonization (NP) (n = 4 or 5 mice per group) with and without supplementation of the drinking water with PABA (1 mg/ml).
Twenty-four hours postinoculation of 7 × 103 CFU/mouse.
Forty-eight hours postinoculation of 5 × 106 CFU/mouse.
Data show log10 values of the mean (standard error of the mean) numbers of S. pneumoniae CFU recovered from spleen or lung homogenates and nasal washes.
P values were calculated using the Kruskal-Wallis test with post hoc tests or the Mann-Whitney U test.
No colonies were recovered after plating neat organ preparations.
ND, not done.
To further confirm that the Δpab mutation has marked effects on virulence, the D39 strain was transformed with the Sp_0665 deletion construct to obtain a D39 Δpab strain. The virulences of the D39 Δpab and wild-type D39 strains were compared in mouse models of sepsis. The D39 Δpab strain was severely attenuated in virulence, with no mice inoculated i.p. with 5 × 104 CFU of the D39 Δpab strain developing fatal disease but 100% of mice inoculated with the wild-type D39 strain or with the D39 Δpab strain and given PABA supplementation in the drinking water developing fatal disease (Fig. 3C). The D39 Δpab strain did cause slowly progressive infection in a proportion of mice when inoculated i.p. at a higher dose of 5 × 106 CFU (Fig. 3C), with only kanamycin-resistant bacteria being recovered from the spleens of dead mice, suggesting that fatal infection was not due to mutant reversion. Overall, the data show that deletion of pabB caused a marked attenuation in S. pneumoniae virulence that can be conditionally restored by addition of PABA to the drinking water.
The results of the growth-in-serum experiments showed that PABA supplementation only partially restored the growth of the Δpab strain. Hence, to accurately assess whether virulence of the Δpab strain is also only partially restored by PABA supplementation, mixed-infection experiments were performed to obtain a CI for the TIGR4 Δpab strain versus the TIGR4 strain with or without PABA supplementation of the mouse drinking water. In the absence of PABA supplementation, no TIGR4 Δpab strain CFU was recovered from mice inoculated intraperitoneally (n = 8) with a 50/50 mix of 7.5 × 103 CFU of the TIGR4 Δpab and TIGR4 strains, giving a CI of less than 0.0002 (SD, 0.0001). In contrast, in mice supplemented with PABA in the drinking water (1 mg/ml, n = 4), approximately 1 in 1,000 recovered bacteria was the TIGR4 Δpab strain, giving a CI of 0.008 (SD, 0.006). These results show that even though PABA supplementation restores virulence to the TIGR4 Δpab strain, this strain is still attenuated in virulence compared to the wild-type strain during mixed infections.
Vaccination with the TIGR4 Δpab strain induces antibodies to S. pneumoniae.
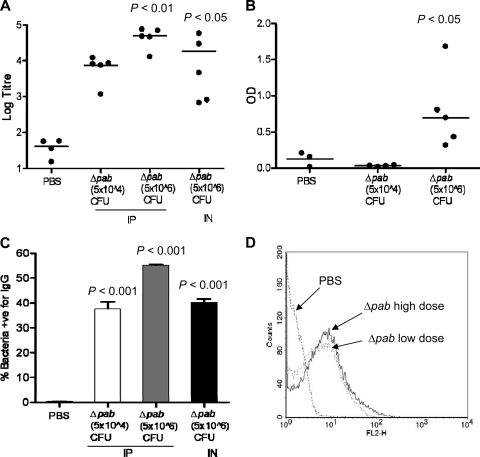
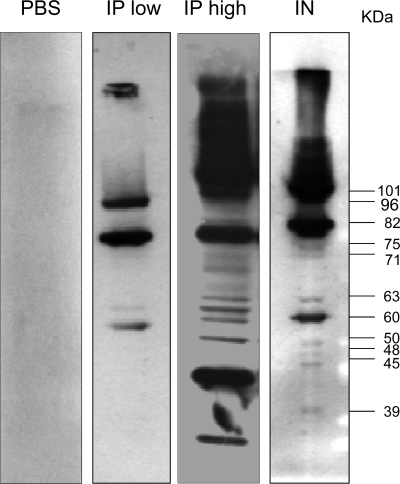
To investigate whether vaccination with the TIGR4 Δpab strain induces protective immunity against S. pneumoniae, groups of 5 mice were vaccinated by either the i.p. (5 × 104 CFU or 5 × 106 CFU of TIGR4 Δpab/mouse) or i.n. (5 × 106 CFU of TIGR4 Δpab/mouse in a 50-μl volume while the mice were under deep anesthesia, ensuring that bacteria are aspirated into the lungs) route, and antibody responses were assessed by ELISAs, immunoblotting, and flow cytometry with sera recovered from mice 4 weeks later. Whole-cell ELISAs for the TIGR4 strain demonstrated significant antibody levels in sera from mice inoculated with the TIGR4 Δpab strain by both the i.p. and i.n. routes (Fig. 4 A). In contrast, no significant antibody response to the capsular serotype 4 antigen was detected (data not shown), and only sera from mice inoculated i.p. with the higher dose of the TIGR4 Δpab strain had significant but low antibody responses to cell wall-associated polysaccharide (Fig. 4B). Flow cytometry analysis showed high levels of IgG binding to live TIGR4 bacteria in sera obtained from mice inoculated either i.p. or i.n. with the TIGR4 Δpab strain (Fig. 4C and D). These data suggest that the antibody responses mainly recognized protein antigens, and immunoblots of TIGR4 whole-cell lysates demonstrated that IgG in serum from mice inoculated with the TIGR4 Δpab strain recognized several protein antigens (Fig. 5). Immunoblots probed with naïve serum showed only minimal binding to S. pneumoniae proteins. Two distinct protein bands with molecular masses of 96 kDa and 82 kDa were present in all three immunoblots probed with immune sera from mice inoculated i.p. (both low and high doses) and i.n. (Fig. 5). Several other proteins were also recognized, with molecular masses ranging from 103 to 32 kDa in the blot treated with immune sera from mice inoculated i.p. with the high dose and from 101 to 39 kDa in the blot treated with immune sera from mice inoculated i.n.
