Abstract
Escherichia coli, a cause of ∼90% of urinary tract infections (UTI), utilizes fimbrial adhesins to colonize the uroepithelium. Pyelonephritis isolate E. coli CFT073 carries 12 fimbrial operons, 5 of which have never been studied. Using multiplex PCR, the prevalence of these 12 and 3 additional fimbrial types was determined for a collection of 303 E. coli isolates (57 human commensal, 32 animal commensal, 54 asymptomatic bacteriuria, 45 complicated UTI, 38 uncomplicated cystitis, and 77 pyelonephritis). The number of fimbrial types per E. coli isolate was distributed bimodally: those with low (3.2 ± 1.1) and those with high (8.3 ± 1.3) numbers of fimbrial types (means ± standard errors of the means). The fimbrial genes ygiL, yadN, yfcV, and c2395 were significantly more prevalent among urine isolates than human commensal isolates. The effect of deletion of Ygi and Yad fimbrial operons on growth, motility, biofilm formation, adherence to immortalized human epithelial cells, and pathogenesis in the mouse model of UTI was examined. Yad fimbriae were necessary for wild-type levels of adherence to a bladder epithelial cell line and for biofilm formation. Deletion of these fimbrial genes increased motility. Ygi fimbriae were necessary for wild-type levels of adherence to a human embryonic kidney cell line, biofilm formation, and in vivo fitness in the urine and kidneys. Complementation of each fimbrial mutant restored wild-type levels of motility, biofilm formation, adherence and, for ygi, in vivo fitness. A double deletion strain, Δygi Δyad, was attenuated in the urine, bladder, and kidneys in the mouse model, demonstrating that these fimbriae contribute to uropathogenesis.
INTRODUCTION
Uropathogenic Escherichia coli (UPEC) is the most prevalent cause of uncomplicated urinary tract infection (UTI) and one of the most common human pathogens (7). To better combat this pathogen, through the development of vaccines or alternative therapeutic approaches, it is imperative to understand the factors required for UPEC to successfully colonize its host. Numerous virulence factors have been determined, including toxins, siderophores, capsule, and adhesins (21–24, 36); however, no core set of virulence factors has been identified that defines all UPEC isolates.
The first recognized virulence factors of UPEC were two fimbrial adhesins, type 1 and P fimbriae (19). Indeed, E. coli encodes both the chaperone-usher fimbriae and type IV pili (53). Chaperone-usher fimbriae are rigid rod-like structures with flexible fibrillar tips that terminate in an adhesin (17, 48). These fimbriae are comprised of main subunits, accessory pilins, and adhesins that are transported to the periplasm through the Sec system. Periplasmic chaperones bind the subunits and ensure proper folding as well as transport the subunits to the outer membrane usher (17, 48). In the outer membrane, a dimer of usher proteins docks each subunit and adds it to a growing fimbria (17).
Chaperone-usher fimbriae are subdivided into clades based on the sequence of the usher proteins, with type 1 fimbriae being the prototypical fimbriae of clade γ1 and P fimbriae being the representative fimbriae of clade π (17). Type 1 fimbriae are established UPEC virulence factors that are encoded on the E. coli K-12 chromosomal backbone. Expression of these fimbriae is phase variable due to the promoter residing on an invertible element and the activity of recombinases that switch the promoter between the on and off positions (3, 30). Type 1 fimbriae bind uroplakin in the bladder, thus mediating colonization of the urinary tract. P fimbriae, encoded on pathogenicity islands in UPEC strains, bind Gal(α1-4)Gal-specific glycosphingolipids on kidney epithelium and are implicated as a virulence factor in pyelonephritic strains (28). Three molecular variants of the tip adhesin of P fimbriae (PapG1, PapG2, and PapG3) confer different binding specificities (20).
Although type IV pili are encoded on the chromosome of both commensal and uropathogenic strains of E. coli, expression has not been studied. Enteropathogenic E. coli, however, expresses type IV pili, designated bundle-forming pili (8). Type IV pili are similar to the type II secretion apparatus, in that the pilus assembles on the periplasmic side of the inner membrane and protrudes through a pore in the outer membrane (8). These structures are comprised of a major subunit, or pilin, and can have several minor pilins incorporated into the pilus, forming a long flexible structure. Several Gram-negative pathogens utilize type IV pili for twitching motility, adherence, invasion, biofilm formation, competence, or autoaggregation (8).
The genome of E. coli CFT073, a prototypical UPEC isolate cultured from the blood and urine of a patient with pyelonephritis, contains 12 putative fimbrial operons (53). Ten of these are chaperone-usher fimbriae, and two are putative type IV pili. The chaperone-usher fimbriae include type 1, P (two operons), F1C, Auf, and F9 fimbriae, as well as the putative fimbriae encoded by the yeh, yad, yfc, and ygi operons. The two putative type IV pili are encoded by c2394-c2395 and ppdD-hofBC (53). While strains isolated from human urine with both P and type 1 fimbriae have been shown to have the greatest capacity for adherence to uroepithelial cells and, in fact, enhance infection in mice (1, 5, 15, 28), urine isolates without either fimbrial type are also adherent in vivo (15). Furthermore, both P and type 1 fimbriae are dispensable when E. coli colonizes the neurogenic bladder of paralyzed patients (16). As well, the majority of E. coli isolates analyzed directly from human urine have the fim switch in the off position (30), and so are not expressing type 1 fimbriae, suggesting the presence of additional adherence mechanisms involved in uropathogenesis. Similarly, P, type 1, and S fimbriae are not required for the adherence of UPEC to an immortalized bladder epithelial cell line, again demonstrating the need to understand the contribution of other fimbriae and adhesins to colonization of epithelial cell surfaces (34).
The purpose of this study was to determine the prevalence of uncharacterized fimbriae in E. coli strains isolated from fecal samples of healthy people and animals and from clinical samples from patients with various presentations of urinary tract infection. We further determined the contributions of two previously uncharacterized UPEC-associated fimbriae, Yad and Ygi, to adherence to host epithelial cells, biofilm formation, motility, and in vivo fitness.
MATERIALS AND METHODS
Strain collection.
A strain collection was assembled that contained 303 E. coli isolates, including human commensal (n = 57) (35, 37), animal commensal isolates from the ECOR collection (n = 32) (37), and strains isolated from individuals with asymptomatic bacteriuria (ABU; n = 54) (14, 37), complicated UTI (n = 45) (31, 52), uncomplicated cystitis (n = 38) (12, 37, 45), and pyelonephritis (n = 77) (35, 37, 51).
Fimbrial typing.
Isolates were tested in duplicate by multiplex PCR using single colonies as template. Purified genomic DNAs from CFT073 (positive for focH, c1936, c2395, ppdD, yadN, yehA, aufA, ygiL, yfcV, and papG2), UTI89 (a cystitis isolate positive for c1936, ppdD, yadN, yehA, aufA, ygiL, yfcV, sfaS, and papG3), 536 (a pyelonephritis isolate positive for c2395, ppdD, yadN, yehA, aufA, yfcV, sfaS, pixC, and papG3), and BN406 (clone positive for draC) were included as positive-control templates. A no-template control was also included. Multiplex PCR was carried out using the Qiagen multiplex PCR kit. Thermocycler conditions were 95°C for 15 min, followed by 30 cycles of 30 s at 94°C, 1.5 min at 63°C, and 1.5 min at 72°C, and then final extension incubation for 10 min at 72°C. Three primer mixes were used: mix 1 (focH, c1936, c2395, ppdD, and yadN), mix 2 (draC, yehA, aufA, ygiL, yfcV, and sfaS), and mix 3 (pixC, papG1, papG2, and papG3).
The CFT073 fimbrial gene sequence for each gene screened in the multiplex PCR study was analyzed with BLAST and the NCBI database for all E. coli sequences. Sequences obtained from this search were aligned using ClustalW, and primers were designed in regions with the highest homology that gave specific binding as determined by in silico PCR (2). Primers for the three fimbrial genes not present in CFT073 (sfaS, pixC, and draC) were designed similarly from alignment of all sequences found in the NCBI database for those genes. The primers used to amplify papG1, papG2, and papG3 were previously reported (20). The multiplex primer mixes were set up so that individual bands were at least 68 bp apart and were tested for cross-reactivity. Primer sequences are available upon request.
RNA isolation, cDNA synthesis, and reverse transcriptase quantitative PCR.
