Abstract
The Treponema denticola outer membrane lipoprotein-protease complex (dentilisin) contributes to periodontal disease by degrading extracellular matrix components and disrupting intercellular host signaling pathways. We recently demonstrated that prcB, located upstream of and cotranscribed with prcA and prtP, encodes a 22-kDa lipoprotein that interacts with PrtP and is required for its activity. Here we further characterize products of the protease locus and their roles in expression, formation, and localization of outer membrane complexes. PrcB migrates in native gels as part of a >400-kDa complex that includes PrtP and PrcA, as well as the major outer sheath protein Msp. PrcB is detectable as a minor constituent of the purified active protease complex, which was previously reported to consist of only PrtP and auxiliary polypeptides PrcA1 and PrcA2. Though it lacks the canonical ribosome binding site present upstream of both prcA and prtP, PrcB is present at levels similar to those of PrtP in whole-cell extracts. Immunofluorescence microscopy demonstrated cell surface exposure of the mature forms of PrtP, PrcA1, PrcB, and Msp. The 16-kDa N-terminal acylated fragment of PrtP (predicted to be released during activation of PrtP) was present in cell extracts but was detected neither in the purified active protease complex nor on the cell surface. PrcA2, detectable on the surface of Msp-deficient cells but not that of wild-type cells, coimmunoprecipitated with Msp. Our results indicate that PrcB is a component of the outer membrane lipoprotein protease complex and that Msp and PrcA2 interaction may mediate formation of a very-high-molecular-weight outer membrane complex.
INTRODUCTION
The Treponema denticola locus encoding the dentilisin protease complex, including PrcB, PrcA, and PrtP, is conserved in oral Treponema spp. (13, 28, 33). In T. denticola, PrcB, PrcA, and PrtP are acylated, as shown by sequence analysis and behavior in phase-partitioned detergent extracts (20, 24). Analysis of defined mutant strains suggests that PrcB and PrcA are required for native expression of PrtP protease activity (24, 31). PrtP is one of only two known lipoproteins in the subtilase family of subtilisin-like serine proteases (44), the other being SphB1, the subtilisin autotransporter that catalyzes maturation of the virulence factor FhaB (filamentous hemagglutinin) at the surface of Bordetella pertussis (14). Other bacterial subtilases involved in pathogenesis include the SubAB cytotoxin of Shiga-toxigenic Escherichia coli (39).
Studies with isogenic T. denticola mutants implicate PrtP protease activity in induction of matrix metalloproteinase 2 (MMP-2)-dependent fibronectin fragmentation in periodontal ligament cells (38), in epithelial tissue penetration due to disruption of intercellular junction integrity (12), and in cleavage of complement regulatory protein factor H subsequent to its binding to T. denticola (35). Bacterial lipoproteins are potent activators of both innate inflammatory responses and apoptosis (1), and surface-exposed spirochetal lipoproteins induce such responses through Toll-like receptor 2 (TLR2) (1, 29). Though the mechanism of TLR2 activation by T. denticola has not been determined (3, 42), T. denticola lipoproteins, such as the components of the dentilisin complex, may act as pathogen-associated microbial patterns (PAMPs) (37) in initiation of TLR-mediated responses (36).
Previous work by Grenier et al. utilized immunogold transmission electron microscopy to detect the T. denticola protease complex on the outer surface of the cell (25) but did not address localization or expression of its individual components. Previous studies by our group and others (5, 24, 32, 34) have identified the protease operon and its products (summarized in Fig. 1). In addition to PrtP, two other proteins are encoded in the protease locus: PrcB (5, 24) and PrcA, whose posttranslational cleavage to PrcA1 and PrcA2 is dependent on PrtP activity (34). PrtP, PrcA, and PrcB are lipoproteins, as shown by sequence analysis and behavior in Triton X-114 extracts. Native PrtP is cleaved at residue 159 (32 and data not shown), which presumably activates the protease and releases a 16-kDa acylated N-terminal fragment of PrtP (herein designated PrtP-N). The mature protease complex has previously been reported to consist only of PrcA1, PrcA2, and activated PrtP (5, 32, 34, 45). We recently reported that PrcB, a lipoprotein encoded upstream of prcA, interacts with PrtP and is required for expression of PrtP and dentilisin activity but is not required for expression of PrcA (24).
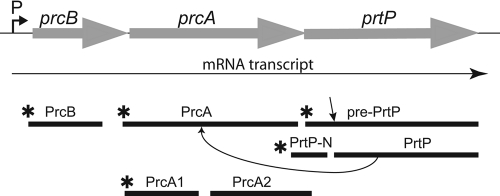
Fig. 1.
Structure and products of the T. denticola protease operon. The three genes of the protease operon (prcB, prcA, and prtP) are shown in gray above the ∼5-kb mRNA transcript. The location of the putative promoter (P) is indicated 5′ to prcB. The predicted lipoprotein products are shown below the transcript, with asterisks indicating acylation. The mature form of PrtP results from cleavage at residue 159, releasing a 16-kDa N-terminal acylated polypeptide. Cleavage of PrcA to PrcA1 and PrcA2 is dependent on PrtP protease activity.
Msp, one of the most highly expressed T. denticola proteins, is a pore-forming outer membrane protein that has lytic effects on erythrocytes and epithelial cells (20) and disrupts regulation of both actin (2, 46) and intracellular calcium (2, 47) in fibroblasts. Like other members of the porin family, native Msp is a detergent-stable trimeric complex. Any mutation in the protease operon that blocks prtP transcription results in greatly reduced Msp expression and absence of Msp trimers (5, 22, 30, 34). We hypothesized that proper expression of Msp requires either PrtP protease activity or specific interaction with a protease complex component.
Here, we present data on expression of proteins encoded in the prcB-prcA-prtP locus, show that PrcB is a component of the protease complex, confirm surface localization of Msp and most of the polypeptide products of the protease locus, and demonstrate protein-protein interaction between Msp and PrcA2. These results provide important clues as to the mechanisms of formation of high-molecular-weight outer membrane complexes in this periodontal pathogen and represent advances in understanding the molecular basis of expression of two of its key virulence factors.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
T. denticola ATCC 35405 (9) and isogenic msp mutant strain MHE (22) were grown in NOS broth medium as previously described (10, 27), with erythromycin (Em) (40 μg ml−1) added as appropriate. Cultures were examined by dark-field microscopy for purity and typical strain morphology.
