Abstract
Phagocytic responses are critical for effective host defense against opportunistic fungal pathogens. Macrophages sample the phagosomal content and orchestrate the innate immune response. Toll-like receptor 9 (TLR9) recognizes unmethylated CpG DNA and is activated by fungal DNA. Here we demonstrate that specific triggering of TLR9 recruitment to the macrophage phagosomal membrane is a conserved feature of fungi of distinct phylogenetic origins, including Candida albicans, Saccharomyces cerevisiae, Malassezia furfur, and Cryptococcus neoformans. The capacity to trigger phagosomal TLR9 recruitment was not affected by a loss of fungal viability or cell wall integrity. TLR9 deficiency has been linked to increased resistance to murine candidiasis and to restriction of fungal growth in vivo. Macrophages lacking TLR9 demonstrate a comparable capacity for phagocytosis and normal phagosomal maturation compared to wild-type macrophages. We now show that TLR9 deficiency increases macrophage tumor necrosis factor alpha (TNF-α) production in response to C. albicans and S. cerevisiae, independent of yeast viability. The increase in TNF-α production was reversible by functional complementation of the TLR9 gene, confirming that TLR9 was responsible for negative modulation of the cytokine response. Consistently, TLR9 deficiency enhanced the macrophage effector response by increasing macrophage nitric oxide production. Moreover, microbicidal activity against C. albicans and S. cerevisiae was more efficient in TLR9 knockout (TLR9KO) macrophages than in wild-type macrophages. In conclusion, our data demonstrate that TLR9 is compartmentalized selectively to fungal phagosomes and negatively modulates macrophage antifungal effector functions. Our data support a model in which orchestration of antifungal innate immunity involves a complex interplay of fungal ligand combinations, host cell machinery rearrangements, and TLR cooperation and antagonism.
INTRODUCTION
The emergence of invasive fungal infections as a major cause of morbidity and mortality in immunosuppressed patients (23) has prompted studies into how fungal pathogens are recognized by the host. Phagocytosis of fungi by macrophages, dendritic cells, and neutrophils promotes fungal clearance. The innate immune system senses fungi through pattern recognition receptors (PRRs), such as C-type lectin receptors (CLRs), Toll-like receptors (TLRs), and NOD-like receptors, that bind to fungal pathogen-associated molecular patterns (PAMPs) (20, 33). Ligation of PRRs by fungal PAMPs results in the induction of downstream intracellular events that elicit the pathogen-specific adaptive immune response (7, 33). Because the fungal cell wall is a dynamic, complex structure, a wide array of PAMPs are displayed at variable concentrations depending on fungal species and morphotype, which results in engagement of multiple host PRRs (27). Cooperation between CLRs and TLRs in the recognition of specific fungal PAMP combinations allows innate immune cells to discriminate between pathogens and fungal morphotypes (12, 29, 34).
The different members of the TLR family recognize a broad range of PAMPs (17, 21) and can be divided into two groups based on their subcellular localization: TLR1, -2, -4, -5, and -6 are expressed at the plasma membrane, whereas the nucleic acid-sensing TLRs, TLR3, -7, -8, and -9, are localized to intracellular compartments (4, 17). Evidence for the importance of TLRs in fungal defense is mounting. The environmental fungi Aspergillus fumigatus and Cryptococcus neoformans and the human commensals Candida albicans and Malassezia furfur are opportunistic human pathogens that can cause invasive infection with morbidity and mortality in immunocompromised persons (23). Human studies revealed that polymorphisms in the TLR4 and TLR9 genes are associated with increased susceptibility to pulmonary aspergillosis (6, 9). In vivo and in vitro studies have implicated TLR2, TLR4, and TLR9 in innate and adaptive immunity to C. albicans and A. fumigatus (26, 33). However, the precise roles of individual TLRs in the orchestration of the complex inflammatory pattern that follows in vivo fungal infection and the cell biological processes that underlie TLR-mediated host-pathogen interactions remain poorly understood.
TLR9 activation is a tightly regulated, multistep process involving TLR9 subcellular trafficking, proteolytic cleavage, and dimerization (11, 18, 19, 30). Recently, we demonstrated that phagocytosis of A. fumigatus by macrophages rapidly induces a dramatic redistribution of TLR9 to the phagosomal membrane, whereas bead-containing phagosomes in the same cell fail to acquire TLR9 (16). To investigate if the capacity to recruit TLR9 to the phagosome is unique to A. fumigatus, we examined TLR9 recruitment after phagocytosis of other clinically relevant fungal pathogens of distinct phylogenetic origins. In the current study, we demonstrate that the ability to trigger TLR9 recruitment is conserved across C. albicans, Saccharomyces cerevisiae, M. furfur, and C. neoformans. The highly conserved nature of the response indicates that TLR9 may serve an important role in macrophage antifungal defense. We sought to determine the impact of TLR9 deficiency on the macrophage antifungal immune response. Compared to wild-type (WT) macrophages, TLR9 knockout (TLR9KO) macrophages had comparable phagocytic capacities for uptake of C. albicans and S. cerevisiae and normal phagosome maturation. We found that TLR9 deficiency increased macrophage tumor necrosis factor alpha (TNF-α) production in response to C. albicans and S. cerevisiae, independent of yeast viability. The increase was reversible by functional complementation of the TLR9 gene, confirming that TLR9 was responsible for modulation of macrophage TNF-α production. Further investigation showed that TLR9 deficiency enhanced the macrophage antifungal effector response by increasing macrophage activation and microbicidal activity against C. albicans and S. cerevisiae.
MATERIALS AND METHODS
Reagents.
All products used for cell culture were from Invitrogen (Carlsbad, CA). All other reagents were purchased from Sigma unless noted otherwise.
Cell lines and cell culture.
RAW 264.7 and HEK293T cells were purchased from ATCC and cultured according to ATCC recommendations. Immortalized bone marrow macrophage cell lines derived from C57BL/6 and TLR9-deficient mice were a gift from Douglas Golenbock (University of Massachusetts Medical School, Worcester, MA) and were cultured and characterized phenotypically by analysis of macrophage surface marker expression and cytokine expression profiles as described previously (15).
Viral transduction.