E. coli CFT073 cultured overnight in LB broth with aeration at 37°C was quantified by spectrophotometry at an optical density at 600 nm (OD600), and 1-ml samples were diluted to 109 CFU/ml. Bacteria were collected by centrifugation, washed once with phosphate-buffered saline (PBS), and resuspended in 20 ml of either sterile human urine (filter sterilized, 0.22-μm pore size; Millipore) or 20 ml fresh LB broth. Bacteria were cultured statically at 37°C, and culture samples (2 ml) were removed during the early, mid-, and late exponential phases, as well as stationary phase. RNA was stabilized with 4 ml RNAprotect (Qiagen), and total RNA was isolated using the RNeasy Mini system (Qiagen) according to the manufacturer's instructions. Total RNA and cDNA sample concentrations were estimated using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific). cDNA was synthesized from total RNA using the SuperScript double-stranded cDNA synthesis system (Invitrogen) according to the manufacturer's instructions. Reverse transcriptase quantitative PCR (qPCR) was performed in a Mx300P thermalcycler (Stratagene), using 30 ng cDNA template, 0.1 μM primers, and Brilliant SYBR green reagents (Stratagene). Data were normalized to gapA and analyzed with MxPro 4.0 software (Stratagene).
Construction and complementation of fimbrial mutants.
Deletion mutants lacking entire fimbrial operons were constructed in strain CFT073 by using the lambda red recombinase system described by Datsenko and Wanner (6). Primers containing sequence homologous to the 5′ and 3′ ends of the target sequence were designed and used to amplify the resistance cassette from either template plasmid pKD3 or pKD4 (encoding chloramphenicol or kanamycin resistance, respectively). Lambda red-mediated recombination was then used to replace the selected fimbrial operons with these PCR products. A double mutant was constructed by first replacing the ygi operon with the Kan cassette from pKD4 and then replacing the yad operon with the Cam cassette from pKD3 by two rounds of lambda red-mediated recombination. Primers homologous to flanking regions of each fimbrial operon were designed for confirmation of replacement.
To complement the single fimbrial operon mutations in CFT073, the entire operons and their endogenous promoters were amplified by PCR, restriction digested, and ligated into pGEN-MCS, a derivative of plasmid pGEN222 (26). For yad, the PCR product and the vector were digested with HindIII and NotI, whereas for ygi, the PCR product and vector were digested with NdeI and BamHI. Plasmid constructs were electroporated into their respective mutants and selected for resistance to ampicillin (100 μg/ml). Constructs were confirmed by isolation of the plasmid DNA and PCR amplification using primers flanking the insertion sites.
Motility assays.
Motility was evaluated for each mutant in soft agar plates (1% tryptone, 0.5% NaCl, and 0.25% agar) and compared to the parental wild-type strain as described previously (27). Briefly, mutants were cultured overnight in LB, used to inoculate 5 ml of sterile LB, and incubated at 37°C with aeration to an OD600 of 1.0 to 1.2. Cultures were standardized to an OD600 of 1.0 and stabbed into the center of soft agar plates by using an inoculating needle. Plates were incubated for 16 h at 30°C, at which time the diameter of motility was measured. Motility of the complemented mutants was examined similarly, but the medium and the soft agar contained ampicillin (100 μg/ml) for maintenance of the plasmid. Wild-type CFT073 and each mutant transformed with the empty plasmid pGEN-MCS were included as controls.
Biofilm assays.
Biofilm formation following static growth for 24 h was examined in 24-well polystyrene plates treated for tissue culture use (a hydrophilic surface) and 96-well nontreated polystyrene plates (hydrophobic surface) at both 30°C and 37°C. Briefly, the wild type or mutants were cultured statically overnight in LB and M9 minimal medium (0.2% glucose or 0.4% glycerol as carbon source). Cultures were diluted 1/100 into fresh LB or M9 medium and cultured statically for 24 h at either 30°C or 37°C in polystyrene plates. Culture fluid was removed by aspiration, and the plates were washed once with sterile distilled H2O (dH2O) to remove unbound bacteria. Plates were then stained with crystal violet (0.7% solution) for 3 min. Quantification was conducted by dissolving the crystal violet in 100% ethanol for 15 min (1 ml for 24 wells; μQuant; BioTek Instruments, Inc.).
Detection of flagella.
Bacteria were cultured in LB for either 4 h or overnight and standardized to an OD600 of 0.35, and 1.5 ml of the standardized suspensions was centrifuged at 4,320 × g, 10 min, 4°C to pellet bacteria. Bacteria were lysed by resuspension in 100 μl dH2O and 20 μl 6× sodium dodecyl sulfate (SDS) sample buffer, followed by boiling for 10 min. Sample lysates (10 μl) were electrophoresed on a 12.5% denaturing SDS-polyacrylamide gel and transferred to a polyvinylidene difluoride membrane (Immobilon-P; Millipore Corp.). Blots were incubated with a 1:40,000 dilution of rabbit polyclonal antiserum to H1 flagella (Statens Serum Institut, Copenhagen, Denmark) followed by a 1:20,000 dilution of peroxidase-conjugated goat anti-rabbit immunoglobulin G (Sigma). Blots were developed using chemiluminescence according to the manufacturer's instructions (Amersham ECL Plus; GE Healthcare Life Sciences).
Detection of type 1 fimbriae.
Type 1 fimbrial expression was determined for all mutants by mannose-sensitive hemagglutination of guinea pig erythrocytes as described previously (44). Briefly, bacteria were cultured statically for 48 h, then passaged and cultured statically for an additional 48 h in LB broth. Approximately 109 CFU were resuspended in 100 μl of PBS (pH 7.4) and serially diluted 2-fold in 96-well round-bottom microtiter plates. An equal volume of a 3% (vol/vol) suspension of guinea pig erythrocytes (Rockland Immunochemicals) was added to each well, with or without α-methyl mannoside (25 μg/ml). Hemagglutination was scored as positive when a diffuse mat of erythrocytes appeared across the bottom of the well and as negative if a tight button of cells was observed.
Adherence assays.
The immortalized bladder epithelial cell line UM-UC-3 (ATCC CRL-1749) and the human embryonic kidney cell line HEK-293 (ATCC CRL-1573) were cultured to confluence in 24-well cell culture plates (Corning) in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum, 1% glutamate, 1% penicillin, and 1% streptomycin (DMEM-PS) at 37°C in a humidified atmosphere with 5% CO2 (37°C with CO2). Epithelial cell cultures were washed once with PBS and then inoculated with 250 μl of 4 × 108 CFU/ml E. coli CFT073 (wild type) or isogenic mutants in DMEM without antibiotics. The inoculum was diluted and spread onto LB plates to determine the initial CFU/well. The infected epithelial cells were incubated at 37°C with CO2 for 30 min and then washed three times with PBS. Epithelial cells, along with any adherent bacteria, were lifted by incubation in 1 ml PBS containing 5 mM EDTA and plated onto LB to enumerate cell-associated bacteria. Adherence was expressed as the final cell-associated CFU/initial CFU/well. Adherence of each mutant was normalized to the wild-type control. All assays were performed in triplicate.
In vivo murine cochallenge model of ascending UTI.
Six- to 8-week-old CBA/J mice were infected transurethrally as previously described (18), with the following modification. Overnight 37°C aerated cultures of wild-type E. coli CFT073 and the fimbrial mutant to be tested, Δyad, Δygi, or Δyad Δygi, were centrifuged (3,500 × g, 30 min, 4°C) to collect bacteria. Bacteria were resuspended in 30 ml PBS and diluted to an OD600 of 4.0 (∼109 CFU/ml). Each mutant was mixed 1:1 with the parental strain, and then 50 μl of this mixture (108 CFU) was introduced to each mouse through a sterile 0.28-mm polyethylene catheter attached to an infusion pump (Harvard Apparatus). Dilutions of the suspension were plated to quantify inocula on LB agar with and without the appropriate antibiotics to differentiate the antibiotic-resistant mutant from the antibiotic-susceptible wild-type strain. At 48 h postinoculation (hpi), mice were euthanized and target organs were removed. To quantify the bacterial load, the target organs were homogenized in 3 ml sterile PBS with a GLH homogenizer (Omni International) and, using an Autoplate 4000 spiral plater (Spiral Biotech), dilutions were plated on LB with and without the appropriate antibiotics. The competitive index (CI) was calculated as follows: mutant(Output CFU/Input CFU)/wild type(Output CFU/Input CFU). The CI was normalized by using the log10 of the CI and then tested for statistical significance by using a two-tailed Wilcoxon signed rank test, where a P value of <0.05 was considered significant.
Statistical analysis.