E. coli JM109 (50) and E. coli Rosetta(DE3)/pLysS (Novagen, Inc., Madison, WI) were used as hosts for cloning and expression of recombinant proteins, respectively. E. coli was grown on LB agar or broth medium with ampicillin (50 μg ml−1), kanamycin (30 μg ml−1), and chloramphenicol (34 μg ml−1) as appropriate. Plasmid vector pSTBlue-1 (Novagen) was used for direct cloning of PCR products, and 6×His-tagged constructs were made in pET30b (Novagen).
Preparation of recombinant antigens for antibody production.
DNA fragments encoding PrcB, the N-terminal region of PrcA (PrcA1 polypeptide), and the N-terminal region of PrtP were amplified by PCR from T. denticola genomic DNA, initially cloned in pSTBlue-1 and maintained in E. coli JM109. For generation of 6×His-tagged constructs, the relevant DNA fragments were transferred to pET30b and then expressed in E. coli Rosetta (DE3)/pLysS as described previously (8, 24). E. coli cultures expressing 6×His-tagged proteins of interest were lysed by treatment with BugBuster (EMD Biosciences), and extracts were further disrupted by sonication. Proteins of interest were purified from cell extracts by nickel affinity chromatography (4). Eluted protein was used to immunize rabbits to generate polyclonal antibodies, as described previously (5).
Preparation of T. denticola extracts.
T. denticola cultures were harvested by centrifugation at 10,000 × g (10 min, 4°C), washed once in phosphate-buffered saline (PBS), and suspended in PBS at an optical density at 600 nm (OD600) of 0.2. Whole-cell lysates were prepared by sonication prior to suspension in sample buffer. For some experiments, Triton X-114 extraction and phase partitioning of outer membrane proteins were performed as described previously for T. pallidum (17), with slight modifications as described by Miao et al. (38). The final detergent-phase extract was then precipitated in acetone and resuspended in electrophoresis sample buffer.
Purification of T. denticola protease complex.
The PrtP protease complex was purified from the detergent phase of the Triton X-114 extracts of T. denticola MHE by preparative sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described by Miao et al. (38).
Polyacrylamide gel electrophoresis.
Standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was done as described previously (21). Whole-cell extracts were prepared by suspending T. denticola cells (1-ml cultures at an optical density at 600 nm of 0.2) in standard SDS-PAGE sample buffer containing β-mercaptoethanol and 2 mM phenylmethylsulfonyl fluoride (PMSF). Prior to electrophoresis, samples were either heated at 100°C for 5 min or held on ice. SDS-polyacrylamide gels were stained with Coomassie blue G-250 or silver nitrate.
Modified two-dimensional (2D) blue native PAGE (BN-PAGE) was conducted as described by Wittig et al. (49), with the following minor modifications. Bacteria were harvested by centrifugation at 5,000 × g for 5 min and resuspended in 1/10 volume solubilization buffer (50 mM imidazole-HCl, 500 mM aminocaproic acid, 0.2% Triton X-100, 1 mM PMSF, pH 7). Following solubilization for 30 min at 4°C and centrifugation at 5,000 × g for 5 min, Coomassie blue G-250 and glycerol were added to the supernatant at final concentrations of 0.25% (wt/vol) and 10% (vol/vol), respectively. Samples (15 μl lane−1) were separated on 4 to 16% native gels (Invitrogen, Carlsbad, CA) at 4°C, starting at 100 V until the sample entered the gel and then with current limited to 15 mA and voltage limited to 500 V in running buffer (50 mM bis-Tris, 50 mM tricine, pH 6.8, with 0.02% Coomassie blue G-250 in the cathode buffer). After the dye front had moved about one-third of the desired total running distance, the cathode buffer was replaced with running buffer lacking dye. Following electrophoresis in the first dimension and noting the position of molecular weight standards, the gel was sliced into 6 horizontal strips. Proteins were eluted from each strip in a Bio-Rad model 422 electroeluter (Bio-Rad, Hercules, CA) following the manufacturer's instructions. For second-dimension electrophoresis, eluted protein samples were heated at 100°C for 5 min and subjected to standard SDS-PAGE.
Immunoblotting.
Standard Western immunoblotting of SDS-polyacrylamide gels was done as described previously (21). Proteins blotted to nitrocellulose membranes were detected with rabbit polyclonal antibodies raised against T. denticola whole cells (26), T. denticola chymotrypsinlike protease (CTLP) complex (25), or recombinant T. denticola proteins, followed by horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (Thermo Scientific, Rockford, IL). With the exception of antibodies against T. denticola whole cells and anti-CTLP, all primary antibodies used in these studies were raised against purified recombinant antigens and validated for specificity against the wild type and appropriate isogenic mutants (data not shown). Protein bands of interest were visualized using SuperSignal West Pico chemiluminescent substrate (Thermo Scientific).
Quantitative Western blotting was done essentially as described by Feissner et al. (19). Dilution series of recombinant PrtP and PrcB were quantitated by comparison with defined quantities of bovine serum albumin (BSA) on Coomassie blue-stained gels and then probed with specific antibodies to determine the approximate detection limit of each antibody. The dilution level of each antibody had previously been determined so as to ensure both specificity and sensitivity. Starting with T. denticola cultures at an optical density at 600 nm of 0.2 (approximately 5 × 108 ml−1), we made dilution series of whole-culture, cell-free supernatant and pelleted cells resuspended in an equal volume of PBS. Relative levels of T. denticola proteins of interest in each preparation were estimated from Western immunoblots probed with a cocktail of relevant antibodies at the predetermined dilutions.
Surface immunolabeling.