Fig. 4.
Antibody responses in serum recovered from mice 4 weeks after vaccination with 5 × 104 or 5 × 106 CFU of the TIGR4 Δpab strain. Control mice were inoculated with PBS only. (A) Whole-cell ELISA IgG titers against the TIGR4 strain; (B) IgG to cell wall polysaccharide expressed as the OD obtained using a 1-in-20 dilution of serum. (A and B) Each data point represents results from an individual mouse, and the bars represent median values. P values were obtained with Kruskal-Wallis tests with post hoc analysis. (C) Mean proportion of TIGR4 bacteria positive for IgG when assessed by flow cytometry after incubation in sera from mice inoculated with TIGR4 Δpab at 5 × 104 CFU i.p. (clear column), TIGR4 Δpab at 5 × 106 CFU i.p. (gray column), or TIGR4 Δpab at 5 × 106 CFU i.n. (black column). P values were obtained using ANOVA tests with post hoc analysis. (D) Example of a flow cytometry histogram for IgG binding to the TIGR4 strain in sera from mice inoculated i.p. with the TIGR4 Δpab strain (dotted line, 5 × 104 CFU; solid line, 5 × 106 CFU) or PBS (dashed line).
Fig. 5.
Immunoblots of wild-type TIGR4 whole-cell lysates probed with 1% serum from mice inoculated with PBS, TIGR4 Δpab at 5 × 104 CFU i.p. (IP low), TIGR4 Δpab at 5 × 106 CFU i.p. (IP high), or TIGR4 Δpab at 5 × 106 CFU i.n. Estimated molecular masses of the proteins detected are indicated to the side of the protein bands.
Identification of antigen targets for serum antibodies from Δpab-vaccinated mice.
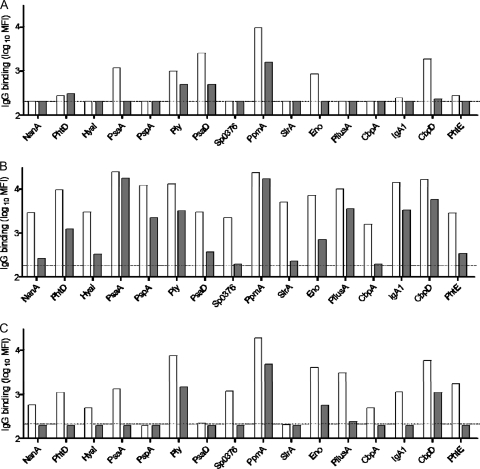
xMAP beads conjugated with 16 recombinant pneumococcal proteins and a Luminex bead assay were used to identify possible pneumococcal proteins recognized by serum IgG from TIGR4 Δpab-vaccinated mice. The number of antigens recognized and/or the strength of the antibody response varied between the three vaccination conditions, with immune sera from mice vaccinated i.p. with a high dose (5 × 106 CFU) showing the strongest antibody response, sera from mice vaccinated with a low dose (5 × 104 CFU) i.p. showing the weakest, and sera from mice vaccinated i.n. into the lungs having an intermediate response (Fig. 6). IgG from the high-dose i.p. and the i.n. vaccinated mice recognized most (i.n.) or all (i.p.) 16 of the tested antigens. PpmA, Ply, and CbpD were dominant antigens for both high-dose i.p. and i.n. vaccinated mice, with sera from high-dose i.p. vaccinated mice also having high levels of IgG against PsaA, PspA, pilus A, and IgA1. Sera from low-dose i.p. vaccinated mice recognized only six antigens: PpmA, PsaA, PsaD, Ply, Eno, and CbpD (Fig. 6). These results demonstrate that vaccination with the TIGR4 Δpab strain induced an effective antibody response to S. pneumoniae and suggest that these antibodies mainly recognized TIGR4 protein antigens rather than capsule or cell wall polysaccharide.