Comparison of proportions between the source groups for individual fimbrial types was conducted using the chi-square test for heterogeneity. If the P value was <0.05, then individual pairwise comparisons were conducted using Fisher's exact test. Comparison of fimbrial type scores between the source groups was conducted with a Kruskal-Wallis one-way analysis of variance, and when this test yielded a P value of <0.05, individual comparisons were conducted using Dunn's multiple comparison test. Cluster analysis was used to visualize the distribution of fimbrial profiles among the source groups represented in the strain collection. A data matrix consisting of strains (rows) and the presence or absence of fimbrial genes (columns) was used to generate a distance matrix. The distance matrix was generated in R (39), utilizing the ecodist package (11) and Jaccard's metric for binary data. An unweighted pair group method with arithmetic averaging dendrogram was constructed in MEGA version 5 (46). A paired t test was used to compare motility diameters, and an unpaired Student's t test was used to compare biofilm formation phenotypes and adherence data.
RESULTS
Prevalence of fimbrial types differs between commensal and UPEC isolates.
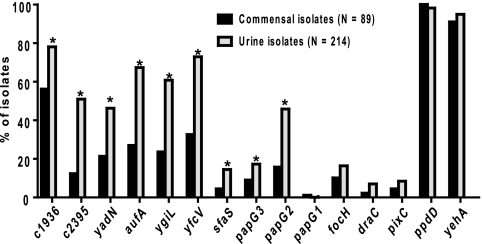
A collection of 303 strains of E. coli, representing isolates from fecal samples, ABU, uncomplicated cystitis, pyelonephritis, and complicated UTI, were screened by multiplex PCR for the presence of 15 fimbrial genes (Table 1). The genes represented 13 fimbriae, 10 found in the genome of E. coli CFT073 and 3 others (S, Pix, and Dr fimbriae) that have been demonstrated to be involved in the uropathogenesis of other UPEC strains (9, 10, 25, 32). Each gene was detected in at least three isolates. Nine fimbrial genes (c2395, yfcV, ygiL, yadN, c1936, papG2, papG3, aufA, and sfaS) (Table 1) were significantly associated with urine isolates compared to commensal strains (Fig. 1; all P values of <0.001). Additionally, sfaS was significantly associated with cystitis and ABU isolates, whereas c2395 and papG2 were significantly associated with pyelonephritis isolates.
Table 1.
Proportion of each clinical source group in which the fimbrial gene was present
| Gene | % of clinical group (n) with gene presenta |
|||||
|---|---|---|---|---|---|---|
| Animal commensal (32) | Human commensal (57) | ABU (54) | Complicated UTI (45) | Cystitis (38) | Pyelonephritis (77) | |
| c1936 | 0.75** | 0.46 | 0.74 | 0.84** | 0.76* | 0.79** |
| c2395 | 0.03 | 0.18 | 0.43** | 0.52*** | 0.39* | 0.62*** |
| yadN | 0.09 | 0.28 | 0.52* | 0.36 | 0.37 | 0.53** |
| aufA | 0.22 | 0.30 | 0.67*** | 0.68*** | 0.58*** | 0.73*** |
| ygiL | 0.19 | 0.26 | 0.57*** | 0.57*** | 0.61** | 0.66*** |
| yfcV | 0.16** | 0.42 | 0.70** | 0.68*** | 0.76*** | 0.77*** |
| sfaS | 0.03 | 0.05 | 0.24* | 0.07 | 0.24* | 0.08 |
| papG3 | 0.09 | 0.09 | 0.26 | 0.02** | 0.29* | 0.14 |
| papG2 | 0.03 | 0.23 | 0.20* | 0.55 | 0.16 | 0.74*** |
| papG1 | 0.00 | 0.02 | 0.00 | 0.02 | 0.00 | 0.01 |
| focH | 0.13 | 0.09 | 0.13 | 0.11 | 0.16 | 0.22 |
| draC | 0.03 | 0.02 | 0.04 | 0.05 | 0.08 | 0.10 |
| pixC | 0.02 | 0.02 | 0.06 | 0.07 | 0.13 | 0.09 |
| ppdD | 1.00 | 1.00 | 1.00 | 0.98 | 1.00 | 0.97 |
| yehA | 0.97 | 0.88 | 0.93 | 0.98 | 0.95 | 0.96 |
As assessed by multiplex PCR. *, P < 0.05; **, P < 0.01; ***, P < 0.001 compared to the human commensal proportion.
Fig. 1.
Percentages of isolates that were positive for each gene when assayed by multiplex PCR. Black bars indicate fecal commensal isolates from either human or animal origin, and gray bars indicate isolates from human urine of patients presenting with uncomplicated cystitis, pyelonephritis, asymptomatic bacteriuria, or complicated urinary tract infection. *, P < 0.001 as determined by Fisher's exact test.
The average number of fimbrial types is lower in commensal strains than in pathogenic E. coli.
Consistent with these prevalence differences for individual fimbrial types, the number of fimbrial types per isolate (Fig. 2) differed significantly between commensals and urinary isolates (P < 0.001). In contrast, the different groups of urine isolates did not differ significantly for number of fimbrial types. Similarly, there was no significant difference in the number of fimbrial types between the animal and human commensal isolates. Pyelonephritis isolates carried the most fimbrial types (mean ± standard error of the mean, 7.5 ± 2.3), whereas the animal commensal isolates had the fewest (3.9 ± 2.1) (Fig. 2).
Fig. 2.
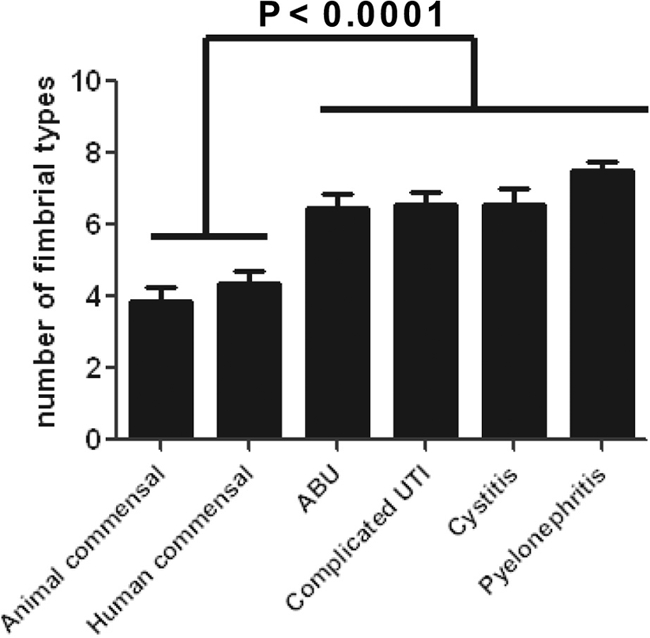
Mean number of fimbrial types per group. The presence or absence of 15 fimbrial genes was assessed by multiplex PCR. The comparison of the mean number of fimbrial types per isolation source group demonstrated that urine isolates have two to three more fimbrial types than commensal isolates. Error bars depict the standard deviations, and P values were calculated by one-way analysis of variance followed by Tukey's multiple comparison test. There was no statistical difference in the number of fimbrial types between the animal and human commensal groups or between the urine isolate groups.
The number of fimbrial types per isolate was distributed bimodally.
Overall, the number of fimbrial types per isolate was distributed bimodally (Fig. 3A), with the resulting low- and high-number populations having means of 3.2 ± 1.1 versus 8.3 ± 1.3 fimbrial types per isolate, respectively. The human commensal isolates were predominantly (72%) in the low-number population, whereas the urine isolates were predominantly (68%) in the high-number populations (Fig. 3B). The average number of fimbrial types per isolate in the low population was significantly less in the human commensal (2.8 ± 1.1 fimbrial types) than the pyelonephritis (3.6 ± 1.0; P < 0.05), cystitis (3.7 ± 1.2; P < 0.05), or complicated UTI (3.9 ± 0.7; P < 0.01) source groups. However, the average number of fimbrial types per isolate in the high population was similar across all source groups.
Fig. 3.