Surface localization of T. denticola proteins was evaluated by immunofluorescence as described previously for Leptospira (16, 23), with minor modifications as follows. Suspensions of live bacteria in PBS (30 μl; OD600 of 0.2) were placed on glass slides and fixed with 3% glutaraldehyde in PBS for 20 min, followed by thorough washing three times with PBS. For experiments requiring membrane permeabilization, fixed slides were incubated with 0.2% Triton X-100 in PBS for 10 min and washed with PBS. The slides were blocked with 5% nonfat dry milk (NFDM) in PBS for 30 min prior to incubation for 1 h at 37°C with specific primary rabbit antibodies, which were diluted at 1:200 in PBS and 1% NFDM. Slides were gently washed with PBS and blocked with 1% BSA before probing for 1 h at 37°C with DyLight549-conjugated goat anti-rabbit IgG (Thermo Scientific). Finally, the slides were washed with PBS, mounted with 90% glycerol, and visualized by dark-field and fluorescence microscopy at ×400 or ×1,000 magnification on a Reichert MicroStar IV microscope.
Immunoprecipitation assay.
Three-day, 10-ml cultures of T. denticola (OD600 of ∼0.5) were harvested by centrifugation at 5,000 × g for 5 min, washed with cold PBS, and incubated for 30 min at 4°C on a roller in 1 ml of Tris-buffered saline (TBS) (pH 7.4) containing 0.5% Triton X-100 and 10 μl protease inhibitor cocktail (Roche, Indianapolis, IN). The suspension was centrifuged at 13,000 × g for 10 min at 4°C to remove unlysed cells. Immunoprecipitation (IP) of T. denticola proteins from 3-day cultures of T. denticola was done as described previously (24) using antibodies raised against Msp, PrcA2, and FlaA and protein A agarose slurry (Thermo Scientific). Immune complexes were eluted with 100 μl of 0.2 M glycine-HCl, pH 2.5. Eluted samples were neutralized with 10 μl of 1 M Tris, pH 8, and processed for SDS-PAGE and immunoblotting. Immunoprecipitated proteins were detected using various rabbit polyclonal antibodies followed by detection using horseradish peroxidase-conjugated Clean-Blot IP detection reagent (Thermo Scientific).
RESULTS
PrcB is present in the purified protease complex.
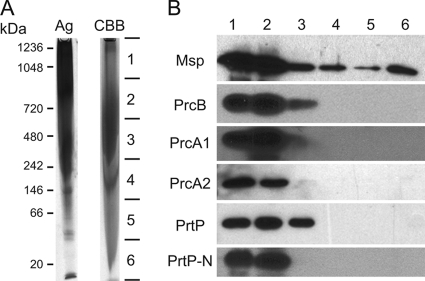
Studies of the PrtP serine protease complex (variously designated CTLP [45], dentilisin [32], and PAP [41] over the past 20 years) have consistently reported that the protease complex is composed of three polypeptides subsequently identified as PrtP (70 kDa), PrcA1 (30 kDa), and PrcA2 (40 kDa) (32, 34). As shown in Fig. 2A, the purified protease complex migrates as a complex of approximately 95 kDa that when boiled resolves to three prominent polypeptides. We recently demonstrated that PrcB, a 22-kDa lipoprotein encoded by the first gene in the protease operon, interacts with PrtP and is required for PrtP activity (24). PrcB has not previously been identified as a component of the active protease complex. To determine whether PrcB is present in the active protease complex, the purified protease complex was subjected to Western immunoassays. As shown in Fig. 2B, specific antibodies react with the three major polypeptides of the protease complex (PrcA1, PrcA2, and PrtP) as expected, and PrcB is also detected in the purified protease complex. We thus conclude that the faint band visible at approximately 22 kDa in Fig. 2A is PrcB.
Fig. 2.
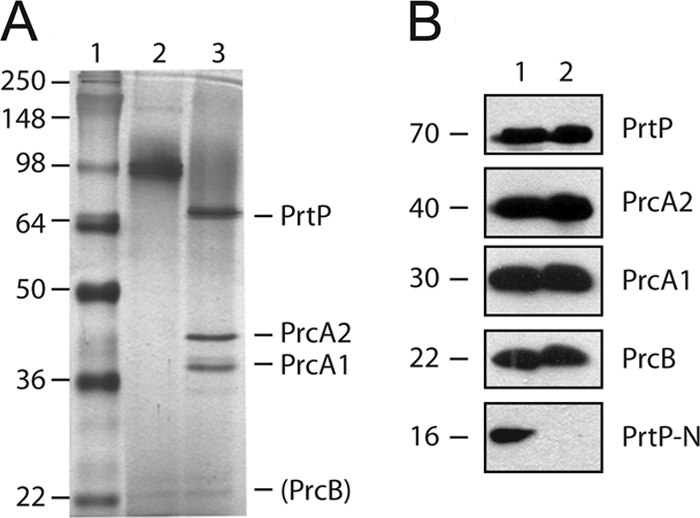
Detection of products of the prcB-prcA-prtP operon in T. denticola and purified protease complex. (A) Silver-stained SDS-polyacrylamide gel of purified protease complex (0.4 μg per lane). Lanes: 1, molecular mass standards (kDa); 2, unheated protease complex; 3, boiled protease complex. The locations of the PrtP, PrcA1, and PrcA2 bands are indicated, as is that of the putative PrcB band. (B) Immunoblots of T. denticola whole-cell extracts (equivalent to 10 μl culture per lane) and purified protease complex (0.4 μg per lane) probed with antibodies specific for PrcB, PrcA1, PrcA2, PrtP N-terminal domain, and mature PrtP. The approximate size of each polypeptide (in kDa) is indicated.
The N-terminal acylated fragment of PrtP is absent in purified protease complex.
PrtP is predicted to be a lipoprotein, but in the active protease complex PrtP is cleaved, presumably autocatalytically, resulting in release or degradation of a 16-kDa N-terminal acylated polypeptide. To determine the fate of this polypeptide, we examined cell extracts and purified protease complex for the presence of the N-terminal portion of PrtP (PrtP-N). As shown in Fig. 2B, PrtP-N was present in cell extracts as a 16-kDa polypeptide but was not detected in the purified protease complex. Consistent with its predicted acylation, PrtP-N segregated to the detergent phase of Triton X-114 extracts of T. denticola cells (data not shown).