Fig. 6.
Results of Luminex IgG binding to various S. pneumoniae cell surface antigens on xMAP beads in immune sera from mice inoculated with TIGR4 Δpab at 5 × 104 CFU i.p. (A), 5 × 106 CFU i.p. (B), or 5 × 106 CFU i.n. (C) assayed by flow cytometry. Clear columns, results obtained with 10% serum; gray columns, results obtained with 1% serum. Pooled sera were assayed in duplicate at serial dilutions, and results are presented as the log10 mean MFI at each dilution. Results are presented after subtracting the background levels in the serum from naïve mice inoculated with PBS and the background levels from blank protein beads.
Vaccination with the Δpab strain is strongly protective against infection with homologous S. pneumoniae strains.
To investigate whether vaccination with the Δpab strain protects against subsequent S. pneumoniae infection, groups of 5 mice vaccinated i.p. with a single dose of 5 × 104 CFU of the TIGR4 Δpab strain were 4 weeks later inoculated i.p. with 103 CFU of the TIGR4 strain. Mice were culled after 24 h, and the numbers of bacterial CFU in the spleen were calculated by plating serial dilutions. Although a median of 4.2 log10 bacteria were recovered from control mice (vaccinated with PBS), no bacteria were recovered from mice that had previously been vaccinated with the TIGR4 Δpab strain (Table 4). Similarly, vaccination by i.n. inoculation of 5 × 106 CFU of the TIGR4 Δpab strain into the lungs while the mice were under deep anesthesia protected against subsequent i.n. challenge with 5 × 106 CFU of the TIGR4 strain, with a median log10 number of CFU of 0 in spleens versus 1.9 for control mice (Table 4). There were also lower numbers of CFU in the lungs, although this result was not statistically significant (Table 4). Hence, both i.p. and i.n. vaccination with the TIGR4 Δpab strain protects against subsequent challenge via the same inoculation route with fully virulent TIGR4 bacteria. Mice vaccinated i.p. with the D39 Δpab strain were also protected against subsequent infection with the D39 strain, with partial protection after vaccination with 5 × 104 CFU and complete protection after vaccination with 5 × 106 CFU detected (Table 4).
Table 4.
Protective effect of vaccination with TIGR4 Δpab or D39 Δpab strain or PBS for mice subsequently challengeda
| Vaccination strain | No. of CFU and vaccination route | Challenge strain and route | Target organ | Median (IQR) log10 CFU/mlb |
P valuec | |
|---|---|---|---|---|---|---|
| PBS control | Δpab strain | |||||
| TIGR4 Δpab | 5 × 104 i.p. | TIGR4 i.p. | Spleen | 4.16 (3.65–4.60) | 0d | <0.001 |
| 5 × 104 i.p. | D39 i.p. | Spleen | 6.38 (6.34–6.38) | 5.46 (1.69–6.36) | 0.191 | |
| Blood | 7.96 (7.93–7.99) | 5.92 (2.34–7.49) | 0.191 | |||
| 5 × 104 i.p. | 0100993 i.p. | Spleen | 5.04 (3.73–5.90) | 5.08 (3.90–5.63) | 0.84 | |
| Blood | 5.60 (5.06–6.11) | 5.42 (5.14–6.36) | 0.84 | |||
| 5 × 106 i.n. | TIGR4 i.n. | Lung | 4.04 (3.24–4.85) | 1.30 (0–4.80)e | 0.125 | |
| Spleen | 1.88 (1.18–3.18) | 0 (0–1.3)e | 0.017 | |||
| D39 Δpab | 5 × 104 i.p. | D39 i.p. | Spleen | 7.83 (4.92–7.99) | 0 (0–3.36) | 0.0159 |
| Blood | 8.57 (3.83–8.57) | 0 (0–4.07) | 0.095 | |||
| 5 × 104 i.p. | 0100993 i.p. | Spleen | 5.04 (3.73–5.90) | 5.08 (2.24–6.05) | 0.90 | |
| Blood | 5.60 (5.06–6.11) | 5.41 (4.03–6.71) | 0.29 | |||
| 5 × 106 i.p. | D39 i.p. | Spleen | 6.05 (4.11–7.12) | 0d | <0.001 | |
| Blood | 6.76 (3.31–8.14) | 0d | <0.001 | |||
The mice were challenged by i.p. inoculation with 1 × 103 or i.n. inoculation with 5 × 106 TIGR4, D39, or 0100993 S. pneumoniae CFU.