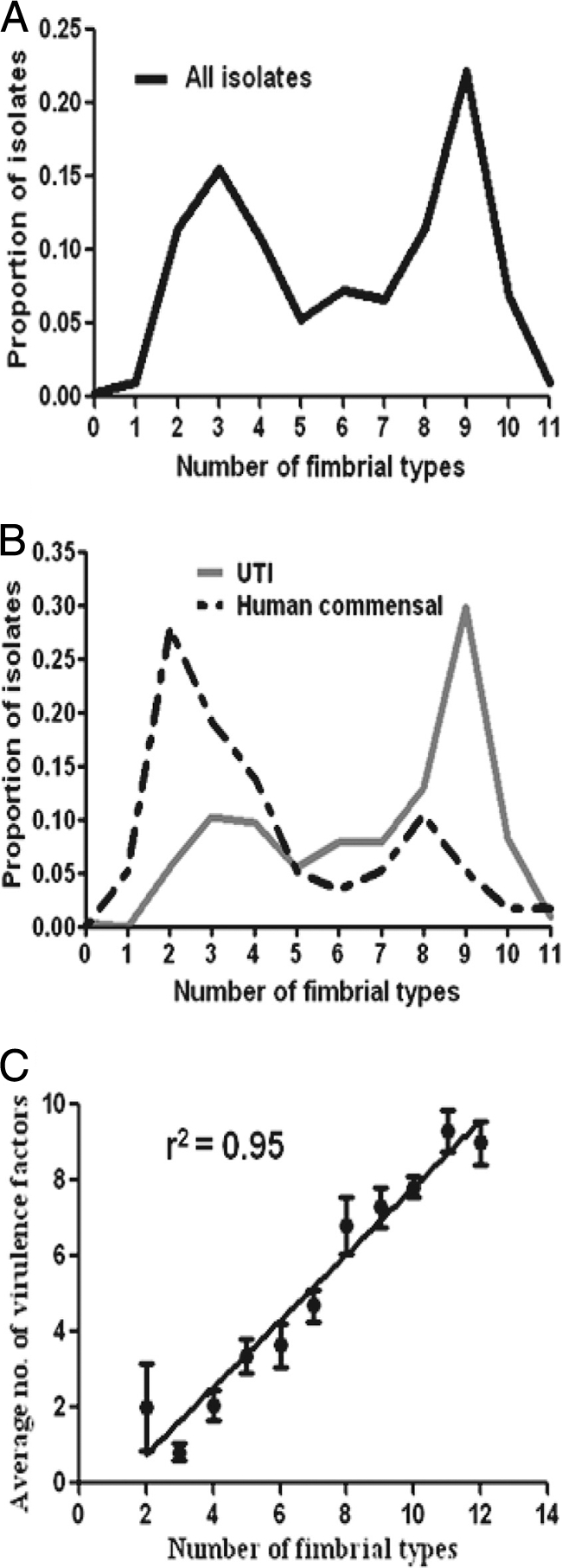
Proportion of isolates with each indicated number of fimbrial types. (A) Distribution of all isolates (n = 303) according to number of fimbrial types. (B) Distribution of UTI (n = 214) versus human commensal (n = 57) isolates according to number of fimbrial types. (C) Comparison of the number of fimbrial types with the number of nonfimbrial virulence genes present in each isolate. Data points represent average numbers of (nonfimbrial) virulence genes for each number of fimbrial types. Error bars represent the standard errors of the means.
The number of fimbrial types correlates with the number of virulence factor genes.
Previously, the prevalence of 15 virulence and fitness genes was determined by multiplex PCR; these genes represented adhesins (fimA, papA, and tosA), iron acquisition systems (chuA, hma, iutA, iroN, fyuA, iha, and ireA), and secreted toxins (hlyA, cnf1, sat, picU, and tsh) (50). A total of 227 of the same isolates were tested in this study, including 57 human fecal, 32 animal fecal, 23 complicated UTI, 37 cystitis, 1 ABU, and 77 pyelonephritis isolates. The number of virulence genes was determined for each isolate and compared to the number of fimbrial types in each isolate (including fimA but excluding papA, since pap is already represented by the papG alleles). The number of fimbriae correlated positively with the mean number of previously determined virulence genes (50) (r2 = 0.951) (Fig. 3C). Correspondingly, the mean number of virulence genes per isolate was significantly lower in the low population (2.0 ± 2.3) than in the high population (7.0 ± 2.7; P < 0.001).
Specific fimbrial profiles are associated with E. coli isolated from different clinical presentations.
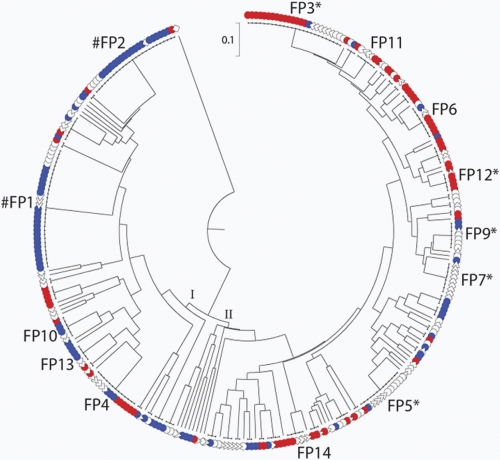
To determine whether distinct combinations of the 15 fimbrial genes could differentiate among the six source groups, aggregate fimbrial profiles were analyzed. The 303 isolates exhibited 104 distinct profiles, which were used to construct a dendrogram depicting their distribution among the six source groups (Fig. 4). The fimbrial profiles distributed into two major clusters, with cluster I comprising primarily fecal isolates and cluster II primarily pyelonephritis isolates. Of the 104 fimbrial profiles, 57 (55%) were unique (represented by a single isolate), 33 (32%) were more prevalent (occurred in 2 to 4 isolates), and 14 (13%) were common (occurred in ≥5 isolates). Of the common profiles (Fig. 4 and Table 2), profile 1 was significantly associated with human commensal strains (P < 0.001). Profile 2 was significantly associated with animal commensal strains (P < 0.001). Profiles 3 and 12 were positively associated with pyelonephritis isolates (P < 0.001 and P < 0.05, respectively). Profile 5 was positively associated with both cystitis and ABU isolates (P < 0.05), whereas profile 9 was associated only with ABU (P < 0.05). Profile 7 was significantly associated with complicated UTI isolates (P < 0.01). This demonstrates that specific combinations of fimbriae are associated with virulent isolates of E. coli, and other fimbrial profiles are associated with commensal isolates. Therefore, the fimbrial profile could potentially predict the severity of disease caused by a specific isolate.
Fig. 4.
Dendrogram depicting the distribution of fimbrial profiles by source group. Among the 303 study isolates, 104 fimbrial profiles were identified. In the dendrogram there are two main clusters, I and II. The most prevalent profiles are labeled by fimbrial profile (FP) number. #, the FP is significantly associated with commensal strains; *, the FP is significantly associated with UPEC strains. Isolate sources: red •, pyelonephritis; blue •, commensal; ○, ABU; ▵, cystitis; ⋄, complicated UTI.
Table 2.
Distribution of fimbrial profiles by source group
| Fimbrial profilea | No. of isolates | Distribution of fimbrial profile by isolation source (n)b |
Constituent genes | |||||
|---|---|---|---|---|---|---|---|---|
| Animal commensal (32) | Human commensal (57) | ABU (54) | Complicated UTI (45) | Cystitis (38) | Pyelonephritis (77) | |||
| 1 | 30 | 7 | 15*** | 5 | 0* | 3 | 0*** | ppdD, yehA |
| 2 | 25 | 13*** | 6 | 4 | 1 | 1 | c1936, ppdD, yehA | |
| 3 | 25 | 1 | 3 | 6 | 15*** | c1936, c2395, ppdD, yadN, yehA, aufA, ygiL, yfcV, papG2 | ||
| 4 | 13 | 3 | 3 | 2 | 5 | ppdD, yehA, papG2 | ||
| 5 | 13 | 1 | 6* | 5* | 1 | c1936, ppdD, yadN, yehA, aufA, ygiL, yfcV, sfaS, papG3 | ||
| 6 | 9 | 1 | 1 | 1 | 1 | 5 | focH, c1936, c2395, ppdD, yadN, yehA, aufA, ygiL, yfcV, papG2 | |
| 7 | 8 | **6 | 2 | c1936, c2395, ppdD, yehA, aufA, ygiL, yfcV | ||||
| 8 | 7 | 1 | 1 | 1 | 2 | 1 | 1 | c1936, ppdD, yehA, pixC |
| 9 | 7 | 2 | 4* | 1 | c1936, c2395, ppdD, yadN, yehA, aufA, ygiL, yfcV | |||
| 10 | 6 | 3 | 2 | 1 | ppdD, yehA, yfcV, papG2 | |||
| 11 | 6 | 1 | 2 | 1 | 2 | c1936, ppdD, yadN, yehA, aufA, ygiL, yfcV, papG2 | ||
| 12 | 5 | 1 | 4* | focH, c1936, c2395, ppdD, yehA, aufA, ygiL, yfcV, papG2 | ||||
| 13 | 5 | 3 | 2 | c1936, ppdD, yehA, papG2 | ||||
| 14 | 5 | 1 | 2 | 2 | c1936, c2395, ppdD, yehA, aufA, ygiL, yfcV, papG2 | |||
Fimbrial profiles were numbered from the most prevalent to the least prevalent.