Retroviral transduction of macrophages with TLR9-green fluorescent protein (TLR9-GFP) was performed as described previously (16). Briefly, HEK293T cells were transfected with plasmids encoding vesicular stomatitis virus glycoprotein G (VSV-G) and Gag-Pol as well as with the retroviral pMSCV vector, encoding mouse TLR9 fused at the C terminus to GFP (pMSCV-TLR9-GFP), which was a gift from Hidde Ploegh (Whitehead Institute for Biomedical Research, Cambridge, MA). Twenty-four and 48 h after transfection, the medium containing viral particles was collected, filtered through a 0.45-μm membrane, and added to RAW 264.7 macrophages or immortalized macrophages. The next day, cells were given fresh medium, and antibiotic selection in 5 μg/ml puromycin was initiated. Lentiviral transduction of macrophages was performed to express CD63-mRFP1 and CD82-mRFP1 as described previously (1, 16).
Fungal strains and growth.
The well-described Candida albicans wild-type strain SC5314 and Aspergillus fumigatus wild-type strain 293 were gifts from Eleftherios Mylonakis (Massachusetts General Hospital, Boston, MA). The wild-type Saccharomyces cerevisiae strain S288c (ATCC 204508) and wild-type Malassezia furfur (ATCC 14521) were obtained from ATCC. Cryptococcus neoformans H99 (serotype A) was used. All fungi were grown fresh for each experiment. A. fumigatus was grown on Sabouraud dextrose agar plates supplemented with 100 μg/ml ampicillin at 30°C for 3 to 5 days and then harvested by scraping and washing three times in phosphate-buffered saline (PBS). M. furfur was plated on Sabouraud dextrose agar plates supplemented with autoclaved olive oil and 100 μg/ml ampicillin and grown at 30°C for 3 to 5 days. M. furfur yeast cells were dislodged from plates by gentle scraping and were resuspended and washed three times in PBS–0.05% Tween. C. albicans and S. cerevisiae yeast cells were grown overnight at 30°C in yeast extract-peptone-dextrose (YPD) medium and then washed three times in PBS. C. neoformans was grown in standard YPD liquid medium. Cells were counted using a hemocytometer and adjusted to the desired organism density prior to use. For preparation of killed yeast cells, cells were resuspended in PBS and either incubated for 10 min at 100°C in a heating block or autoclaved at 121°F for 20 min. Control plating experiments confirmed that these treatments resulted in nonviable cells.
Phagocytosis assay.
After overnight growth, C. albicans or S. cerevisiae was labeled using Alexa Fluor 488 (Invitrogen, Carlsbad, CA) as described previously (16). Wild-type and TLR9KO macrophages were incubated with the indicated fungi at a multiplicity of infection (MOI) of 5:1. After washing with PBS, cells were fixed with 1% paraformaldehyde and washed again with PBS. Cells were assessed for green fluorescence on a FACSCalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ). Flow cytometry analysis was performed using FlowJo software (Tree Star Inc., Ashland, OR).
TNF-α and nitric oxide production assays.
Yeast cells were introduced to macrophages at the indicated ratios and were incubated for 6 h for TNF-α analysis. TNF-α production in the supernatant was measured using a DuoSet mouse TNF-α kit (R&D Systems) per the manufacturer's instructions. For nitric oxide production analysis, macrophages were first treated overnight with 10 ng/ml gamma interferon (IFN-γ; eBiosciences) and then exposed to yeast cells for 36 h. Supernatant was removed and tested for nitrite by using a Griess reagent system (Molecular Probes, Eugene, OR) according to the manufacturer's instructions.
XTT and CFU assays.
XTT (2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide) assay for assessment of fungal cell damage was performed as described previously (22, 31). Briefly, macrophages were plated and exposed to yeast cells in quadruplicate at the indicated ratios. Yeast-free and macrophage-free control wells were included in quadruplicate for all experiments. After coincubation with S. cerevisiae for 6 h or with C. albicans for 2 h, cells were subjected to hypotonic lysis by three gentle washes and a 30-min incubation with sterile distilled water. To facilitate cell lysis, membranes were disrupted mechanically by vigorous pipetting. After centrifugation at 1,500 × g for 3 min, supernatants were carefully removed. YPD medium containing 400 μg/ml of XTT and 50 μg/ml of coenzyme Q was added, and the plates were incubated for 2 h at 37°C. The optical density at 450 nm (OD450) was measured, and OD650 measurements were used to check for plate irregularities. Data were expressed as percentages of surviving fungal cells, determined according to the following formula: [(OD450 of yeast incubated with macrophages − OD450 of macrophages alone)/(OD450 of yeast alone − OD450 of medium alone)] × 100%. For the CFU assay, macrophages were exposed to S. cerevisiae at the indicated ratios overnight. After three washes with PBS, cells were subjected to lysis with 0.2% Triton X-100. Serial dilutions from each well were made in distilled water and plated in triplicate onto YPD agar plates. The number of CFU was determined manually after 48 h of incubation at room temperature.
Statistical analysis.
Standard deviations (SD) were determined using Microsoft Excel. In consultation with the MGH Biostatistics Center, we calculated errors based on the propagation of errors associated with each measurement. For comparisons of two groups, means ± SD were analyzed by two-tailed, unpaired Student's t test. Calculations were performed using an online statistical software package (http://www.graphpad.com/quickcalcs/ErrorProp1.cfm; GraphPad). Data were considered significantly different if the P value was ≤0.05.
Confocal microscopy.
Spinning disk confocal microscopy was performed on cells plated in complete medium in a chambered cover glass (Lab-Tek/Nunc, ThermoScientific, Rochester, NY) in a temperature-regulated environmental chamber. Fluorescence fungal surface labeling and live-cell imaging were performed as described previously (16). Briefly, cells were imaged on a Nikon Ti-E inverted microscope equipped with a CSU-X1 confocal head (Yokogawa). A coherent, 4-W, continuous-wave laser was used as an excitation light source to produce excitation at a wavelength of 488 nm. To acquire high-quality fluorescence images, a high-magnification, high-numerical-aperture (NA) objective was used (100×, 1.49 NA, oil immersion; Nikon). A halogen light source and air condenser (0.52 NA) were used for bright-field illumination, and a polarizer (MEN51941; Nikon) and Wollaston prisms (MBH76190; Nikon) were used to acquire differential imaging contrast (DIC) images. Images were acquired using an electron-multiplying charge-coupled device (EM-CCD) camera (C9100-13; Hamamatsu) and MetaMorph software (Molecular Devices, Downingtown, PA).