Data show log10 values of the median (interquartile ratio [IQR]) numbers of S. pneumoniae CFU recovered from blood, spleen, or lung homogenates (n = 5 or 6) at 24 h (i.p. route) or 48 h (i.n. route) after challenge.
P values refer to the comparison of the results for PBS and Δpab strain-vaccinated mice (Mann-Whitney U tests).
No colonies were recovered from any mice after plating neat organ preparations.
Data in parentheses are minimum and maximum values and not interquartile ratios.
Vaccination with the TIGR4 Δpab strain provides minimal or no protection against systemic infection with the D39 or 0100993 S. pneumoniae strain.
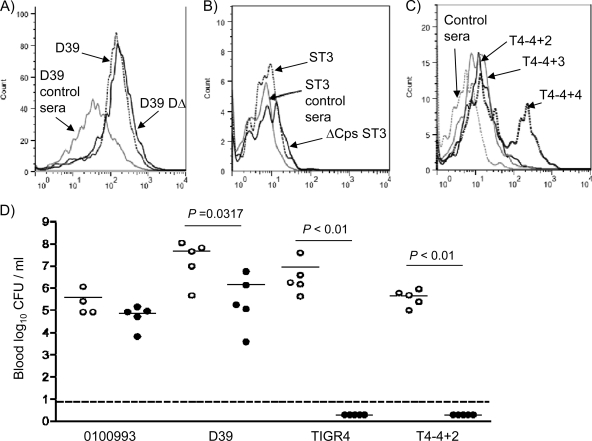
To investigate whether vaccination with Δpab strains protects against heterologous infection, mice inoculated i.p. with 5 × 104 CFU of the TIGR4 Δpab or D39 Δpab strain were challenged with the D39 and 0100993 strains. Prior vaccination with TIGR4 Δpab caused a trend toward lower numbers of bacterial CFU in spleen and blood when mice were challenged with the D39 strain, but this was not statistically significant (Table 4). Vaccination with either the TIGR4 Δpab or D39 Δpab strain did not protect against infection with the 0100993 strain (Table 4). Flow cytometry demonstrated that after incubation in sera from TIGR4 Δpab-vaccinated mice, there was significant IgG binding to live D39 bacteria but only minimal IgG binding to strain 01000993 bacteria, even unencapsulated strains (Fig. 7 A to C). To assess whether protection could be induced by higher-dose vaccination, mice were vaccinated with 5 × 105 CFU of the TIGR4 Δpab strain twice 10 days apart before challenge 4 weeks later with the TIGR4, D39, and 0100993 strains. Despite complete protection against the homologous challenge with the TIGR4 strain, vaccination with the TIGR4 Δpab strain caused a reduction in the numbers of CFU in blood only when challenge was with the D39 strain and no protection against the 0100993 strain (Fig. 7D). To identify whether the different capsular serotypes prevented protection against heterologous strains after vaccination with TIGR4 Δpab, mice were also challenged with TIGR4 capsular-switch strains expressing the serotype 2 or 3 capsule (37, 40). The TIGR4 serotype 3 capsular-switch strain was unable to establish infection in the PBS-treated mice after i.p. inoculation, preventing assessment of the role of the serotype 3 capsule (data not shown). The TIGR4 serotype 2 capsular-switch strain established significant infection in the PBS-treated mice but was unable to establish infection in TIGR4 Δpab-vaccinated mice (Fig. 7D). Hence, failure of vaccination with the TIGR4 Δpab strain to protect against the D39 strain was not due to the different capsular serotype but was dependent on strain background.
Fig. 7.
Protection against heterologous strains after vaccination with TIGR4 Δpab. (A to C) Examples of flow cytometry histograms for IgG binding in serum from mice inoculated i.p. with the TIGR4 Δpab strain (5 × 106 CFU) to the D39 (dotted line) and unencapsulated D39 (D39DΔ; solid line) strains (A), the 0100993 (ST3; dotted line) and unencapsulated 0100993 (ΔCps ST3; solid line) strains (B), and the TIGR4 capsular-switch strains expressing capsular serotypes 4 (T4-4+4), 2 (T4-4+2), and 3 (T4-4+3) or PBS (control sera; dashed line) (C). (D) Log10 numbers of bacterial CFU/ml in blood recovered 24 h after challenge with 103 CFU of the 0100993, D39, TIGR4, and TIGR4 capsular-switch expressing serotype 2 (T4-4+2) strains. Mice were previously vaccinated i.p. with 5 × 105 CFU of the TIGR4 Δpab strain (black circles) or with PBS as controls (clear circles) twice, 10 days apart, with the last dose given 4 weeks before challenge. Each symbol represents the result from one mouse, and the P values were generated using Mann-Whitney U tests. Dotted line, lower limit of detection for bacteria in blood (no colonies were cultured from the blood of TIGR4 Δpab-vaccinated mice inoculated with the TIGR4 or T4-4+2 capsular-switch strain).