Numbers in parentheses are the total numbers in each source group. *, P < 0.05; **, P < 0.01; ***, P <0.001 (by Fisher's exact test for isolate source compared to all other source groups).
Phylogenetic distribution of fimbrial types.
E. coli are divided into five major phylogenetic groups, A, B1, B2, D, and E, with UPEC belonging primarily to groups B2 and D; commensal E. coli phylogroups vary according to the local environment of the host population (4, 47). The mean number of fimbrial types in each phylogenetic group, as expected, increased from A (2.3 ± 0.5), B1 (3.7 ± 0.9), D (4.4 ± 0.7), to B2 (8.4 ± 1.4). All isolates in the population with a low number of fimbrial types (n = 54) were found in phylogenetic groups A, B1, and D. The isolates in the high fimbrial type group (n = 18) were mostly B2, with two in D and three from B1. By examining the distribution of fimbrial profiles within the ECOR collection, we correlated fimbrial profiles with phylogenetic group. Among the 72 ECOR collection strains, 31 fimbrial profiles were identified. With respect to the phylogenetic groups, groups A and B1 exhibited a total of only 6 and 5 profiles, respectively. Group A was dominated by fimbrial profile 1, and group B was dominated by profile 2. In contrast, groups B2 and D were very diverse, exhibiting 11 and 10 profiles, respectively, none of which accounted for more than 14% (group B2) or 25% (group D) of isolates within the group.
Fimbrial expression changes with growth phase in urine and LB.
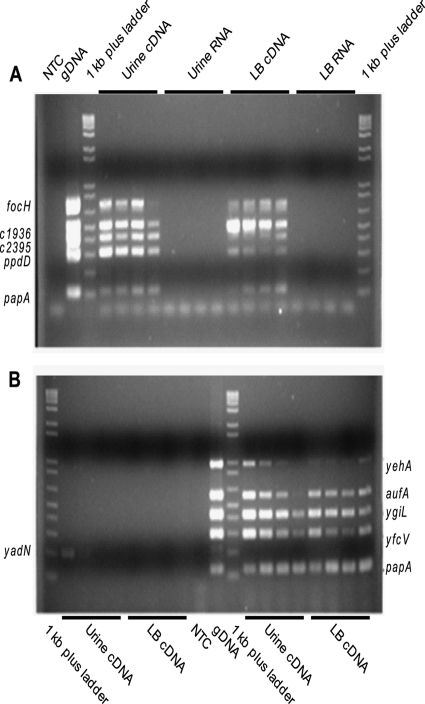
Using multiplex PCR on cDNA derived from E. coli CFT073 cultured statically in urine or LB medium and sampled over time, the expression of 10 fimbrial types was examined. In human urine, all of the assessed fimbrial genes were expressed in early exponential phase (Fig. 5). The Ygi, Yfc, Yad, Yeh, Auf, and F1C fimbrial genes were expressed early, but expression decreased over time. The two putative type IV pili, represented by ppdD and c2395, were highly expressed without variation over time. In contrast, in LB, the Yeh, Auf, and F1C fimbrial genes were expressed throughout growth with no temporal variation, but the two putative type IV pili increased in expression over time. Thus, expression patterns of fimbriae for bacteria cultured in urine differed from those cultured in LB. However, all fimbrial genes present in E. coli CFT073 were transcribed in both urine and LB, suggesting that the uncharacterized fimbriae could be physiologically relevant for study in the context of urinary tract infection. Further analysis by reverse transcriptase quantitative PCR demonstrated that expression levels of the fimbrial genes yadN (2.4-fold ± 0.6-fold), ygiL (1.7-fold ± 0.4-fold), yfcV (2.2-fold ± 1.1-fold), and c2395 (2.7-fold ± 1.1-fold) were not significantly different in urine compared to LB at mid-exponential phase, again demonstrating that these fimbriae are biologically relevant to study for involvement in uropathogenesis.
Fig. 5.
Multiplex PCR of fimbrial genes using cDNA from E. coli CFT073 cultured in urine or LB as template. (A) Multiplex PCR of focH, c1936, c2395, ppdD, and papA transcripts from bacteria isolated in early (lanes 4, 8, 12, and 16), mid- (lanes 5, 9, 13, and 17), and late exponential phase (lanes 6, 10, 14, and 18), as well as stationary phase (lanes 7, 11, 15, and 19). NTC, no-template control; +, positive genomic DNA control. (B) PCR of yadN (lanes 2 to 9) and multiplex PCR of yehA, aufA, ygiL, yfcV, and papA (lanes 10 to 20) transcripts from bacteria isolated in early (lanes 2, 6, 13, and 17), mid- (lanes 3, 7, 14, and 18), and late exponential phase (lanes 4, 8, 15, and 19), as well as stationary phase (lanes 5, 9, 16, and 20).
Adherence to biotic surfaces.
Two fimbrial gene clusters (ygi and yad) were selected for further analysis, since these operons were significantly enriched in urinary isolates (ygi, 61% of urinary versus 24% of fecal isolates; yad, 46% urinary versus 21% fecal isolates). Three fimbrial mutants, Δyad, Δygi, and Δyad Δygi, were constructed and found to have no growth defects in LB medium, M9 minimal salts medium, or urine compared to the wild type (data not shown). Since type 1 fimbriae promote adherence and biofilms and can inhibit motility, each fimbrial knockout was tested for type 1 fimbrial production by mannose-sensitive hemagglutination prior to phenotypic analysis. All three mutants hemagglutinated guinea pig erythrocytes in a mannose-sensitive manner, consistent with expression of type 1 fimbriae at titers similar to the wild type (data not shown). Therefore, the observed phenotypes were likely due to an effect of the fimbrial knockout per se, not a change in type 1 fimbrial expression.
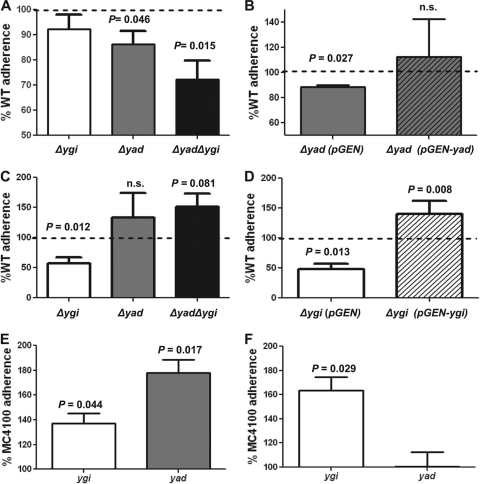
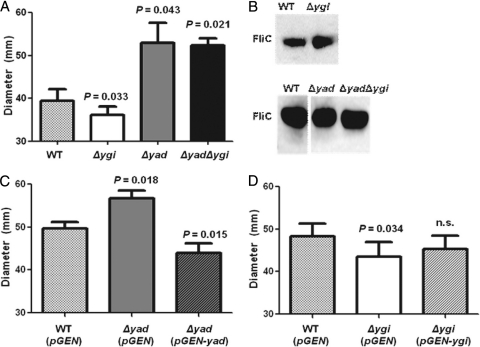
The contributions of ygi or the yad operons to adherence were assessed by utilizing the immortalized epithelial cell lines UM-UC-3 (human bladder) and HEK-293 (human embryonic kidney). Compared to the wild type, while not dramatically different, the percentage of the inoculum that was adherent was significantly decreased for the Δyad mutant (86.2 ± 5.2% of wild type; P = 0.046 [Fig. 6A]) or the double mutant Δyad Δygi (72.2 ± 7.6%; P = 0.015 [Fig. 6A]) on UM-UC-3 cells, but not HEK-293 cells. Complementation with a yad-containing plasmid restored adherence to wild-type levels (Fig. 6B). Δygi was reduced in adherence to HEK-293 cells by 42.6% (Fig. 6C) but was not significantly different from the wild type in adherence to UM-UC-3 cells. Complementation with a ygi-containing plasmid restored adherence to wild-type levels (P = 0.17) (Fig. 6D). This suggests that Yad fimbriae contribute to adherence to bladder epithelial cells, whereas Ygi fimbriae contribute to kidney colonization.