Very-high-molecular-mass outer membrane complexes of T. denticola.
The protease complex and oligomeric Msp protein migrate as complexes of approximately 95 and 150 kDa, respectively, when unheated samples are subjected to standard SDS-PAGE. Both complexes have also been reported as having much higher apparent-molecular-weight forms, as detected by gelatin zymography (PrtP [41]) and Western immunoblotting (Msp [26, 48]). To determine the native form and composition of the protease complex, we subjected T. denticola lysates to modified two-dimensional blue native PAGE, in which protein fractions eluted from the first-dimension native gel were separated in the second dimension by standard SDS-PAGE and immunoblotted. As shown in Fig. 3B, both PrtP and PrcB were represented equally in fractions from 250 kDa to >1,000 kDa and were not detected in lower-molecular-mass fractions. PrcA1 and PrcA2 were detected in fractions of >700 kDa but not in lower-molecular-mass fractions. In comparison, Msp was detected primarily at >700 kDa and was also present at much lower levels in subsequent fractions.
Fig. 3.
High-molecular-mass protein complexes of T. denticola. (A) T. denticola extract separated by BN-PAGE and stained with silver nitrate (Ag) or Coomassie blue (CBB). Molecular mass standards and horizontal gel slices 1 to 6 are indicated on the left and right, respectively. (B) Immunoblots of proteins eluted from BN-PAGE fractions. Lane numbering indicates BN-PAGE fractions 1 to 6, as shown in panel A. PrtP-N, PrtP, PrcA1, PrcA2, PrcB, and Msp were detected with specific rabbit polyclonal antibodies.
Relative expression levels of protease complex proteins.
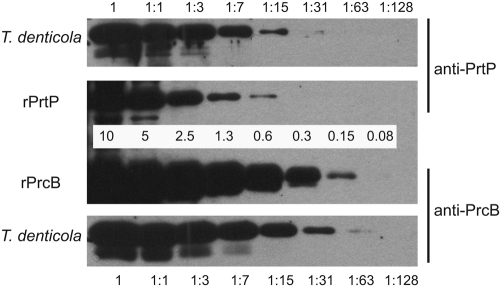
It is evident that the purified protease complex contains PrtP and PrcA polypeptides in a 1:1 molar ratio (Fig. 2A). Although PrcB is detected in Western blots, it appears to be present at very low levels relative to those of PrcA and PrtP in the purified protease complex. We hypothesized that this might be due to inefficient translation initiation of PrcB relative to that of PrcA and PrtP from the polycistronic prcB-prcA-prtP operon. Unlike prtP and prcA, the prcB coding region is not preceded by a canonical Shine-Dalgarno (SD) sequence, suggesting that PrcB protein might be translated and expressed at lower efficiency than PrtP and PrcA. The deduced mRNA sequence 5′ to prcB contains several poly-U sequences that could mediate translation initiation by an SD-independent mechanism (6). Because it is not possible to directly determine relative protein expression levels from immunoblots using different antibodies, we performed semiquantitative Western immunoassays using a cocktail of antibodies raised against PrcB and PrtP whose relative sensitivity was determined using known quantities of the respective recombinant proteins. The antibodies detected approximately 0.15 ng (anti-PrcB) and 0.6 ng (anti-PrtP) of recombinant target protein (Fig. 4, middle). By probing a serially diluted T. denticola cell extract with the same cocktail of antibodies (Fig. 4, top and bottom), we calculated a PrtP-PrcB ratio (wt/wt) of 2.5 to 3.2. Taking into account the ratio between their molecular masses (approximately 70 kDa and 22 kDa, respectively), we concluded that PrtP and PrcB are expressed at a molar ratio not markedly different from 1:1.
Fig. 4.
Quantitative Western blot analysis of PrtP and PrcB. Detection limits of the antibodies were determined by probing a calibrated mixture of recombinant PrtP (rPrtP) and recombinant PrcB (rPrcB) with a cocktail of anti-PrtP and anti-PrcB antibodies. The amounts of each recombinant protein in the calibration blot (in ng per lane) are shown in the middle. A serially diluted T. denticola culture was probed with a cocktail of anti-PrtP and anti-PrcB antibodies. The dilution series for T. denticola culture is shown for the top and bottom panels.
Localization of T. denticola protease complex and Msp.
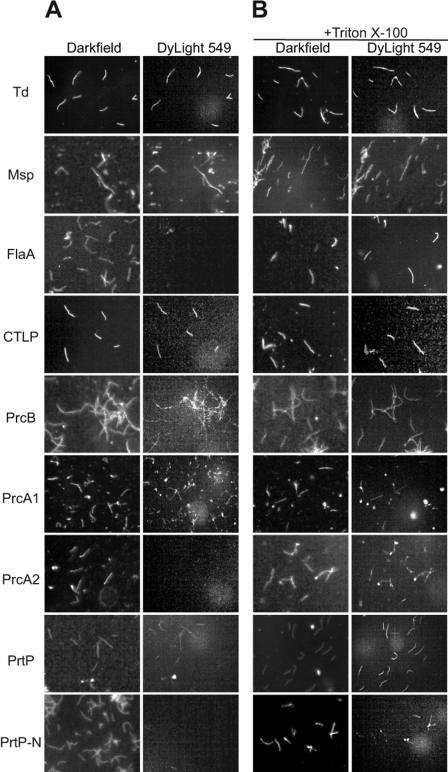
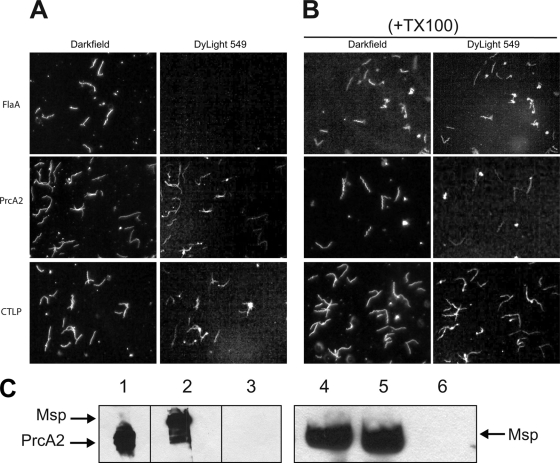
Grenier et al. used antibodies raised against gel-purified protease complex (CTLP complex) in immunogold transmission electron microscopy to detect the protease complex on the T. denticola outer membrane (25). Determination of surface localization of Msp has been controversial, with studies reporting significant extracellular exposure on the outer membrane or periplasmic localization, depending on the detection methodology and reagents utilized (7, 20, 48). To confirm that both the protease complex and the Msp complex are surface exposed on intact cells, we performed immunofluorescence microscopy of intact and detergent-permeabilized T. denticola cells. Both intact and permeabilized T. denticola cells were probed with antibodies raised against whole cells, the CTLP complex, Msp, and the periplasmic FlaA protein. As shown in Fig. 5, both the protease complex and Msp were clearly labeled on intact cells, while FlaA was labeled only on permeabilized cells. Parallel studies using T. denticola cells immobilized in low-melting-point agarose droplets prepared by the method of Cox et al. (15) yielded the same results (data not shown).