Transmission electron microscopy (TEM).
Yeast cells were fixed with 2.0% glutaraldehyde with 1.0% (vol/vol) dimethyl sulfoxide (DMSO). They were then poststained with a mixture of 0.1% ruthenium red (EMS) plus 1.0% osmium tetroxide (EMS) in cacodylate buffer for 1 h at room temperature. Cells were pelleted again, rinsed once with buffer, and then stabilized in 2.0% agarose for ease of handling. The pellets were dehydrated through a graded series of ethanol and embedded in Eponate resin (Ted Pella, Redding, CA) at 60°C overnight. Thin sections were cut on a Leica EM UC7 microtome and collected on Formvar-coated slot grids. Sections were examined in a JEOL 1011 transmission electron microscope at 80 kV, and images were collected using an AMT digital imaging system (Advanced Microscopy Techniques, Danvers, MA).
RESULTS
The fungal component triggering TLR9 recruitment is conserved across fungal taxonomic groups.
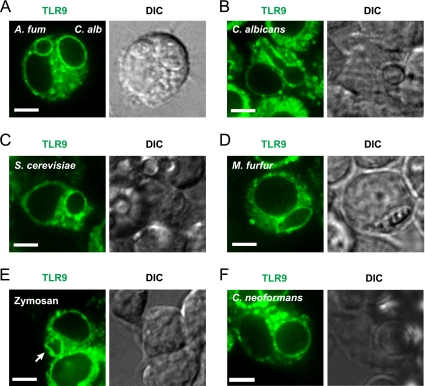
Previously, we demonstrated that TLR9 is robustly and specifically recruited to phagosomes containing A. fumigatus but not to bead-containing phagosomes, suggesting a role for TLR9 recruitment in innate immune responses to this invasive opportunistic human pathogen (16). However, the identity of the component of A. fumigatus that triggers accumulation of TLR9 on the phagosomal membrane remains to be identified. The fungal cell wall is a dynamic, complex structure with a strong immunostimulatory capacity (20). The main cell wall components differ between the major taxonomic groups of fungi, and the wall composition changes throughout cell division and conversion to different morphotypes. To investigate if the capacity to recruit TLR9 is unique to A. fumigatus or if this is a conserved feature across fungi of distinct phylogenetic origins, we examined TLR9 recruitment after phagocytosis of the fungal pathogens C. albicans, S. cerevisiae, Malassezia furfur, and Cryptococcus neoformans (23). C. albicans is the leading cause of invasive fungal infections (14), and TLR9 has been implicated in immune responses to C. albicans in vivo (5) and in vitro (24). We examined the fate of TLR9 after phagocytosis of WT C. albicans yeast cells by using a stable RAW macrophage cell line expressing TLR9-GFP and spinning disk confocal microscopy for live-cell imaging (16). We observed robust recruitment of TLR9 to the C. albicans phagosome within 1 h (Fig. 1A). In order to compare the level of TLR9 recruitment to what we observed previously for A. fumigatus phagosomes (16), we coexposed the macrophages to C. albicans yeast cells and A. fumigatus resting conidia and examined individual mammalian cells that had taken up both pathogens. The levels of TLR9 recruitment were comparable between C. albicans- and A. fumigatus-containing phagosomes (Fig. 1A) and phagosomes in macrophages which had been exposed to C. albicans alone (data not shown). As described previously (16), we confirmed that fungi resided within intracellular compartments by obtaining DIC images (Fig. 1A) and by serial imaging of the entire volume of the phagosome to confirm that TLR9 recruitment was uniform (see Movie S1 in the supplemental material). We examined cells for up to 4 h after phagocytosis. TLR9 accumulation on the phagosomal membrane was retained during hypha formation and was stable over the time observed (Fig. 1B). Interestingly, we also observed robust recruitment of TLR9 to the monomorphic organism S. cerevisiae (Fig. 1C), an ascomycete, demonstrating that TLR9 recruitment is independent of the pathogen's ability to switch between morphotypes and of cell wall surface changes related to filament formation. We next examined recruitment to the lipophilic yeast M. furfur, which is part of the human cutaneous commensal flora and is able to cause skin and systemic disease in predisposed individuals (3). In sharp contrast to those of A. fumigatus, C. albicans, and S. cerevisiae, the M. furfur cell wall contains a lipid-rich layer around the yeast cell that is poorly characterized but appears to be involved in inhibition of phagocytosis and killing (3). Strikingly, TLR9 was also recruited to M. furfur phagosomes (Fig. 1D), indicating that the lipid-rich layer did not shield the fungal component responsible for triggering TLR9 recruitment. We observed that TLR9 recruitment was still retained when cells were exposed to zymosan, a cell wall fragment of S. cerevisiae (Fig. 1E). In order to facilitate identification of its intracellular location, zymosan was fluorescently labeled (image not shown). Finally, we exposed macrophages to C. neoformans, a basidiomycete (Fig. 1F). Similar to other fungi, C. neoformans recruited TLR9 to the fungus-containing phagosome. Collectively, these observations support the notion that phagocytosed fungal content dictates recruitment of TLR9 to the phagosomal membrane. Our data demonstrate that the capacity to trigger TLR9 recruitment is a highly conserved feature of fungi of different taxonomic groups.
Fig. 1.
Fungi from different taxonomic groups trigger phagosomal recruitment of TLR9. (A to F) Confocal microscopy of RAW macrophages expressing TLR9-GFP (green). One focal plane is shown. Bar, 5 μm. (A) TLR9 is robustly recruited to C. albicans and A. fumigatus phagosomes. RAW cells were incubated with A. fumigatus resting conidia and C. albicans yeast cells for 1 h. A representative macrophage that has phagocytosed one C. albicans yeast cell (right) and one A. fumigatus resting conidium (left) is shown, demonstrating that both phagosomes have acquired TLR9. The DIC image demonstrates the presence of the two fungal organisms within the cell. (B) TLR9 accumulation on C. albicans phagosomes is retained during hypha formation. RAW cells were exposed to C. albicans yeast cells and incubated for 4 h. (C and D) TLR9 is recruited to S. cerevisiae (C)- and M. furfur (D)-containing phagosomes. RAW cells were incubated with yeast cells for 1 h. (E) Zymosan recruits TLR9 to the phagosome. The white arrow indicates the phagocytosed zymosan particle. RAW cells were incubated with zymosan for 30 min. (F) C. neoformans recruits TLR9 to the fungal phagosome. Representative images are shown in all panels.