Fig. 6.
Adherence of Yad and Ygi fimbrial mutants to the uroepithelial cell lines UM-UC-3 (bladder [A, B, and E]) and HEK-293 (human embryonic kidney [C, D, and F]). (B and D) Restoration of adherence to wild-type levels upon complementation of each isogenic fimbrial operon deletion. (E and F) Expression of Yad and Ygi fimbriae in a nonfimbriated strain, E. coli MC4100, promotes adherence. The data are the averages of four independent experiments, normalized to the wild-type control. Error bars represent the standard errors of the means, and the dashed lines represent wild-type adherence levels.
Since the double mutant Δyad Δygi was not significantly different from adherence of the wild type to HEK-293 cells, expression of the other fimbriae in E. coli CFT073 may be affected by the absence of both the yad and ygi operons. To test this, multiplex PCR was conducted on cDNA derived from RNA isolated from the wild type, Δyad, Δygi, and Δyad Δygi. No significant differences in the expression of the nonmutated fimbrial genes were observed. This could indicate that fimbrial expression is perturbed at translation; however, this has yet to be tested.
To further demonstrate that the Yad and Ygi fimbriae contribute to adherence to uroepithelial cells, the operons were cloned into pGEN and expressed from their natural promoters in the nonfimbriated strain, E. coli MC4100, transformed with these plasmids. Adherence to UM-UC-3 and HEK-293 cells was assessed and compared to E. coli MC4100 carrying the empty pGEN vector. As expected from the reduced adherence of the fimbrial mutants, the addition of Yad fimbrial genes increased adherence to the bladder epithelial cell line UM-UC-3 (178.0 ± 17.9%; P = 0.017 [Fig. 6E]). E. coli MC4100, transformed with Ygi fimbrial genes, also adhered in higher numbers to the bladder epithelial cell line (137.3 ± 13.9%; P = 0.044 [Fig. 6E]). When the adherence to the kidney epithelial cell line HEK-293 was assessed, the expression of Ygi fimbrial genes dramatically increased adherence (163.5 ± 19.1%; P = 0.029 [Fig. 6F]), whereas expression of Yad fimbriae had no effect (100.6 ± 20.6%; P = 0.963).
Biofilm formation is reduced in Yad and Ygi fimbrial deletions.
Biofilms are important in the pathogenesis of UTI due to their formation in the proximal and distal tubules of the kidneys during pyelonephritis (33). Fimbriae, such as type 1 and F9 fimbriae, are involved in the production of biofilms during infection (38, 40, 49), and yadC, one of the genes in the yad operon, has been found to be upregulated 8.8-fold in E. coli CFT073 biofilms compared to planktonically grown cells (13). Therefore, we hypothesized that Yad and Ygi fimbriae may contribute to this process. In E. coli CFT073, Δyad, Δygi, and Δyad Δygi were tested for defects in biofilm formation in 24-well polystyrene tissue culture plates at 37°C and 30°C. At 30°C, no difference was observed for any of the fimbrial knockouts; at 37°C, decreased biofilm production, compared to the wild type, was observed with all 3 knockouts: Δyad (22% lower; P = 0.005), Δygi (31% lower; P = 0.001), and Δyad Δygi (39% lower; P = 0.004). When the two fimbrial operons were expressed in trans in the mutant strains, the wild-type biofilm phenotype was restored [Δyad(pGEN-yad), 98% of wild type, P = 0.91; Δygi(pGEN-ygi), 82% of wild type, P = 0.20]. This defect was dependent on the hydrophilicity of the polystyrene: when the plate was not treated for tissue culture (the surface was hydrophobic), the fimbrial operon deletions no longer affected biofilm formation (data not shown).
Motility is affected by deletion of the yad and ygi operons.
Adherence and motility are thought to be regulated inversely during infection, so that a bacterium that is expressing flagella is not expressing fimbriae, and vice versa (29). Type 1, S, and P fimbriae have been shown to regulate motility by affecting the transcription of flagellar genes (29, 42–44). Therefore, we tested each fimbrial mutant for motility dysfunction. After 16 h, Δyad had increased swimming motility compared to the wild type (1.33-fold increase; P = 0.043 [Fig. 7A]), whereas Δygi was slightly less motile (∼10% decrease; P = 0.033 [Fig. 7A]). Interestingly, the motility of the Δyad Δygi knockout was not significantly different from the Δyad mutant, demonstrating that the Δyad phenotype is dominant to that of Δygi. Motility was restored to wild-type levels by complementation for Δygi (Fig. 7D), and the yad-complemented strain was significantly less motile than the wild-type vector control (P < 0.001) (Fig. 7C).
Fig. 7.
Motility of Yad and Ygi fimbrial mutants compared to the wild type. Motility was determined by measuring the diameter on swim plates at 16 h postinoculation. Data are the averages of four independent experiments, and error bars indicate standard errors of the means; data were analyzed by using a paired t test. (A) Motility of isogenic mutants. (B) Western blot of isogenic mutants, demonstrating that the amount of flagellin in the mutants was not significantly different than wild-type levels. (C) Complementation of Δyad restores (i.e., reduces) motility. (D) Complementation of Δygi restores motility.
To determine if the effect of fimbrial gene deletion on motility was due to a change in the expression of flagella, Western blotting against FliC (flagellin) was conducted. Compared to the wild type, neither Δyad, Δygi, nor Δyad Δygi produced a significantly different amount of FliC protein (Fig. 7B), suggesting that although the fimbrial mutants cause a change in motility, this is not due to an effect on the production of flagella.
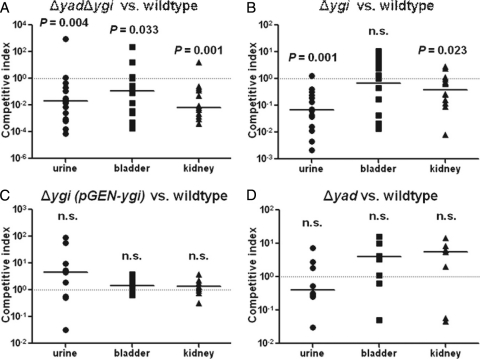
Yad and Ygi fimbriae contribute to colonization in the murine model of ascending UTI.
Since adherence and motility are both important aspects of virulence, and the deletion of the Yad and Ygi fimbrial gene clusters affects these states, we hypothesized that these two fimbriae contribute to uropathogenesis. To test this hypothesis, we examined the potential of the double fimbrial knockout, Δyad Δygi, to colonize the murine urinary tract in competition with wild-type E. coli CFT073 (Fig. 8A). The double fimbrial mutant was outcompeted by the wild type in the urine (16.4-fold; P = 0.004), bladder (10.7-fold; P = 0.03), and kidneys (18.4-fold; P < 0.001) (Fig. 8A). When the individual Δygi mutant was tested in competition with the wild type, Δygi was again outcompeted in the urine (12.1-fold; P < 0.001) and the kidneys (4.1-fold; P = 0.02), but not the bladder (P = 0.89) (Fig. 8B). This result is consistent with the adherence data, which showed that Ygi is involved in adherence to kidney epithelial cells but not bladder epithelial cells. Complementation with a ygi-containing plasmid restored fitness in vivo (Fig. 8C). When the Δyad mutant was tested in competition with the wild type in vivo, it was not significantly outcompeted, suggesting that this fimbrial type is not essential for colonization of the host (Fig. 8D). Since the double fimbrial deletion strain, Δyad Δygi, had decreased fitness in the bladder and neither single operon deletion was deficient in bladder colonization, this suggests that the two fimbriae may be redundant during colonization of the bladder. Therefore, only when both fimbrial genes are deleted is a phenotype observed during bladder colonization.
Fig. 8.
In vivo cochallenge of fimbrial mutants with the wild type in the murine model of ascending UTI. (A) Δyad Δygi is outcompeted by the wild type in the urine, bladder, and kidneys. (B) Δygi is outcompeted by the wild type in the urine and the kidneys. (C) Δygi complemented with pGEN-ygi is restored to wild-type levels of colonization in vivo. (D) Δyad colonizes the mouse urinary tract in similar numbers to the wild type. n.s., not statistically significant.