Fig. 5.
Localization of T. denticola major surface protein and protease complex polypeptides. A surface localization immunofluorescence assay was performed with T. denticola ATCC 35405 with or without membrane permeabilization, using polyclonal antibodies raised against the indicated proteins or polypeptides followed by DyLight549-conjugated goat anti-rabbit IgG. T. denticola cells were visualized by dark-field microscopy and by fluorescence microscopy. Photomicrographs from representative experiments are shown. (A) Intact cells. (B) Permeabilized cells.
Localization of protease complex polypeptides.
Aside from its composition and surface localization, little is known of the structure of the native protease complex or its presentation on the cell surface. To determine the antibody accessibility of individual protease complex polypeptides, we performed immunofluorescence assays of intact and detergent-permeabilized T. denticola ATCC 35405. As shown in Fig. 5, PrtP, PrcA1, and PrcB were detected on the surface of intact cells, while PrcA2 and PrtP-N were detected only on permeabilized cells. Because PrcA2 can be detected in culture supernatants (data not shown) and is present in the active protease complex (Fig. 2), it seemed likely that absence of anti-PrcA2 antibody reactivity on intact cells was due to masking of PrcA2 by the conformation of the protease complex or other surface components. Interestingly, PrcA2 was readily visualized on the surface of Msp-deficient isogenic mutant T. denticola MHE (Fig. 6A), indicating that absence of Msp rescues the antibody accessibility of PrcA2 on the T. denticola surface. Using the agarose droplet method of Cox et al. (15), similar results were obtained for PrcA2 in both T. denticola ATCC 35405 and MHE (data not shown). As for PrtP-N (the 16-kDa acylated N-terminal fragment cleaved from active PrtP), it is also possible that conformation of the protease complex may contribute to lack of antibody reactivity. Alternatively, PrtP-N may be localized to the inner leaflet of the outer membrane.
Fig. 6.
Evidence for interaction between Msp and PrcA2. (A and B) Surface localization immunofluorescence assay using T. denticola MHE (isogenic msp-deficient mutant of ATCC 35405) without (A) or with (B) membrane permeabilization, using polyclonal antibodies raised against the indicated proteins or polypeptides followed by DyLight549-conjugated goat anti-rabbit IgG. T. denticola cells were visualized by dark-field microscopy and by fluorescence microscopy. Photomicrographs from representative experiments are shown. (C) Coimmunoprecipitation of Msp and PrcA2 from T. denticola ATCC 35405 cell extracts. Lanes 1 to 3 show immunoblots of an anti-Msp immunoprecipitate probed with antibodies raised against PrcA2 (lane 1), Msp (lane 2), and FlaA (lane 3). Lanes 4 (anti-PrcA2 immunoprecipitate), 5 (anti-Msp immunoprecipitate), and 6 (anti-FlaA immunoprecipitate) show an immunoblot probed with anti-Msp antibodies. The locations of Msp (53 kDa) and PrcA2 (40 kDa) are indicated.
Coimmunoprecipitation of Msp and PrcA2.
To confirm the interaction between Msp and PrcA2 suggested by the immunofluorescence assay showing cell surface binding of PrcA2-specific antibodies only on the Msp-deficient mutant (Fig. 6A), we performed immunoprecipitation assays using antibodies raised against recombinant Msp, PrcA2, and FlaA polypeptides. As shown in Fig. 6B, both anti-Msp and anti-PrcA2 pulled down both Msp and PrcA2, while anti-FlaA immunoprecipitated neither Msp nor PrcA2. Antibodies raised against components of the protease complex other than PrcA2 did not coimmunoprecipitate Msp from T. denticola lysates (data not shown). These results provide strong evidence of specific interaction between Msp and PrcA2 that likely contributes to the formation of the very-high-molecular-weight outer membrane complex consisting of Msp and the dentilisin protease complex.
DISCUSSION
The T. denticola protease locus encodes three lipoproteins, at least two of which undergo further posttranslational cleavage. This study is the first to examine the fate of all five polypeptide products of the protease locus. In addition to PrcB, these include the products of PrcA (PrcA1, acylated N-terminal 30-kDa polypeptide; PrcA2, C-terminal 40-kDa polypeptide) and PrtP (PrtP, mature 65-kDa polypeptide; PrtP-N, N-terminal acylated 16-kDa polypeptide). All five polypeptides were detected as part of a very-high-molecular-weight complex in BN-PAGE, and all but PrtP-N were detected in the purified active protease complex. All but PrtP-N and PrcA2 were visualized on the surface of intact T. denticola ATCC 35405, though PrcA2 was detected on the surface of a T. denticola msp mutant strain.