Wild-type and TLR9-deficient macrophages exhibit comparable phagocytosis of fungi and similar phagosomal maturation.
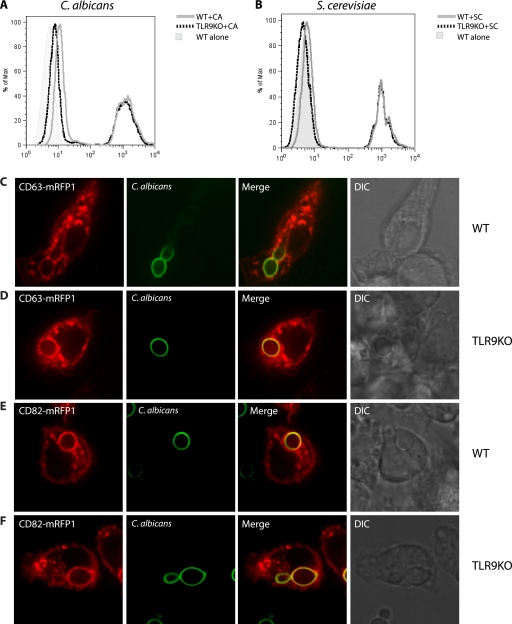
To explore the role of TLR9 in immunity to fungal pathogens, we used cell lines generated from bone marrow-derived macrophages derived from C57BL/6 (WT) or TLR9−/− (TLR9KO) mice. To ensure that phagocytosis was not impaired by the loss of TLR9, we incubated both WT and TLR9KO macrophages with either fluorescent C. albicans or S. cerevisiae for 2 h and then assessed the number of cells that had taken up fungi by flow cytometry. The numbers of WT and TLR9KO macrophages that had taken up either type of fungi were comparable (35% versus 31% for C. albicans [Fig. 2A] and 44% versus 40% for S. cerevisiae [Fig. 2B]), indicating that the selective deficiency of TLR9 does not affect uptake of fungi by macrophages. We next sought to determine if phagosomal maturation was impaired in TLR9KO macrophages. We expressed either CD63-mRFP1 or CD82-mRFP1 by lentiviral transduction in both WT and TLR9KO macrophages. We have previously shown that both tetraspanins are specifically recruited to fungal phagosomes, with distinct kinetics (1, 2). When fluorescent C. albicans was added to these cells, we used spinning disk confocal microscopy to assess the rate of appearance of these tetraspanins on the fungal phagosome. CD63-mRFP1 was recruited to C. albicans phagosomes in both WT and TLR9KO macrophages (Fig. 2C and D, respectively). Moreover, the rates of appearance of CD63-mRFP1 on fungal phagosomes were similar in WT and TLRKO macrophages. CD82-mRFP1 was also recruited to C. albicans phagosomes in both WT and TLR9KO macrophages (Fig. 2E and F, respectively). Again, the rates of appearance of CD82-mRFP1 on fungal phagosomes were similar in WT and TLRKO macrophages. These data indicate that WT and TLR9KO macrophages have comparable levels of phagocytosis of fungi and that the rate of phagosomal maturation is not affected by the selective loss of TLR9.
Fig. 2.
Wild-type and TLR9-deficient macrophages exhibit comparable phagocytosis of C. albicans and S. cerevisiae and similar phagosomal maturation. WT and TLR9KO macrophages were incubated with fluorescent C. albicans (A) or S. cerevisiae (B) at an MOI of 5:1 for 2 h. Cells were washed with PBS to remove extracellular yeast. Cells were then analyzed by flow cytometry. WT (C) and TLR9KO (D) macrophages expressing CD63-mRFP1 were exposed to C. albicans at an MOI of 1:1 and imaged immediately using spinning disk confocal microscopy. WT (E) and TLR9KO (F) macrophages expressing CD82-mRFP1 were exposed to C. albicans at an MOI of 1:1 and imaged immediately using spinning disk confocal microscopy.
TLR9 deficiency leads to increased macrophage TNF-α production in response to C. albicans and S. cerevisiae.
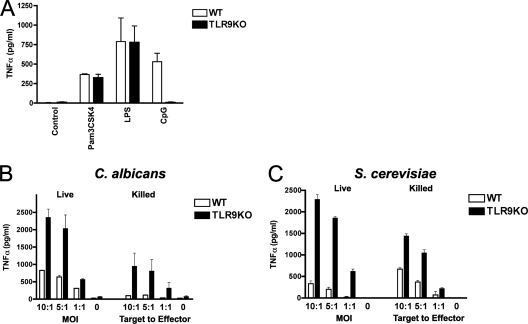
The role of TLR9 in antifungal defense is poorly understood. In vivo experiments suggested a detrimental effect of TLR9 in defense against C. albicans, as TLR9-deficient mice were highly resistant to candidiasis and showed more potent antifungal effector activity than WT mice (5). Our observation that induction of phagosomal TLR9 recruitment is a highly conserved response also suggests that TLR9 serves an important role in antifungal defense. To investigate how TLR9 modulates macrophage antifungal immune responses, we used immortalized bone marrow macrophages from WT and TLR9KO mice. As anticipated, TNF-α production in response to CpG was absent in TLR9KO macrophages, while the responses to the TLR2 ligand Pam3CSK4 and the TLR4 ligand lipopolysaccharide (LPS) were normal (Fig. 3A). Surprisingly, when we exposed macrophages to live C. albicans yeast cells at an MOI of 1:1, we found that TLR9KO cells produced significantly more TNF-α than WT cells (P < 0.003) (Fig. 3B). The increased TNF-α response in TLR9KO compared to WT macrophages was consistently significant for different MOIs (P < 0.04), and the difference was sustained when we used heat-killed (HK) or UV-treated C. albicans (data not shown). We next exposed cells to live and HK S. cerevisiae and again found that TLR9 deficiency resulted in increased TNF-α production (P < 0.01) (Fig. 3C). This difference between TLR9KO and WT cells was independent of yeast viability, although overall TNF-α production in response to HK yeast was lower than that in response to live yeast. The fungal cell wall contains a multitude of TLR ligands that can activate macrophages (20). Both TLR2 and TLR4 have been shown to play an important role in the recognition of fungi, either alone or in cooperation with other pattern recognition receptors, and they enhance innate effector functions (26, 33). While TLR9 stimulation through CpG induces TNF-α production, our data indicate that in the context of innate fungal recognition, TLR9 suppresses TNF-α production, possibly through cross talk with other pattern recognition receptors.