DISCUSSION
As well as being a central branch point in the biosynthesis of aromatic amino acids such as tryptophan, tyrosine, and phenylalanine, chorismate is necessary for the synthesis of PABA and therefore for folic acid, an essential cofactor for the biosyntheses of purines, pyrimidines, etc. (23, 38). Many bacteria and plants have the ability to synthesize PABA, but higher organisms do not (3, 34), which makes the PABA biosynthetic pathway an attractive target for new vaccination and therapeutic strategies (18, 36). Genome-wide searches with S. pneumoniae TIGR4 and the BLAST program revealed 2 genes whose products have high degrees of similarity to PabB and PabA of E. coli, components I and II of PABA synthase (also called 4-amino-4-deoxychorismate [ADC] synthase), respectively, which catalyze the formation of ADC from chorismate in the presence of glutamine (42). In bacteria, PABA biosynthesis is generally believed to proceed via a two-step process requiring the presence of two enzymes, ADC synthase and ADC lyase (3, 28, 34, 39, 42). ADC lyase is encoded by the gene pabC and cleaves ADC to generate PABA and pyruvate (8). However, we found no match to pabC in the S. pneumoniae genome using BLAST searches. Wegkamp et al. reported a pabBC fusion gene in Lactococcus lactis, Streptococcus pyogenes, and Helicobacter pylori (39) with homology to both pabB and pabC of E. coli. The high homology (56% identity and 73% similarity over 573 amino acid residues) of the S. pyogenes pabBC fusion gene to the annotated pabB of S. pneumoniae TIGR4 suggests that a pabBC fusion gene is also likely to be present in S. pneumoniae. Whether this gene encodes a single enzyme which synthesizes PABA in one step is not known. We created Δpab deletion mutants in the TIGR4 and D39 strain backgrounds by replacing the annotated pabB gene with a kanamycin resistance marker in frame. In vitro growth experiments in serum revealed a profound lack of growth of the TIGR4 Δpab strain that was partially restored by supplementing with external PABA, confirming that deletion of Sp_0665 created PABA-dependent auxotrophic mutant strains. However, in the nutrient-rich undefined medium THY, growth of the Δpab strains was normal. Hava and Camilli have previously identified that disruption of pabB affects S. pneumoniae virulence (9), and consistent with these data, we found that both the TIGR4 Δpab and D39 Δpab strains were severely but not completely attenuated in virulence in mouse models of disease. Inoculation with a lower dose of Δpab bacteria failed to cause disease, although a higher dose, given i.p., of the D39 Δpab strain caused delayed but fatal infection in a proportion of mice. This perhaps reflects slow bacterial growth due to direct acquisition of some folate from the host or low PABA availability from the diet. Strikingly, PABA supplementation of the mouse drinking water restored the virulence of both the TIGR4 Δpab and D39 Δpab strains, with the numbers of bacterial CFU in target organs and rates of development of fatal infection being similar to those for the wild-type strains. PABA auxotrophic mutations causing a similar phenotype have previously been reported in Gram-negative bacterial pathogens, such as E. coli (8, 17), Salmonella enterica serovar Typhi (36), and Shigella flexneri (6), and in the fungal pathogen A. fumigatus (5). Using highly sensitive mixed-infection experiments, the virulence of the TIGR4 Δpab strain was still attenuated compared to that of the wild-type TIGR4 strain when mouse drinking water was supplemented with PABA. This may be because even in vitro exogenous complementation with PABA did not fully restore growth of the TIGR4 Δpab strain in serum to wild-type levels, perhaps due to unexpected effects of the mutation unrelated to loss of pabB.
The PABA-dependent conditional virulence of the Δpab mutant makes it an attractive tool for studying host-pathogen interactions. For example, the inability of this strain to undergo significant replication within the host while retaining normal interactions with complement and neutrophils suggests that it could be used to assess the rate of clearance of S. pneumoniae from different sites of infection and under different host conditions. Furthermore, studying the effects of specific mutations on the host inflammatory response is often confounded by large differentials in numbers of bacterial CFU in vivo between the wild-type control and the mutant strains under investigation, which themselves create differences in inflammatory responses. This problem could be averted by using a Δpab S. pneumoniae strain background, preventing replication of both the mutant strain under investigation and the control strain so that differences in the inflammatory response are solely due to the effects of the mutation.