DISCUSSION
Isolates of uropathogenic E. coli have been categorized primarily by the clinical syndrome from which they were isolated; however, UPEC strains vary significantly when the presence or absence of virulence factor genes are compared between isolates. Fimbriae represent one class of virulence factor necessary for colonizing the urinary tract. While some fimbriae are associated with uropathogenic strains, including the Dr, S, P, and F1C fimbriae, several uncharacterized fimbriae have also been annotated in the genome of E. coli CFT073, a prototypical pyelonephritis strain (35, 53). By screening a collection of pathogenic and commensal isolates, the presence of 9 fimbrial types was found to be significantly associated with uropathogenic strains, including four previously uncharacterized fimbriae: the putative type IV pilus (c2394/5) and three putative chaperone-usher fimbriae, Yad, Ygi, and Yfc. A previous study identified two genes in the yad operon, yadL and yadM, as markers of E. coli strains that can move from the rectum to the vagina to the urinary tract (54), suggesting that Yad fimbriae may be involved in UTI pathogenesis.
When comparing the prevalence of each fimbrial type, the ABU and cystitis subgroups carried similar fimbriae, as did the pyelonephritis and complicated UTI subgroups. When comparing the fimbrial profiles of individual strains, 7 of the 14 common profiles were significantly distributed by source group. Therefore, combinations of fimbriae are associated with specific disease states. For example, fimbrial profile 1 (yehA and ppdD) is associated with human commensal isolates, while fimbrial profile 3 (c1936, c2395, ppdD, yadN, yehA, aufA, ygiL, yfcV, and papG2) is associated with pyelonephritis isolates. Therefore, we propose that one can predict the ability of an unknown strain to infect the urinary tract based on fimbrial profiling.
Although no single fimbrial type defined all UPEC isolates, the number of fimbrial genes in a strain clearly correlated with the ability to cause disease. Two populations of E. coli were characterized in our study: the low- and high-fimbriated populations. The low population had on average 3.2 ± 1.1 fimbriae, few other virulence factors (2.0 ± 2.3), and was primarily isolated from fecal samples. In contrast, the high population had on average 8.3 ± 1.3 fimbriae, more virulence factors (7.0 ± 2.7), and was primarily associated with UPEC isolates. This demonstrates that, although strains have been previously categorized by isolation source, this is an artificial grouping. Rather, the high and low fimbrial populations can be used to separate the strains by pathotype, that is, the ability to cause disease based on virulence determinants. While the number of fimbriae separates E. coli isolates into two populations that correspond to isolation source, the number of virulence factors per isolate does not (data not shown). Nevertheless, the average number of virulence factors is significantly different between fecal isolates (3.7 ± 0.43) and UPEC isolates (5.9 ± 0.2). The strong linear correlation between the number of fimbriae and the number of virulence factors present further demonstrated that the number of fimbriae in an isolate could be used to predict uropathogenesis.
Two previously uncharacterized fimbriae, Yad and Ygi, were found to be more prevalent in UPEC than commensal strains and effected virulence-related phenotypes, including motility, biofilm formation, adherence to immortalized human epithelial cells, and in vivo pathogenesis. While both Yad and Ygi fimbriae are involved in biofilm formation to hydrophilic surfaces, the two fimbria types demonstrated different tissue specificities. Yad fimbriae were necessary for wild-type levels of adherence to cultured bladder epithelial cells, and Ygi fimbriae were necessary for full adherence to immortalized kidney cells. In a nonfimbriated strain, E. coli MC4100, expression of Yad fimbriae increased adherence to bladder epithelial cells, whereas Ygi fimbriae increased adherence to both bladder and kidney epithelial cells. While P fimbriae seem to be responsible for the majority of kidney colonization, several other fimbriae can mediate kidney colonization, such as F1C, Dr, and S fimbriae (1). Here we demonstrated that Ygi fimbriae are another factor necessary for wild-type levels of kidney colonization in the murine model of ascending UTI. While Yad fimbriae seem to be involved in binding bladder epithelial cells in vitro, there was no appreciable difference in colonization of the bladder in vivo compared to wild-type E. coli CFT073. This may be due to the presence of type 1 fimbriae and the 10 other fimbriae encoded in the genome of E. coli CFT073 potentially compensating for the loss of Yad. Interestingly, the double fimbrial mutant, Δyad Δygi, was deficient compared to the wild type in colonizing the bladder as well as the kidneys, suggesting that different fimbrial types act in concert to colonize the bladder. This hypothesis is also supported by the ability of both Yad and Ygi fimbriae to promote adherence to the bladder epithelial cell line when expressed in E. coli MC4100.
UPEC reciprocally regulates the expression of flagella and fimbriae, thereby controlling the transition between motile and sessile lifestyles (29, 41, 42, 44). The expression of some fimbriae, such as type 1, P, and S fimbriae, inhibits motility, either by production of a transcriptional regulator that affects the expression of FlhDC, the master regulator of flagellar synthesis (P and S fimbriae [42, 43]) or by an unknown mechanism (type 1 fimbriae [29, 41]). Here we have shown that another fimbrial type, Yad, also represses motility, but not by affecting flagellar expression. The mechanism by which Yad fimbriae inhibit motility may be strictly mechanical; however, this remains to be examined. Ygi seems to induce motility when expressed, but not by modulating the amount of flagellin. However, the effect of Ygi fimbriae on motility was so slight that it may not be biologically relevant.
This study demonstrates that the heretofore-uncharacterized fimbrial operons in E. coli CFT073 may represent an overlooked set of virulence factors. The number of fimbrial types in a strain is a strong predictor of uropathogenesis, and two previously uncharacterized fimbriae, Yad and Ygi, contribute to colonization of the host. These fimbriae are potentially meaningful targets for therapeutics, as Yad and Ygi fimbriae are present in most uropathogenic E. coli and are not expressed by commensal strains. Future studies should be conducted to determine if fimbriation is a predictor of virulence in a broader strain collection of commensal and pathogenic isolates. This work has unveiled the importance of the number of fimbriae for UPEC, and this could potentially be a common theme for all extraintestinal E. coli pathogens.
ACKNOWLEDGMENTS
This work was supported in part by Public Health Service grants AI43363 and AI59722 from the National Institutes of Health and by the Office of Research and Development, Medical Research Service, Department of Veterans Affairs.
We personally thank Sara Smith for her help with the animal studies and Bogdan Nowicki for providing E. coli strain BN406 for a draC positive control.
Footnotes
Published ahead of print on 12 September 2011.