Fig. 3.
TLR9 deficiency leads to increased macrophage TNF-α production in response to C. albicans and S. cerevisiae. (A) Immortalized bone marrow-derived TLR9KO macrophages are unresponsive to CpG but retain responsiveness to other TLR agonists. WT and TLR9KO cells were incubated with 10 ng/ml Pam3CSK4, 1 ng/ml LPS, or 1 μM CpG for 4 h, and supernatants were analyzed for TNF-α by enzyme-linked immunosorbent assay (ELISA). (B and C) TLR9KO macrophages produce significantly more TNF-α than WT macrophages in response to C. albicans (B) and S. cerevisiae (C). WT and TLR9KO macrophages were exposed to live or heat-killed yeast cells at the indicated MOI or target-to-effector-cell ratio, respectively, and incubated for 6 h. Supernatants were analyzed for TNF-α by ELISA. Data represent means and SD and are representative of at least four independent experiments. P values were <0.04 (B) or <0.01 (C) for comparing cytokine secretion by WT and TLR9KO macrophages at all indicated ratios.
TLR9 is responsible for modulation of TNF-α production in WT macrophages.
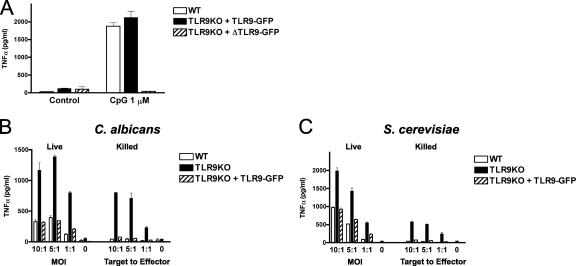
In order to confirm that the increase in fungus-induced TNF-α production was the result of the absence of TLR9, we sought to restore TLR9 gene function in TLR9KO macrophages to reconstitute the WT phenotype. We transduced TLR9KO macrophages with either a GFP-tagged version of wild-type TLR9 (TLR9-GFP) or a GFP-tagged deletion mutant lacking N-terminal TLR9 proteolytic cleavage residues 441 to 470 (ΔTLR9-GFP), which is uncleavable (30) and incapable of normal intracellular trafficking (16). As expected, TLR9-GFP restored the WT phenotype in response to CpG, whereas ΔTLR9-GFP-transduced control cells remained unresponsive to CpG (Fig. 4A). Importantly, functional complementation of TLR9 decreased the TNF-α production level to that in WT cells in response to both live and HK C. albicans (Fig. 4B) and S. cerevisiae (Fig. 4C). These data confirm that TLR9 is responsible for modulation of TNF-α production in WT macrophages.
Fig. 4.
TLR9 is responsible for modulation of TNF-α production in WT macrophages. (A) Functional complementation of TLR9KO macrophages with TLR9-GFP restores the WT phenotype in response to CpG. TLR9KO macrophages were transduced with either a GFP-tagged version of wild-type TLR9 (TLR9-GFP) or a GFP-tagged deletion mutant lacking N-terminal TLR9 proteolytic cleavage residues 441 to 470 (ΔTLR9-GFP). (B and C) Functional complementation of TLR9 decreases the TNF-α level to the WT level in response to both C. albicans (B) and S. cerevisiae (C) yeast cells. Macrophages were exposed to live or autoclaved yeast cells at the indicated MOI or target-to-effector-cell ratio, respectively, and incubated for 6 h. Supernatants were analyzed for TNF-α by ELISA. Data represent means and SD of cytokine concentrations and are representative of three independent experiments.
The fungal component responsible for TLR9 recruitment and activation is sustained after loss of fungal cell wall structural integrity.
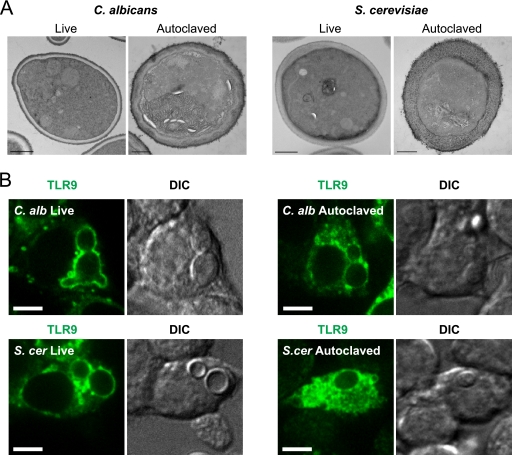
We next wished to study if the observed TLR9-mediated cytokine-suppressive effect was dependent on the intactness of the fungal cell wall surface and ultrastructure. The yeast cell wall is composed of the three main components chitin, β-glucans, and mannoproteins and consists of two layers, with chitin predominating in the inner layer and mannoproteins predominating in the outer cell wall (20, 28). Autoclaving yeast cells is a method often used to disrupt cell wall integrity and to release mannoproteins from the yeast cell wall (8, 13) Interestingly, when we measured TNF-α production in response to autoclaved C. albicans (Fig. 4B) and S. cerevisiae (Fig. 4C) yeast cells, we found that the relative increase in TNF-α production by TLR9KO cells was sustained and reversible by complementation with TLR9-GFP, indicating that the fungal cell wall component responsible for TLR9 activation was unaffected. For morphological examination of the yeast cell wall ultrastructure, we performed TEM on both live and autoclaved C. albicans and S. cerevisiae yeast cells (Fig. 5A). The images confirmed that the characteristic fungal cell wall architecture that is present in live yeast cells is disrupted after autoclave treatment. Confocal imaging revealed that in spite of the loss of the outer cell wall layer, autoclaved C. albicans and S. cerevisiae yeast cells still retained the capacity to recruit TLR9 to the phagosome (Fig. 5B), although less robustly than their live counterparts. Our data clearly demonstrate that the fungal component that triggers TLR9 recruitment and TLR9-mediated suppression of TNF-α production is sustained after removal of the outer cell wall mannoproteins and loss of cell wall integrity.