The attenuated virulence of the Δpab TIGR4 S. pneumoniae strain allowed us to assess the potential efficacy of systemic administration of a live attenuated vaccine against S. pneumoniae. We used i.p. inoculation of the Δpab strain, a standard approach for establishing the potential efficacy of new S. pneumoniae vaccine candidates (7, 14, 33). To assess whether the site of vaccination was important, we also investigated the efficacy of i.n. inoculation of the TIGR4 Δpab strain into the lungs. Neither vaccination route is practical in humans, but the results for those routes can provide proof of principle that can be further developed using the subcutaneous, sublingual, or intramuscular vaccination route. Despite rapid clearance when given systemically, the Δpab mutant induced powerful antibody responses against TIGR4 but, surprisingly, not against the capsule or the cell wall polysaccharide. This suggested that most of the antibody response was targeted against protein antigens, and this was confirmed by immunoblots of bacterial whole-cell lysates and the xMAP bead assay measuring IgG binding to multiple protein antigens. The xMAP bead assays contained 16 different antigens (35), and strikingly, after inoculation with the Δpab strain, serum IgG against all these antigens was detectable, although responses to certain antigens (e.g., PsaA and PpmA) seemed to be dominant. There were only negligible IgG responses to these antigens detectable in sera from controls. The dominant antigens identified from the xMAP bead assay may correspond to the dominant antigens seen on the immunoblots of whole-cell lysates, as PsaA has an estimated molecular mass of 33.7 kDa, and PpmA, CbpD, Ply, and PspA have estimated molecular masses of 35, 50.3, 52.5, and 62 kDa, respectively.
The antigens included in the xMAP bead assays were largely pneumococcal surface proteins, many of which are conserved between strains and known to protect against invasive S. pneumoniae infections in animal models when used as protein vaccines (1, 2, 4, 7, 21, 24). However, although vaccination with Δpab strains induced strong protection against subsequent infection with the homologous wild-type S. pneumoniae strain, vaccination with the Δpab strain failed, surprisingly, to induce biologically significantly protection against infection with heterologous strains, even after a two-dose vaccination schedule. One potential explanation for a lack of cross-protection could be variable access to target antigens due to differences in capsular properties between serotypes. However, there was strong IgG binding to the D39 strain in sera from TIGR4 Δpab-vaccinated mice, and conversely, although vaccination with the TIGR4 Δpab strongly protected against infection with a capsular-switch TIGR4 strain expressing a serotype 2 capsule, there was reduced IgG binding to this strain compared to that to the TIGR4 strain. These data demonstrate that serotype-dependent effects on access to subcapsular antigens do not explain failure of protection against heterologous strains. Instead, our results indicate that although after vaccination with the TIGR4 Δpab strain there are antibody responses to a range of antigens, perhaps antibody responses to only select antigens are functionally important and actually mediate protection. Protective efficacy could then vary between strains if these important antigens are present in only a subset of strains (e.g., pilus A), show allelic variation (e.g., PspA), or perhaps vary markedly in their expression between strains. Furthermore, if antibody efficacy depends on blocking target protein function (as shown for antibody to PsrP and Ply) (33), then the effects on bacterial virulence may also vary between strains. Alternatively, protection may be mediated by cellular immune responses rather than by the antibody to protein antigens. However, Th17-mediated adaptive immune responses protect against mucosal infection (22, 23, 44) rather than systemic disease, and it is likely that antibody responses have an important role for Δpab-induced systemic immunity. Our results contrast with published data suggesting that nasopharyngeal colonization with S. pneumoniae mutant strains can cross-protect against heterologous strains (31, 32). The reasons for these differences are not clear but could be related to the route of inoculation (nasopharyngeal versus systemic), the prolonged duration of exposure to the vaccine strain during colonization compared to rapid clearance after i.p. vaccination, or bacterial/mouse genetic factors. In addition, both Roche et al. (32) and Richards et al. (31) used a pneumonia challenge model, which is less stringent than the sepsis model used in this work and therefore more likely to demonstrate cross-protective immunity. Further investigation of the factors influencing whether antibody responses to live S. pneumoniae induce cross-protective responses against heterologous strains is necessary.
To conclude, in this study we have created a PABA auxotrophic mutant S. pneumoniae strain that has conditional virulence in mice, which may make this strain a useful tool for investigating host-bacterial interactions. We have used the Δpab strain to assess the potential efficacy of a live attenuated S. pneumoniae vaccine. However, despite inducing antibody responses to a number of conserved protein antigens and strong protection against challenge with homologous strains expressing different capsular serotypes, vaccination with the Δpab bacteria failed to significantly protect against infection with heterologous strains. Overall, our data suggest that parenteral administration of a live attenuated vaccine is not an attractive approach for preventing S. pneumoniae infections due to the potential difficulty in inducing protection against a broad range of strains.
ACKNOWLEDGMENTS
This work was undertaken at UCLH/UCL, which received a proportion of funding from the Department of Health's NIHR Biomedical Research Centre's funding scheme, and was supported by the UCL Charities funding and grants from the Wellcome Trust (grant reference 076442) and the Medical Research Council (grants G0700829 and G0600410).
We thank Tim Mitchell (University of Glasgow) and Peter Hermans (Radboud University Nijmegen Medical Centre) for providing most of the proteins included in the Luminex bead assay, and Marc Lipsitch (Harvard) and Krzysztof Trzcinski (UMC Utrecht) for providing the TIGR4 capsular-switch strains.