We show here that T denticola PrcB is part of the PrtP lipoprotein-serine protease complex. PrcB has not previously been identified as a component of the active protease complex. Studies of the PrtP protease complex (variously designated CTLP [45], dentilisin [32], and PAP [41] over the past 20 years) have consistently reported that the protease complex is composed of three polypeptides, identified as PrtP (65 to 72 kDa), PrcA1 (30 kDa), and PrcA2 (40 kDa). Interestingly, closer examination of a Coomassie blue-stained gel image of the purified protease components in one previous study (32) and a silver-stained gel image in another (24) reveals a very faintly staining band at a molecular weight similar to that of PrcB. Our results indicate that prior failures to detect PrcB in the purified active protease complex are likely due to detergent sensitivity of the PrcB-PrtP interaction. These results suggest that, while analysis of the active protease complex has been greatly facilitated by its stability in the presence of SDS, this feature also helped perpetuate an absence of attention to the likelihood that other proteins might also be part of the protease complex.
We show here that T. denticola PrcB and PrtP proteins are expressed at similar levels. While the methodology utilized falls within the rather amorphous category of “semiquantitative” assays, it was sufficient to show that there is not a large difference between protein expression levels of PrcB and PrtP. This is significant for several reasons. Expression levels of PrcA and PrtP appear to be equal based on gel electrophoresis of the purified protease complex. This is consistent both with the presence of conserved SD sequences optimally localized 5′ to the ATG start codons of both prcA and prtP and with a model of the protease complex that assumes a 1:1:1 molar ratio among PrtP, PrcA1, and PrcA2. As noted in our previous study (24), we determined that the current T. denticola genome annotation misidentifies the PrcB N-terminal residue and that PrcB is a 22-kDa lipoprotein. The genome annotation identifies PrcB as a 17-kDa periplasmic protein whose translation is dependent on a poorly conserved SD sequence that our results showed to be within the PrcB coding region. Because there is (i) no identifiable SD sequence 5′ to the actual start codon and (ii) very little PrcB in the purified protease complex, we first hypothesized that PrcB might be expressed at very low levels relative to PrcA and PrtP. This would have been consistent with a model in which PrcB functions as a molecular chaperone in assembly of the PrcA1-PrcA2-PrtP complex, dependent on a transient association between PrcB and PrtP. Our results show that PrcB is expressed at levels not greatly different from that of PrtP, suggesting that the noncanonical translation initiation signal 5′ to the PrcB start codon is relatively efficient at binding the ribosomal translation machinery. It is worth noting that the 5′ untranslated regions (UTRs) of prokaryotic genes are highly diverse and that non-SD translation initiation signals are quite common (11). Furthermore, our results support the hypothesis that PrcB is an integral component of the protease complex. This is consistent with our previous report of coimmunoprecipitation of PrcB and PrtP (24). Further studies are in progress to define the interacting domains of PrcB and PrtP and to further characterize transcription and translation of the protease operon.
While PrtP is very rare among subtilases in that it is acylated, it appears to conform to this enzyme class in other respects. For instance, like most subtilases, it is synthesized as a proenzyme that is subsequently secreted and then activated by cleavage of the propeptide (43). Our assumption is that cleavage of PrtP-N is an autocatalytic event. We previously reported that PrcA cleavage to PrcA1 and PrcA2 is dependent on PrtP activity (34). We are continuing to examine the role of PrtP activity in native protease complex expression and processing.
Cellular localization of membrane-associated proteins in spirochetes has been an ongoing issue. Controversies in this area have been most robust in the T. pallidum research field, due both to the relative lability of spirochete outer membranes and to the extreme paucity of confirmed surface-exposed proteins in this organism (40). Using methods designed to distinguish between proteins exposed on the cell surface and proteins localized to the periplasm or inner leaflet of the outer membrane (15, 16), we demonstrated that both the protease complex and the Msp complex are exposed on intact T. denticola cells. While surface localization of the protease complex has previously been reported by immunoelectron microscopy (25), the cellular localization of Msp has been somewhat controversial. T. denticola Msp is a porin-like outer membrane protein whose predicted secondary structure consists primarily of transmembrane beta-sheets with at least one and likely several significant surface-exposed domains (18, 21). However, a study by Caimano et al. proposed that Msp has little or no surface exposure. These conclusions were based on immunofluorescence studies using a poorly characterized rat antiserum raised against an undocumented recombinant Msp fusion polypeptide (7). In our studies, we used rabbit anti-Msp antibodies raised against the full-length mature Msp polypeptide (21). Our results using these antibodies (Fig. 5 and 6) combined with identical results using antibodies raised against native oligomeric Msp purified from T. denticola (data not shown) definitively establish the significant surface exposure of Msp on the outer membrane of intact T. denticola. We can only infer that the differences between the prior study and the present results are due to differences in antibody specificity, specifically in ability to recognize surface-exposed Msp epitopes on intact T. denticola cells.
Having demonstrated Msp surface localization, we then used Msp as a positive control for cell surface localization and the periplasmic protein FlaA as a control for subsurface or periplasmic localization. While Msp was visualized both on intact cells and on detergent-permeabilized cells, FlaA was detectable only on detergent-permeabilized cells. The protease complex is clearly localized on the T. denticola cell surface, but not all of the polypeptide products of the protease operon were detectable on intact cells. PrcB, PrcA1, and the mature form of PrtP are exposed on intact T. denticola cells. Interestingly, by these methods, neither PrcA2 nor PrtP-N was recognized on intact wild-type T. denticola, but PrcA2 was readily visualized on the surface of an Msp-deficient isogenic mutant strain.
The very-high-molecular-weight complex detected by 2D BN-PAGE and immunoblotting contained both Msp and the proteins comprising the protease complex. We hypothesized that the very-high-molecular-weight complex seen in BN-PAGE is composed of multiple copies of the Msp and PrtP complexes. While the possibility that other proteins may be part of the complex cannot be ruled out, we have no evidence of this. The presence of this very-high-molecular-weight complex suggested that there might be specific protein-protein interactions between Msp and one or more components of the protease complex. This was further supported by modified 2D BN-PAGE and immunoblotting of msp-deficient T. denticola MHE, in which the anti-PrtP-reactive very-high-molecular-weight complex resolved at approximately 150 to 200 kDa smaller than with the wild-type strain (data not shown). Using standard immunoprecipitation and immunoblotting, we demonstrated specific binding between Msp and PrcA2 in preparations of gently lysed T. denticola cells. This interaction between Msp and PrcA2 may help to explain the defective Msp phenotypes of isogenic T. denticola strains carrying mutations in the protease locus that affect expression of PrcA (5, 22, 30, 34). Studies of the role of the protease complex in expression and oligomerization of Msp are ongoing in our laboratory.