Fig. 5.
The fungal component responsible for triggering TLR9 recruitment is sustained after loss of fungal cell wall structural integrity. (A) The cell wall ultrastructure of live C. albicans and S. cerevisiae is disrupted by autoclave treatment. Transmission electron microscopy images show live and autoclaved C. albicans (left) and S. cerevisiae (right). Bar, 500 nm. (B) TLR9 recruitment to C. albicans and S. cerevisiae phagosomes is retained after autoclave treatment. Confocal microscopy images of RAW macrophages expressing TLR9-GFP (green) are shown with their DIC counterparts. RAW cells were incubated with live or autoclaved C. albicans or S. cerevisiae yeast cells for 1 h. Representative images are shown in all panels. One focal plane is shown. Bar, 5 μm.
TLR9 deficiency enhances the macrophage antifungal effector response by increasing macrophage activation and microbicidal activity against C. albicans and S. cerevisiae.
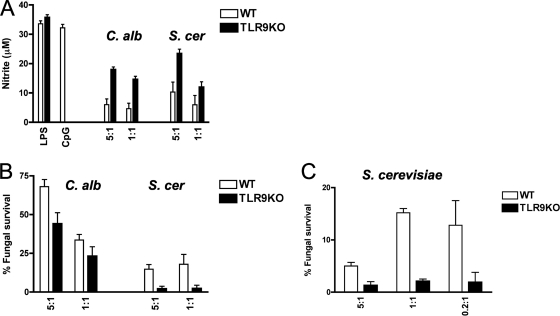
In order to gain more insight into the functional consequences of TLR9 activation in response to fungal infection, we next analyzed nitric oxide production in response to C. albicans and S. cerevisiae as a measure of macrophage activation. We found that TLR9KO cells produced more nitric oxide than WT cells in response to heat-killed C. albicans (P < 0.0001) and S. cerevisiae (P < 0.03), while the responses to LPS were equivalent (Fig. 6A), indicating that TLR9 specifically suppresses macrophage activation in response to fungi. We next assessed the effect of TLR9 on macrophage fungicidal activity by measuring fungal cell damage after exposure to macrophages, using an XTT assay. Because C. albicans hypha formation and phagosome escape resulted in macrophage cell death within 6 h, we coincubated macrophages with live C. albicans yeast for 2 h and then lysed the macrophages. We found that TLR9KO macrophages had significantly more potent antifungal activity, resulting in only 44% survival of C. albicans, compared to almost 70% survival after exposure to WT macrophages (P < 0.02) (Fig. 6B). Consistently, TLR9KO macrophages also showed more potent antifungal activity against S. cerevisiae, resulting in only 2% surviving fungal cells after 6 h of exposure, compared to 18% after exposure to WT macrophages (P < 0.003) (Fig. 6B). These data further support the notion that TLR9 suppresses macrophage activation and microbicidal activity in response to fungi. To confirm the XTT data using an independent assay, we exposed macrophages to S. cerevisiae overnight and then assessed fungal survival by measuring CFU. In agreement with the XTT assay data, TLR9KO macrophages showed significantly more potent antifungal activity against S. cerevisiae than WT macrophages did (P < 0.02) (Fig. 6C). Collectively, these data support a model in which TLR9 deficiency enhances the macrophage antifungal effector response by increasing macrophage activation, cytokine production, and microbicidal activity.
Fig. 6.
TLR9 deficiency leads to increased macrophage activation and microbicidal activity against C. albicans and S. cerevisiae. (A) TLR9 deficiency results in increased nitric oxide production in response to C. albicans and S. cerevisiae. WT and TLR9KO macrophages were pretreated with 10 ng/ml IFN-γ overnight and then exposed to heat-killed C. albicans or S. cerevisiae yeast cells at the indicated target-to-effector-cell ratio or to 0.1 μM CpG or 1 ng/ml LPS for 36 h. Supernatants were analyzed for nitrite by Griess assay as a measure of macrophage activation. Data represent means and SD of cytokine concentrations and are representative of three independent experiments. In comparing WT and TLR9KO macrophages, P values were <0.0001 for C. albicans and <0.03 for S. cerevisiae at all indicated target-to-effector-cell ratios. (B) TLR9 deficiency results in increased fungal cell damage of C. albicans and S. cerevisiae. WT and TLR9KO macrophages were incubated with C. albicans yeast cells for 2 h or with S. cerevisiae for 6 h. Fungal cell survival was then measured by XTT assay. Data represent means and SD for at least four independent experiments, each tested in triplicate. In comparing WT and TLR9KO macrophages, P values were <0.02 for C. albicans and <0.003 for S. cerevisiae at all indicated target-to-effector-cell ratios. (C) TLR9KO macrophages show more potent fungicidal activity against S. cerevisiae. WT and TLR9KO macrophages were incubated overnight and then assessed for fungal survival by measuring CFU. Data represent means and SD for at least four independent experiments, each tested in triplicate. P values were <0.02 for comparing WT and TLR9KO macrophages at all indicated target-to-effector-cell ratios.
DISCUSSION
In recent years, evidence has mounted for the importance of TLRs in antifungal defense (7, 20, 26, 33). However, it remains unclear how the dynamics of host-pathogen interaction contribute to TLR function and regulation during fungal pathogen uptake and whether fungal stimulation of multiple TLRs leads to synergism or antagonism. We recently reported that TLR9 specifically accumulates at the fungal phagosome after phagocytosis of A. fumigatus. We have now demonstrated that the capacity to induce phagosomal recruitment of TLR9 is conserved across clinically relevant pathogens from distinct fungal taxonomic groups, including C. albicans, S. cerevisiae, M. furfur, and C. neoformans. The functional implications of TLR9 activation in antifungal immunity are still poorly understood. We found that TLR9 deficiency enhanced the macrophage antifungal effector response by increasing macrophage TNF-α production, activation, and microbicidal activity against C. albicans and S. cerevisiae. To our knowledge, this report is the first to demonstrate that TLR9 negatively modulates macrophage antifungal immunity. Our data support a model in which orchestration of antifungal innate immunity involves a complex interplay of fungal PAMP combinations, host cell machinery rearrangements, and TLR cooperation and antagonism.