REFERENCES
- 1. Antao E. M., Wieler L. H., Ewers C. 2009. Adhesive threads of extraintestinal pathogenic Escherichia coli. Gut Pathog. 1: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bikandi J., San Millan R., Rementeria A., Garaizar J. 2004. In silico analysis of complete bacterial genomes: PCR, AFLP-PCR and endonuclease restriction. Bioinformatics 20: 798–799 [DOI] [PubMed] [Google Scholar]
- 3. Bryan A., et al. 2006. Regulation of type 1 fimbriae by unlinked FimB- and FimE-like recombinases in uropathogenic Escherichia coli strain CFT073. Infect. Immun. 74: 1072–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clermont O., et al. 2011. Animal and human pathogenic Escherichia coli strains share common genetic backgrounds. Infect. Genet. Evol. 11: 654–662 [DOI] [PubMed] [Google Scholar]
- 5. Connell I., et al. 1996. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc. Natl. Acad. Sci. U. S. A. 93: 9827–9832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Foxman B. 2003. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Dis. Mon. 49: 53–70 [DOI] [PubMed] [Google Scholar]
- 8. Fronzes R., Remaut H., Waksman G. 2008. Architectures and biogenesis of non-flagellar protein appendages in Gram-negative bacteria. EMBO J. 27: 2271–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goluszko P., et al. 1997. Development of experimental model of chronic pyelonephritis with Escherichia coli O75:K5:H-bearing Dr fimbriae: mutation in the dra region prevented tubulointerstitial nephritis. J. Clin Invest. 99: 1662–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Goluszko P., et al. 1997. Dr fimbriae operon of uropathogenic Escherichia coli mediate microtubule-dependent invasion to the HeLa epithelial cell line. J. Infect. Dis. 176: 158–167 [DOI] [PubMed] [Google Scholar]
- 11. Goslee S. C., Urban D. L. 2007. The ecodist package for dissimilarity-based analysis of ecological data. J. Stat. Softw. 22: 1–19 [Google Scholar]
- 12. Hagan E. C., Lloyd A. L., Rasko D. A., Faerber G. J., Mobley H. L. 2010. Escherichia coli global gene expression in urine from women with urinary tract infection. PLoS Pathog. 6: e1001187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hancock V., Vejborg R. M., Klemm P. 2010. Functional genomics of probiotic Escherichia coli Nissle 1917 and 83972, and UPEC strain CFT073: comparison of transcriptomes, growth and biofilm formation. Mol. Genet. Genomics 284: 437–454 [DOI] [PubMed] [Google Scholar]
- 14. Hooton T. M., et al. 2000. A prospective study of asymptomatic bacteriuria in sexually active young women. N. Engl. J. Med. 343: 992–997 [DOI] [PubMed] [Google Scholar]
- 15. Hopkins W. J., Jensen J. L., Uehling D. T., Balish E. 1986. In vitro and in vivo adherence of uropathogenic Escherichia coli strains. J. Urol. 135: 1319–1321 [DOI] [PubMed] [Google Scholar]
- 16. Hull R. A., et al. 2002. Role of type 1 fimbria- and P fimbria-specific adherence in colonization of the neurogenic human bladder by Escherichia coli. Infect. Immun. 70: 6481–6484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jarrell K. F. 2009. Pili and flagella: current research and future trends. Caister Academic Press, Norfolk, United Kingdom: [Google Scholar]
- 18. Johnson D. E., Lockatell C. V., Hall-Craigs M., Mobley H. L., Warren J. W. 1987. Uropathogenicity in rats and mice of Providencia stuartii from long-term catheterized patients. J. Urol. 138: 632–635 [DOI] [PubMed] [Google Scholar]
- 19. Johnson J. R. 1991. Virulence factors in Escherichia coli urinary tract infection. Clin. Microbiol. Rev. 4: 80–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Johnson J. R., Brown J. J. 1996. A novel multiply primed polymerase chain reaction assay for identification of variant papG genes encoding the Gal(α1-4)Gal-binding PapG adhesins of Escherichia coli. J. Infect. Dis. 173: 920–926 [DOI] [PubMed] [Google Scholar]
- 21. Johnson J. R., et al. 2006. Experimental mouse lethality of Escherichia coli isolates, in relation to accessory traits, phylogenetic group, and ecological source. J. Infect. Dis. 194: 1141–1150 [DOI] [PubMed] [Google Scholar]
- 22. Johnson J. R., et al. 2005. Extended virulence genotypes and phylogenetic background of Escherichia coli isolates from patients with cystitis, pyelonephritis, or prostatitis. J. Infect. Dis. 191: 46–50 [DOI] [PubMed] [Google Scholar]
- 23. Johnson J. R., Owens K., Gajewski A., Kuskowski M. A. 2005. Bacterial characteristics in relation to clinical source of Escherichia coli isolates from women with acute cystitis or pyelonephritis and uninfected women. J. Clin. Microbiol. 43: 6064–6072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kaper J. B., Nataro J. P., Mobley H. L. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2: 123–140 [DOI] [PubMed] [Google Scholar]
- 25. Kreft B., et al. 1995. S fimbriae of uropathogenic Escherichia coli bind to primary human renal proximal tubular epithelial cells but do not induce expression of intercellular adhesion molecule 1. Infect. Immun. 63: 3235–3238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lane M. C., Alteri C. J., Smith S. N., Mobley H. L. 2007. Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc. Natl. Acad. Sci. U. S. A. 104: 16669–16674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lane M. C., et al. 2005. Role of motility in the colonization of uropathogenic Escherichia coli in the urinary tract. Infect. Immun. 73: 7644–7656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lane M. C., Mobley H. L. 2007. Role of P-fimbrial-mediated adherence in pyelonephritis and persistence of uropathogenic Escherichia coli (UPEC) in the mammalian kidney. Kidney Int. 72: 19–25 [DOI] [PubMed] [Google Scholar]
- 29. Lane M. C., Simms A. N., Mobley H. L. 2007. complex interplay between type 1 fimbrial expression and flagellum-mediated motility of uropathogenic Escherichia coli. J. Bacteriol. 189: 5523–5533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lim J. K., et al. 1998. In vivo phase variation of Escherichia coli type 1 fimbrial genes in women with urinary tract infection. Infect. Immun. 66: 3303–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lomberg H., Hellstrom M., Jodal U., Svanborg Eden C. 1989. Secretor state and renal scarring in girls with recurrent pyelonephritis. FEMS Microbiol. Immunol. 1: 371–375 [DOI] [PubMed] [Google Scholar]
- 32. Lugering A., Benz I., Knochenhauer S., Ruffing M., Schmidt M. A. 2003. The Pix pilus adhesin of the uropathogenic Escherichia coli strain X2194 (O2:K−:H6) is related to Pap pili but exhibits a truncated regulatory region. Microbiology 149: 1387–1397 [DOI] [PubMed] [Google Scholar]
- 33. Melican K., et al. 2011. Uropathogenic Escherichia coli P and type 1 fimbriae act in synergy in a living host to facilitate renal colonization leading to nephron obstruction. PLoS Pathog. 7: e1001298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Miyazaki J., et al. 2002. Type 1, P and S fimbriae, and afimbrial adhesin I are not essential for uropathogenic Escherichia coli to adhere to and invade bladder epithelial cells. FEMS Immunol. Med. Microbiol. 33: 23–26 [DOI] [PubMed] [Google Scholar]
- 35. Mobley H. L., et al. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect. Immun. 58: 1281–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nielubowicz G. R., Mobley H. L. 2010. Host-pathogen interactions in urinary tract infection. Nat. Rev. Urol. 7: 430–441 [DOI] [PubMed] [Google Scholar]
- 37. Ochman H., Selander R. K. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157: 690–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pratt L. A., Kolter R. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30: 285–293 [DOI] [PubMed] [Google Scholar]
- 39. R Development Core Team 2010. R: a language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: [Google Scholar]
- 40. Schembri M. A., Klemm P. 2001. Biofilm formation in a hydrodynamic environment by novel fimh variants and ramifications for virulence. Infect. Immun. 69: 1322–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Simms A. N., Mobley H. L. 2008. Multiple genes repress motility in uropathogenic Escherichia coli constitutively expressing type 1 fimbriae. J. Bacteriol. 190: 3747–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Simms A. N., Mobley H. L. 2008. PapX, a P fimbrial operon-encoded inhibitor of motility in uropathogenic Escherichia coli. Infect. Immun. 76: 4833–4841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sjostrom A. E., et al. 2009. The SfaXII protein from newborn meningitis E. coli is involved in regulation of motility and type 1 fimbriae expression. Microb. Pathog. 46: 243–252 [DOI] [PubMed] [Google Scholar]
- 44. Snyder J. A., et al. 2005. Coordinate expression of fimbriae in uropathogenic Escherichia coli. Infect. Immun. 73: 7588–7596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stapleton A., Moseley S., Stamm W. E. 1991. Urovirulence determinants in Escherichia coli isolates causing first-episode and recurrent cystitis in women. J. Infect. Dis. 163: 773–779 [DOI] [PubMed] [Google Scholar]
- 46. Tamura K., et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tenaillon O., Skurnik D., Picard B., Denamur E. 2010. The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 8: 207–217 [DOI] [PubMed] [Google Scholar]
- 48. Thanassi D. G., Hultgren S. J. 2000. Assembly of complex organelles: pilus biogenesis in gram-negative bacteria as a model system. Methods 20: 111–126 [DOI] [PubMed] [Google Scholar]
- 49. Ulett G. C., Mabbett A. N., Fung K. C., Webb R. I., Schembri M. A. 2007. The role of F9 fimbriae of uropathogenic Escherichia coli in biofilm formation. Microbiology 153: 2321–2331 [DOI] [PubMed] [Google Scholar]
- 50. Vigil P. D., et al. 2011. Presence of putative repeat-in-toxin gene tosA in Escherichia coli predicts successful colonization of the urinary tract. mBio 2: e00066–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Warren J. W., et al. 1983. A randomized, controlled trial of cefoperazone vs. cefamandole-tobramycin in the treatment of putative, severe infections with gram-negative bacilli. Rev. Infect. Dis. 5 (Suppl. 1): S173–S180 [DOI] [PubMed] [Google Scholar]
- 52. Warren J. W., Tenney J. H., Hoopes J. M., Muncie H. L., Anthony W. C. 1982. A prospective microbiologic study of bacteriuria in patients with chronic indwelling urethral catheters. J. Infect. Dis. 146: 719–723 [DOI] [PubMed] [Google Scholar]
- 53. Welch R. A., et al. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99: 17020–17024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Xie J., Foxman B., Zhang L., Marrs C. F. 2006. Molecular epidemiologic identification of Escherichia coli genes that are potentially involved in movement of the organism from the intestinal tract to the vagina and bladder. J. Clin. Microbiol. 44: 2434–2441 [DOI] [PMC free article] [PubMed] [Google Scholar]