In summary, we have shown that PrcB is a component of the outer membrane lipoprotein protease (dentilisin) complex and is expressed at levels equivalent to those of other components of the complex. Having clearly demonstrated significant surface exposure of Msp, we showed that interaction between PrcA2 and Msp likely mediates formation of the very-high-molecular-weight outer membrane complex and may be required for proper expression of oligomeric Msp. These results provide important clues as to the mechanisms of formation of high-molecular-weight outer membrane complexes in this periodontal pathogen and represent advances in understanding the molecular basis of expression of two of its key virulence factors.
ACKNOWLEDGMENT
This work was supported by Public Health Service grant DE018221 (National Institute of Dental and Craniofacial Research).
Footnotes
Published ahead of print on 10 October 2011.
REFERENCES
- 1. Aliprantis A. O., et al. 1999. Cell activation and apoptosis by bacterial lipoproteins through Toll-like receptor-2. Science 285:736–739 [DOI] [PubMed] [Google Scholar]
- 2. Amin M., et al. 2004. Induction of de novo subcortical actin filament assembly by Treponema denticola major outer sheath protein. Infect. Immun. 72:3650–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Asai Y., Jinno T., Ogawa T. 2003. Oral treponemes and their outer membrane extracts activate human gingival epithelial cells through toll-like receptor 2. Infect. Immun. 71:717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ausubel F. M., et al. (ed.). 1995. Current protocols in molecular biology, vol. 1 Wiley Interscience, New York, NY [Google Scholar]
- 5. Bian X.-L., Wang H.-T., Ning Y., Lee S. Y., Fenno J. C. 2005. Mutagenesis of a novel gene in the prcA-prtP protease locus affects expression of Treponema denticola membrane complexes. Infect. Immun. 73:1252–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boni I. V., Isaeva D. M., Musychenko M. L., Tzareva N. V. 1991. Ribosome-messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic Acids Res. 19:155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Caimano M. J., Bourell K. W., Bannister T. D., Cox D. L., Radolf J. D. 1999. The Treponema denticola major sheath protein is predominantly periplasmic and has only limited surface exposure. Infect. Immun. 67:4072–4083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Capone R., Wang H. T., Sweier D. G., Lopatin D., Fenno J. C. 2008. Human serum antibodies recognize Treponema denticola Msp and PrtP protease complex proteins. Oral Microbiol. Immunol. 23:165–169 [DOI] [PubMed] [Google Scholar]
- 9. Chan E. C., et al. 1993. Treponema denticola (ex Brumpt 1925) sp. nov., nom. rev., and identification of new spirochete isolates from periodontal pockets. Int. J. Syst. Bacteriol. 43:196–203 [DOI] [PubMed] [Google Scholar]
- 10. Chan E. C. S., DeCiccio A., McLaughlin R., Klitorinos A., Siboo R. 1997. An inexpensive solid medium for obtaining colony-forming units of oral spirochetes. Oral Microbiol. Immunol. 12:372–376 [DOI] [PubMed] [Google Scholar]
- 11. Chang B., Halgamuge S., Tang S. L. 2006. Analysis of SD sequences in completed microbial genomes: non-SD-led genes are as common as SD-led genes. Gene 373:90–99 [DOI] [PubMed] [Google Scholar]
- 12. Chi B., Qi M., Kuramitsu H. K. 2003. Role of dentilisin in Treponema denticola epithelial cell layer penetration. Res. Microbiol. 154:637–643 [DOI] [PubMed] [Google Scholar]
- 13. Correia F. F., et al. 2003. Two paralogous families of a two-gene subtilisin operon are widely distributed in oral treponemes. J. Bacteriol. 185:6860–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Coutte L., et al. 2003. Surface anchoring of bacterial subtilisin important for maturation function. Mol. Microbiol. 49:529–539 [DOI] [PubMed] [Google Scholar]
- 15. Cox D. L., Akins D. R., Porcella S. F., Norgard M. V., Radolf J. D. 1995. Treponema pallidum in gel microdroplets: a novel strategy for investigation of treponemal molecular architecture. Mol. Microbiol. 15:1151–1164 [DOI] [PubMed] [Google Scholar]
- 16. Cullen P. A., et al. 2005. Surfaceome of Leptospira spp. Infect. Immun. 73:4853–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cunningham T. M., Walker E. M., Miller J. N., Lovett M. A. 1988. Selective release of the Treponema pallidum outer membrane and associated polypeptides with Triton X-114. J. Bacteriol. 170:5789–5796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Edwards A. M., Jenkinson H. F., Woodward M. J., Dymock D. 2005. Binding properties and adhesion-mediating regions of the major sheath protein of Treponema denticola ATCC 35405. Infect. Immun. 73:2891–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Feissner R., Xiang Y., Kranz R. G. 2003. Chemiluminescent-based methods to detect subpicomole levels of c-type cytochromes. Anal. Biochem. 315:90–94 [DOI] [PubMed] [Google Scholar]
- 20. Fenno J. C., et al. 1998. Cytopathic effects of the major surface protein (Msp) and the chymotrypsinlike protease (CTLP) of Treponema denticola. Infect. Immun. 66:1869–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fenno J. C., Müller K.-H., McBride B. C. 1996. Sequence analysis, expression and binding activity of recombinant major outer sheath protein (Msp) of Treponema denticola. J. Bacteriol. 178:2489–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fenno J. C., Wong G. W. K., Hannam P. M., McBride B. C. 1998. Mutagenesis of outer membrane virulence determinants of the oral spirochete Treponema denticola. FEMS Microbiol. Lett. 163:209–215 [DOI] [PubMed] [Google Scholar]