Though the best-known ligand for TLR9 is unmethylated bacterial and viral CpG-rich DNA, TLR9 has also been implicated in recognition of fungal DNA and induction of proinflammatory cytokines (24, 25, 32). However, these studies were performed using purified fungal DNA and likely reflect the minority of fungus-derived TLR9 ligands during fungal infection. In vivo studies evaluating the impact of TLR9 deficiency in a model of disseminated candidiasis yielded unclear results, with one group showing no difference in mortality and organ fungal growth between TLR9KO and WT mice (35) and another group showing that TLR9 deficiency significantly increased resistance to mucosal candidiasis and significantly reduced organ fungal growth (5). In vitro experiments using peritoneal TLR9KO macrophages and human peripheral blood mononuclear cells (PBMC) treated with a TLR9 antagonist suggested a mild defect in live C. albicans-induced interleukin-10 (IL-10) production (35). Conversely, blockage of TLR9 in human PBMC resulted in increased TNF-α and IL-6 production (35). These data are in line with our finding that TLR9 deficiency in macrophages increased TNF-α production in response to C. albicans and S. cerevisiae. Moreover, functional complementation of the TLR9 gene confirmed that suppression of TNF-α production was mediated by TLR9. As anticipated, the TLR9-mediated negative modulation of TNF-α production was only partial and recapitulated the WT phenotype. These data support the notion that the final cytokine response after fungal uptake is the result of both synergistic and antagonistic signals relayed by multiple PRRs.
The impact of TLR9 deficiency on intracellular macrophage antifungal effector function is illustrated by two lines of evidence. First, nitric oxide production in response to C. albicans and S. cerevisiae was elevated in TLR9KO macrophages, indicating increased macrophage activation. In addition, TLR9 deficiency resulted in higher macrophage microbicidal activity. Although TLR9KO macrophages caused more fungal cell damage than WT macrophages, as measured by metabolic activity in recovered C. albicans after 2 h of exposure, both macrophage lines were unable to contain C. albicans in vitro due to hypha formation, phagosomal escape, and macrophage killing. However, significantly increased microbicidal activity against S. cerevisiae was apparent even after prolonged incubation. Collectively, these data demonstrate that TLR9 deficiency enhances the antifungal effector functions of macrophages.
Remarkably, the dramatic rearrangement of TLR9 to the fungal phagosome was highly conserved in response to fungi from distinct taxonomic groups, and the fungal component triggering TLR9 recruitment was present despite a loss of fungal cell viability and cell wall architecture. Selective compartmentalization of pattern recognition receptors to the fungal phagosome was first described for TLR2 (34). TLR2 is robustly recruited to zymosan-containing phagosomes, where together with TLR6, it discriminates between pathogens (29, 34). We have now extended our previous observation that TLR9 is specifically recruited to A. fumigatus phagosomes (16) to include phagosomes containing C. albicans yeast or hyphae, M. furfur, S. cerevisiae, C. neoformans, or zymosan. Our data suggest that similar to the case for TLR2, TLR9 recruitment serves to effectively sample the contents of the phagosome and subsequently determine the nature of the pathogen. It is important to emphasize that TLR9 is recruited from an intracellular compartment, whereas TLR2 is recruited from the plasma membrane, and therefore phagosomal recruitment may not be mediated by the same mechanism. Moreover, the fungal factor triggering TLR9 recruitment and the fungal TLR9 ligand may not be the same. TLR9 recruitment could be induced by engagement and activation of another PRR by a ligand on the fungal cell wall while the fungal DNA that will engage TLR9 is still shielded. Alternatively, a fungal component may constitute an as yet unidentified ligand for TLR9, as the discovery that malarial hemozoin acts as a direct ligand for TLR9 demonstrated that TLR9 activation is not limited to nucleic acids (10). Future studies are needed to define the identities of the fungal components responsible for triggering TLR9 recruitment and activation.
In conclusion, our data demonstrate that TLR9 is selectively compartmentalized to fungal phagosomes and negatively modulates macrophage antifungal effector functions. While this observation may partially explain the increased in vivo resistance to candidiasis observed in TLR9KO mice by Bellochio et al. (5), the seemingly detrimental role of TLR9 in antifungal immunity is both surprising and puzzling. Negative modulation of immune responses by TLR9 has been described for autoimmunity, where TLR9 deletion does not ameliorate but rather exacerbates pathology in murine lupus models (9). Clearly, a better understanding of the complex web of synergistic and antagonistic signals that orchestrate immunity during the course of a fungal infection has both clinical and therapeutic potential.
Supplementary Material
ACKNOWLEDGMENTS
We thank Douglas Golenbock, Eleftherios Mylonakis, and Hidde Ploegh for kindly providing us with reagents and Robert Wheeler for helpful advice. We thank David Schoenfeld of the MGH Biostatistics Center for his assistance.
Electron microscopy was performed by Mary McKee in the Microscopy Core of the Center for Systems Biology/Program in Membrane Biology, Massachusetts General Hospital, Boston, MA, which is partially supported by an Inflammatory Bowel Disease grant (DK43351) and a Boston Area Diabetes and Endocrinology Research Center award (DK57521). J.M.V. was supported in part by MGH Department of Medicine internal funds, MGH ECOR funds, and the NIAID, NIH (1R01AI092084-01A1).
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
Published ahead of print on 26 September 2011.