Footnotes
Published ahead of print on 26 September 2011.
REFERENCES
- 1. Audouy S. A., et al. 2007. Development of lactococcal GEM-based pneumococcal vaccines. Vaccine 25:2497–2506 [DOI] [PubMed] [Google Scholar]
- 2. Balachandran P., Brooks-Walter A., Virolainen-Julkunen A., Hollingshead S. K., Briles D. E. 2002. Role of pneumococcal surface protein C in nasopharyngeal carriage and pneumonia and its ability to elicit protection against carriage of Streptococcus pneumoniae. Infect. Immun. 70:2526–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bermingham A., Derrick J. P. 2002. The folic acid biosynthesis pathway in bacteria: evaluation of potential for antibacterial drug discovery. Bioessays 24:637–648 [DOI] [PubMed] [Google Scholar]
- 4. Briles D. E., et al. 2003. Immunizations with pneumococcal surface protein A and pneumolysin are protective against pneumonia in a murine model of pulmonary infection with Streptococcus pneumoniae. J. Infect. Dis. 188:339–348 [DOI] [PubMed] [Google Scholar]
- 5. Brown J. S., et al. 2000. Signature-tagged and directed mutagenesis identify PABA synthetase as essential for Aspergillus fumigatus pathogenicity. Mol. Microbiol. 36:1371–1380 [DOI] [PubMed] [Google Scholar]
- 6. Cersini A., Salvia A. M., Bernardini M. L. 1998. Intracellular multiplication and virulence of Shigella flexneri auxotrophic mutants. Infect. Immun. 66:549–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gianfaldoni C., et al. 2007. Streptococcus pneumoniae pilus subunits protect mice against lethal challenge. Infect. Immun. 75:1059–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Green J. M., Merkel W. K., Nichols B. P. 1992. Characterization and sequence of Escherichia coli pabC, the gene encoding aminodeoxychorismate lyase, a pyridoxal phosphate-containing enzyme. J. Bacteriol. 174:5317–5323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hava D. L., Camilli A. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45:1389–1406 [PMC free article] [PubMed] [Google Scholar]
- 10. Havarstein L. S., Coomaraswamy G., Morrison D. A. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U. S. A. 92:11140–11144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hohmann E. L., Oletta C. A., Killeen K. P., Miller S. I. 1996. phoP/phoQ-deleted Salmonella typhi (Ty800) is a safe and immunogenic single-dose typhoid fever vaccine in volunteers. J. Infect. Dis. 173:1408–1414 [DOI] [PubMed] [Google Scholar]
- 12. Huss A., Scott P., Stuck A. E., Trotter C., Egger M. 2009. Efficacy of pneumococcal vaccination in adults: a meta-analysis. CMAJ 180:48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hyams C., et al. 2010. Streptococcus pneumoniae resistance to complement-mediated immunity is dependent on the capsular serotype. Infect. Immun. 78:716–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ivady B. 2010. Pneumococcal conjugate vaccines in the prevention of childhood pneumonia. Acta Microbiol. Immunol. Hung. 57:1–13 [DOI] [PubMed] [Google Scholar]
- 15. Jomaa M., et al. 2005. Antibodies to the iron uptake ABC transporter lipoproteins PiaA and PiuA promote opsonophagocytosis of Streptococcus pneumoniae. Infect. Immun. 73:6852–6859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Khandavilli S., et al. 2008. Maturation of Streptococcus pneumoniae lipoproteins by a type II signal peptidase is required for ABC transporter function and full virulence. Mol. Microbiol. 67:541–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Komano T., Utsumi R., Kawamukai M. 1991. Functional analysis of the fic gene involved in regulation of cell division. Res. Microbiol. 142:269–277 [DOI] [PubMed] [Google Scholar]
- 18. Lacks S. A., Greenberg B., Lopez P. 1995. A cluster of four genes encoding enzymes for five steps in the folate biosynthetic pathway of Streptococcus pneumoniae. J. Bacteriol. 177:66–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lau G. W., et al. 2001. A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol. Microbiol. 40:555–571 [DOI] [PubMed] [Google Scholar]
- 20. Lexau C. A., et al. 2005. Changing epidemiology of invasive pneumococcal disease among older adults in the era of pediatric pneumococcal conjugate vaccine. JAMA 294:2043–2051 [DOI] [PubMed] [Google Scholar]
- 21. Long J. P., Tong H. H., DeMaria T. F. 2004. Immunization with native or recombinant Streptococcus pneumoniae neuraminidase affords protection in the chinchilla otitis media model. Infect. Immun. 72:4309–4313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lu Y. J., et al. 2010. Options for inactivation, adjuvant, and route of topical administration of a killed, unencapsulated pneumococcal whole-cell vaccine. Clin. Vaccine Immunol. 17:1005–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McConkey G. A., et al. 2004. Annotating the Plasmodium genome and the enigma of the shikimate pathway. Trends Parasitol. 20:60–65 [DOI] [PubMed] [Google Scholar]