- 23. Figueira C. P., et al. 2011. Heterologous expression of pathogen-specific genes ligA and ligB in the saprophyte Leptospira biflexa confers enhanced adhesion to cultured cells and fibronectin. BMC Microbiol. 11:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Godovikova V., et al. 2010. Treponema denticola PrcB is required for expression and activity of the PrcA-PrtP (dentilisin) complex. J. Bacteriol. 192:3337–3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grenier D., Uitto V.-J., McBride B. C. 1990. Cellular location of a Treponema denticola chymotrypsinlike protease and importance of the protease in migration through the basement membrane. Infect. Immun. 58:347–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haapasalo M., Müller K.-H., Uitto V.-J., Leung W. K., McBride B. C. 1992. Characterization, cloning, and binding properties of the major 53-kilodalton Treponema denticola surface antigen. Infect. Immun. 60:2058–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Haapasalo M., Singh U., McBride B. C., Uitto V.-J. 1991. Sulfhydryl-dependent attachment of Treponema denticola to laminin and other proteins. Infect. Immun. 59:4230–4237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Heuner K., Bergmann I., Heckenbach K., Gobel U. B. 2001. Proteolytic activity among various oral Treponema species and cloning of a prtP-like gene of Treponema socranskii subsp. socranskii(1). FEMS Microbiol. Lett. 201:169–176 [DOI] [PubMed] [Google Scholar]
- 29. Hirschfeld M., et al. 1999. Cutting edge: inflammatory signaling by Borrelia burgdorferi lipoproteins is mediated by toll-like receptor 2. J. Immunol. 163:2382–2386 [PubMed] [Google Scholar]
- 30. Ishihara K., Kuramitsu H. K., Miura T., Okuda K. 1998. Dentilisin activity affects the organization of the outer sheath of Treponema denticola. J. Bacteriol. 180:3837–3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ishihara K., Kuramitsu H. K., Okuda K. 2004. A 43-kDa protein of Treponema denticola is essential for dentilisin activity. FEMS Microbiol. Lett. 232:181–188 [DOI] [PubMed] [Google Scholar]
- 32. Ishihara K., Miura T., Kuramitsu H. K., Okuda K. 1996. Characterization of the Treponema denticola prtP gene encoding a prolyl-phenylalanine-specific protease (dentilisin). Infect. Immun. 64:5178–5186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Klein E. A., Dewhirst F. E. 2006. Dentilisin is part of a conserved three-gene operon. J. Dent. Res. 85:2126 [Google Scholar]
- 34. Lee S. Y., et al. 2002. Cleavage of Treponema denticola PrcA polypeptide to yield protease complex-associated proteins PrcA1 and PrcA2 is dependent on PrtP. J. Bacteriol. 184:3864–3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McDowell J. V., Huang B., Fenno J. C., Marconi R. T. 2009. Analysis of a unique interaction between the complement regulatory protein factor H and the periodontal pathogen Treponema denticola. Infect. Immun. 77:1417–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Medzhitov R., Janeway C., Jr 2000. Innate immune recognition: mechanisms and pathways. Immunol. Rev. 173:89–97 [DOI] [PubMed] [Google Scholar]
- 37. Medzhitov R., Janeway C., Jr 2000. The Toll receptor family and microbial recognition. Trends Microbiol. 8:452–456 [DOI] [PubMed] [Google Scholar]
- 38. Miao D., Fenno J. C., Timm J. C., Joo N. E., Kapila Y. L. 2011. Treponema denticola chymotrypsin-like protease (dentilisin) induces matrix metalloproteinase-2-dependent fibronectin fragmentation in periodontal ligament cells. Infect. Immun. 79:806–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Paton A. W., Srimanote P., Talbot U. M., Wang H., Paton J. C. 2004. A new family of potent AB(5) cytotoxins produced by Shiga toxigenic Escherichia coli. J. Exp. Med. 200:35–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Radolf J. D., Norgard M. V., Schulz W. W. 1989. Outer membrane ultrastructure explains the limited antigenicity of virulent Treponema pallidum. Proc. Natl. Acad. Sci. U. S. A. 86:2051–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rosen G., Naor R., Sela M. N. 1999. Multiple forms of the major phenylalanine specific protease in Treponema denticola. J. Periodontal Res. 34:269–276 [DOI] [PubMed] [Google Scholar]
- 42. Rosen G., et al. 1999. Activation of murine macrophages by lipoprotein and lipooligosaccharide of Treponema denticola. Infect. Immun. 67:1180–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Siezen R. J. 1996. Subtilases: subtilisin-like serine proteases. Adv. Exp. Med. Biol. 379:75–93 [DOI] [PubMed] [Google Scholar]
- 44. Siezen R. J., Leunissen J. A. 1997. Subtilases: the superfamily of subtilisin-like serine proteases. Protein Sci. 6:501–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Uitto V.-J., Grenier D., Chan E. C., McBride B. C. 1988. Isolation of a chymotrypsinlike enzyme from Treponema denticola. Infect. Immun. 56:2717–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Visser M. B., Koh A., Glogauer M., Ellen R. P. 2011. Treponema denticola major outer sheath protein induces actin assembly at free barbed ends by a PIP2-dependent uncapping mechanism in fibroblasts. PLoS One 6:e23736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang Q., Ko K. S., Kapus A., McCulloch C. A., Ellen R. P. 2001. A spirochete surface protein uncouples store-operated calcium channels in fibroblasts: a novel cytotoxic mechanism. J. Biol. Chem. 276:23056–23064 [DOI] [PubMed] [Google Scholar]
- 48. Weinberg A., Holt S. C. 1991. Chemical and biological activities of a 64-kilodalton outer sheath protein from Treponema denticola strains. J. Bacteriol. 173:6935–6947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wittig I., Braun H. P., Schagger H. 2006. Blue native PAGE. Nat. Protoc. 1:418–428 [DOI] [PubMed] [Google Scholar]
- 50. Yanisch-Perron C., Vieira J., Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119 [DOI] [PubMed] [Google Scholar]