REFERENCES
- 1. Artavanis-Tsakonas K., et al. 2011. The tetraspanin CD82 is specifically recruited to fungal and bacterial phagosomes prior to acidification. Infect. Immun. 79: 1098–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Artavanis-Tsakonas K., Love J. C., Ploegh H. L., Vyas J. M. 2006. Recruitment of CD63 to Cryptococcus neoformans phagosomes requires acidification. Proc. Natl. Acad. Sci. U. S. A. 103: 15945–15950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ashbee H. R., Evans E. G. 2002. Immunology of diseases associated with Malassezia species. Clin. Microbiol. Rev. 15: 21–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barton G. M., Kagan J. C. 2009. A cell biological view of Toll-like receptor function: regulation through compartmentalization. Nat. Rev. Immunol. 9: 535–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bellocchio S., et al. 2004. The contribution of the Toll-like/IL-1 receptor superfamily to innate and adaptive immunity to fungal pathogens in vivo. J. Immunol. 172: 3059–3069 [DOI] [PubMed] [Google Scholar]
- 6. Bochud P. Y., et al. 2008. Toll-like receptor 4 polymorphisms and aspergillosis in stem-cell transplantation. N. Engl. J. Med. 359: 1766–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brown G. D. 2010. How fungi have shaped our understanding of mammalian immunology. Cell Host Microbe 7: 9–11 [DOI] [PubMed] [Google Scholar]
- 8. Cabib E., Roberts R., Bowers B. 1982. Synthesis of the yeast cell wall and its regulation. Annu. Rev. Biochem. 51: 763–793 [DOI] [PubMed] [Google Scholar]
- 9. Carvalho A., et al. 2008. Polymorphisms in Toll-like receptor genes and susceptibility to pulmonary aspergillosis. J. Infect. Dis. 197: 618–621 [DOI] [PubMed] [Google Scholar]
- 10. Coban C., et al. 2010. Immunogenicity of whole-parasite vaccines against Plasmodium falciparum involves malarial hemozoin and host TLR9. Cell Host Microbe 7: 50–61 [DOI] [PubMed] [Google Scholar]
- 11. Ewald S. E., et al. 2008. The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature 456: 658–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gersuk G. M., Underhill D. M., Zhu L., Marr K. A. 2006. Dectin-1 and TLRs permit macrophages to distinguish between different Aspergillus fumigatus cellular states. J. Immunol. 176: 3717–3724 [DOI] [PubMed] [Google Scholar]
- 13. Harris L. K., Franson R. C. 1991. [1-14C]oleate-labeled autoclaved yeast: a membranous substrate for measuring phospholipase A2 activity in vitro. Anal. Biochem. 193: 191–196 [DOI] [PubMed] [Google Scholar]
- 14. Horn D. L., et al. 2009. Epidemiology and outcomes of candidemia in 2019 patients: data from the prospective antifungal therapy alliance registry. Clin. Infect. Dis. 48: 1695–1703 [DOI] [PubMed] [Google Scholar]
- 15. Hornung V., et al. 2008. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol. 9: 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kasperkovitz P. V., Cardenas M. L., Vyas J. M. 2010. TLR9 is actively recruited to Aspergillus fumigatus phagosomes and requires the N-terminal proteolytic cleavage domain for proper intracellular trafficking. J. Immunol. 185: 7614–7622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kawai T., Akira S. 2010. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11: 373–384 [DOI] [PubMed] [Google Scholar]
- 18. Latz E., et al. 2004. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat. Immunol. 5: 190–198 [DOI] [PubMed] [Google Scholar]
- 19. Latz E., et al. 2007. Ligand-induced conformational changes allosterically activate Toll-like receptor 9. Nat. Immunol. 8: 772–779 [DOI] [PubMed] [Google Scholar]
- 20. Levitz S. M. 2010. Innate recognition of fungal cell walls. PLoS Pathog. 6: e1000758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Medzhitov R., Janeway C. A., Jr 1997. Innate immunity: the virtues of a nonclonal system of recognition. Cell 91: 295–298 [DOI] [PubMed] [Google Scholar]
- 22. Meshulam T., Levitz S. M., Christin L., Diamond R. D. 1995. A simplified new assay for assessment of fungal cell damage with the tetrazolium dye, (2,3)-bis-(2-methoxy-4-nitro-5-sulphenyl)-(2H)-tetrazolium-5-carboxanilide (XTT). J. Infect. Dis. 172: 1153–1156 [DOI] [PubMed] [Google Scholar]
- 23. Miceli M. H., Diaz J. A., Lee S. A. 2011. Emerging opportunistic yeast infections. Lancet Infect. Dis. 11: 142–151 [DOI] [PubMed] [Google Scholar]
- 24. Miyazato A., et al. 2009. Toll-like receptor 9-dependent activation of myeloid dendritic cells by deoxynucleic acids from Candida albicans. Infect. Immun. 77: 3056–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nakamura K., et al. 2008. Deoxynucleic acids from Cryptococcus neoformans activate myeloid dendritic cells via a TLR9-dependent pathway. J. Immunol. 180: 4067–4074 [DOI] [PubMed] [Google Scholar]
- 26. Netea M. G., Ferwerda G., van der Graaf C.A., Van der Meer J. W., Kullberg B. J. 2006. Recognition of fungal pathogens by Toll-like receptors. Curr. Pharm. Des. 12: 4195–4201 [DOI] [PubMed] [Google Scholar]
- 27. Netea M. G., et al. 2006. Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and Toll-like receptors. J. Clin. Invest. 116: 1642–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Osumi M. 1998. The ultrastructure of yeast: cell wall structure and formation. Micron 29: 207–233 [DOI] [PubMed] [Google Scholar]
- 29. Ozinsky A., et al. 2000. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between Toll-like receptors. Proc. Natl. Acad. Sci. U. S. A. 97: 13766–13771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Park B., et al. 2008. Proteolytic cleavage in an endolysosomal compartment is required for activation of Toll-like receptor 9. Nat. Immunol. 9: 1407–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ramirez-Ortiz Z. G., et al. 2011. A nonredundant role for plasmacytoid dendritic cells in host defense against the human fungal pathogen Aspergillus fumigatus. Cell Host Microbe 9: 415–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ramirez-Ortiz Z. G., et al. 2008. Toll-like receptor 9-dependent immune activation by unmethylated CpG motifs in Aspergillus fumigatus DNA. Infect. Immun. 76: 2123–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Romani L. 2011. Immunity to fungal infections. Nat. Rev. Immunol. 11: 275–288 [DOI] [PubMed] [Google Scholar]
- 34. Underhill D. M., et al. 1999. The Toll-like receptor 2 is recruited to macrophage phagosomes and discriminates between pathogens. Nature 401: 811–815 [DOI] [PubMed] [Google Scholar]
- 35. van de Veerdonk F. L., et al. 2008. Redundant role of TLR9 for anti-Candida host defense. Immunobiology 213: 613–620 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.