- 24. Melin M., et al. 2010. Interaction of pneumococcal histidine triad proteins with human complement. Infect. Immun. 78:2089–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mera R., Miller L. A., Fritsche T. R., Jones R. N. 2008. Serotype replacement and multiple resistance in Streptococcus pneumoniae after the introduction of the conjugate pneumococcal vaccine. Microb. Drug Resist. 14:101–107 [DOI] [PubMed] [Google Scholar]
- 26. Mogensen T. H. 2009. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 22:240–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Morona J. K., Miller D. C., Morona R., Paton J. C. 2004. The effect that mutations in the conserved capsular polysaccharide biosynthesis genes cpsA, cpsB, and cpsD have on virulence of Streptococcus pneumoniae. J. Infect. Dis. 189:1905–1913 [DOI] [PubMed] [Google Scholar]
- 28. Nichols B. P., Seibold A. M., Doktor S. Z. 1989. para-Aminobenzoate synthesis from chorismate occurs in two steps. J. Biol. Chem. 264:8597–8601 [PubMed] [Google Scholar]
- 29. O'Brien K. L., et al. 2009. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374:893–902 [DOI] [PubMed] [Google Scholar]
- 30. Pearce B. J., Iannelli F., Pozzi G. 2002. Construction of new unencapsulated (rough) strains of Streptococcus pneumoniae. Res. Microbiol. 153:243–247 [DOI] [PubMed] [Google Scholar]
- 31. Richards L., Ferreira D. M., Miyaji E. N., Andrew P. W., Kadioglu A. 2010. The immunising effect of pneumococcal nasopharyngeal colonisation; protection against future colonisation and fatal invasive disease. Immunobiology 215:251–263 [DOI] [PubMed] [Google Scholar]
- 32. Roche A. M., King S. J., Weiser J. N. 2007. Live attenuated Streptococcus pneumoniae strains induce serotype-independent mucosal and systemic protection in mice. Infect. Immun. 75:2469–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rose L., et al. 2008. Antibodies against PsrP, a novel Streptococcus pneumoniae adhesin, block adhesion and protect mice against pneumococcal challenge. J. Infect. Dis. 198:375–383 [DOI] [PubMed] [Google Scholar]
- 34. Schadt H. S., Schadt S., Oldach F., Sussmuth R. D. 2009. 2-Amino-2-deoxyisochorismate is a key intermediate in Bacillus subtilis p-aminobenzoic acid biosynthesis. J. Am. Chem. Soc. 131:3481–3483 [DOI] [PubMed] [Google Scholar]
- 35. Shoma S., et al. 2011. Development of a multiplexed bead-based immunoassay for the simultaneous detection of antibodies to 17 pneumococcal proteins. Eur. J. Clin. Microbiol. Infect. Dis. 30:521–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stocker B. A. 1988. Auxotrophic Salmonella typhi as live vaccine. Vaccine 6:141–145 [DOI] [PubMed] [Google Scholar]
- 37. Trzcinski K., Thompson C. M., Lipsitch M. 2003. Construction of otherwise isogenic serotype 6B, 7F, 14, and 19F capsular variants of Streptococcus pneumoniae strain TIGR4. Appl. Environ. Microbiol. 69:7364–7370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Walsh C. T., Liu J., Rusnak F., Sakaitani M. 1990. Molecular studies on enzymes in chorismate metabolism and the enterobactin biosynthetic pathway. Chem. Rev. 90:1105–1129 [Google Scholar]
- 39. Wegkamp A., van Oorschot W., de Vos W. M., Smid E. J. 2007. Characterization of the role of para-aminobenzoic acid biosynthesis in folate production by Lactococcus lactis. Appl. Environ. Microbiol. 73:2673–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Weinberger D. M., et al. 2009. Pneumococcal capsular polysaccharide structure predicts serotype prevalence. PLoS Pathog. 5:e1000476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wernette C. M., et al. 2003. Enzyme-linked immunosorbent assay for quantitation of human antibodies to pneumococcal polysaccharides. Clin. Diagn. Lab. Immunol. 10:514–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ye Q. Z., Liu J., Walsh C. T. 1990. p-Aminobenzoate synthesis in Escherichia coli: purification and characterization of PabB as aminodeoxychorismate synthase and enzyme X as aminodeoxychorismate lyase. Proc. Natl. Acad. Sci. U. S. A. 87:9391–9395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Young D. B. 2003. Building a better tuberculosis vaccine. Nat. Med. 9:503–504 [DOI] [PubMed] [Google Scholar]
- 44. Zhang Z., Clarke T. B., Weiser J. N. 2003. Cellular effectors mediating Th17-dependent clearance of pneumococcal colonization in mice. J. Clin. Invest. 119:1899–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]