Abstract
Evolutionary adaptation of Pseudomonas aeruginosa to the cystic fibrosis lung is limited by genetic variation, which depends on rates of horizontal gene transfer and mutation supply. Because each may increase following secondary infection or mutator emergence, we sought to ascertain the incidence of secondary infection and genetic variability in populations containing or lacking mutators. Forty-nine strains collected over 3 years from 16 patients were phenotyped for antibiotic resistance and mutator status and were genotyped by repetitive-sequence PCR (rep-PCR), pulsed-field gel electrophoresis (PFGE), and multilocus sequence typing (MLST). Though phenotypic and genetic polymorphisms were widespread and clustered more strongly within than between longitudinal series, their distribution revealed instances of secondary infection. Sequence data, however, indicated that interlineage recombination predated initial strain isolation. Mutator series were more likely to be multiply antibiotic resistant, but not necessarily more variable in their nucleotide sequences, than nonmutators. One mutator and one nonmutator series were sequenced at mismatch repair loci and analyzed for gene content using DNA microarrays. Both were wild type with respect to mutL, but mutators carried an 8-bp mutS deletion causing a frameshift mutation. Both series lacked 126 genes encoding pilins, siderophores, and virulence factors whose inactivation has been linked to adaptation during chronic infection. Mutators exhibited loss of severalfold more genes having functions related to mobile elements, motility, and attachment. A 105-kb, 86-gene deletion was observed in one nonmutator that resulted in loss of virulence factors related to pyoverdine synthesis and elements of the multidrug efflux regulon. Diminished DNA repair activity may facilitate but not be absolutely required for rapid evolutionary change.
INTRODUCTION
Pseudomonas aeruginosa is a primary agent of chronic endobronchial infection in the cystic fibrosis (CF) lung after early childhood. CF patients commonly succumb to clonal descendants of P. aeruginosa strains acquired early in life (10, 60, 91). Because chronic infections can last for years, P. aeruginosa populations undergo thousands of generations challenged by antibiotic treatment, iron depletion (95), hydrophobicity (73), immune system surveillance, microbial competitors, and the changing topography of the CF lung (36, 56, 77). P. aeruginosa adapts physiologically to these challenges via complex regulatory networks that coordinate expression of its large genome. Approximately one in 10 genes in the sequenced PAO1 lab strain are regulatory, many of which influence transport, catabolism, and efflux of organic compounds, including antibiotics (81). The adaptability of P. aeruginosa is also evident in the high ratio of paralogous gene groups represented in its genome (76), a feature that may enable the species to explore adaptive space by neofunctionalization.
P. aeruginosa exhibits remarkable genome plasticity. In naturally occurring isolates, the genome size ranges between 5.2 Mbp and 7 Mbp (87) and consists of a core genome, conserved both in organization and orientation, and various accessory elements prone to recombination (92). These elements range in size from 1 to 200 kbp and account for 15 to 30% of the genomic diversity observed among clonal isolates (46, 47). Such observations have given rise to the distributed-genome hypothesis (DGH), which posits that each P. aeruginosa clone contains a unique collection of genes drawn from a hypothetical “pangenome” that encompasses all genes in the global P. aeruginosa population (76). Because these genes are unequally distributed among clones, P. aeruginosa exhibits phenotypic variability with respect to virulence, metabolic repertoire, and patterns of gene regulation (92). Studies of clonal diversity indicate that within any lineage the P. aeruginosa genome is malleable, frequently given to local changes that result in single-base substitutions, insertions, or deletions (indels) and regional changes that result in deletions, duplications, and inversions (69, 70, 72). In the CF lung such mutations appear more likely to be propagated by vertical gene transfer (VGT) than by horizontal gene transfer (HGT) (80, 93), although superinfection (57) and between-patient cross-infection (41) have both been reported.
When ecological or genetic factors limit HGT between bacterial lineages, the rate of adaptive evolutionary change depends on the rate of mutation supply (5). If the effective population size is low and selection pressure is high, strains with elevated mutation rates may be competitively advantaged (5), especially if the population is not well adapted and/or if adaptation requires sequential selection of multiple beneficial mutations (18). “Mutator” phenotypes may manifest transiently following a stress-induced increase in the expression of recombination enzymes and/or error-prone polymerases (4, 12). Conversely, they may manifest constitutively and heritably as a result of mutations that cause unregulated expression of these or other enzymes involved in DNA proofreading and repair (32, 61, 64). Because heritable mutators commonly arise from defects in methyl-directed mismatch repair (MMR) (e.g., mutS and mutL), they are more susceptible not only to single-base-pair substitutions but also to homologous recombination (references 18 and 63 and references therein), making them susceptible to genome rearrangement and HGT (68).
Oliver et al. (61) reported a high prevalence of heritable P. aeruginosa mutators in chronic CF infections (approximately 37%) but not in acute P. aeruginosa pulmonary infections of non-CF patients. Because mutators exhibit mutation frequencies that exceed wild-type frequencies by an order(s) of magnitude, more deleterious than beneficial mutations are likely to accumulate in their descendant lineages. As a result, one would expect mutators to come under purifying selection soon after they arise (32). The high incidence of mutators in CF patients with chronic respiratory infection (CRI) suggests that these strains not only persist by “hitchhiking” with beneficial mutations but also are peculiarly adept at doing so in the CF lung owing to environmental complexity (62) and/or to an increased incidence of compensatory mutations that offset fitness costs of drug resistance (65). Interestingly, laboratory-cultured P. aeruginosa biofilm microcolonies become highly enriched in mutators (13, 60), and drug-resistant P. aeruginosa mutator populations resist invasion by drug-sensitive nonmutators, even in the absence of selection (65). Presently, our understanding of the relative contribution of mutators to adaptive genetic variation in CF CRI consists chiefly of a limited number of studies that link them statistically to an increased incidence of multiple antibiotic resistance (MAR) (21, 30), decreased lung function (21, 89), and the emergence of CRI-specific adaptive genotypes (58).
To increase our understanding of the role that mutators may play in producing adaptive variation, we investigated the phenotypic and genotypic diversity in 49 clinical P. aeruginosa isolates from 16 CF patients treated over a 3-year period at Necker Children's Hospital in Paris, France. Each isolate was characterized phenotypically with respect to mucoidy, sensitivity to multiple classes of antibiotics, and frequency with which it spontaneously produced rifampin-resistant mutants and then was characterized genetically via multilocus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE)/repetitive-sequence PCR (rep-PCR) genome fingerprinting (15, 27, 84). Two age-matched series, similar in duration but different in mutator incidence, were sequenced at the mutL and mutS loci and subjected to detailed genomic analysis using Affymetrix PAO1 microarrays (93). Our overall goals were to test the hypotheses that P. aeruginosa evolved clonally within individual patients in the absence of secondary infection and recombination and that genetic diversity, estimated as either nucleotide polymorphism or structural variation, was greater in populations from which mutators were isolated than in populations from which they were not.
MATERIALS AND METHODS
Strain collection.
P. aeruginosa isolates were obtained from the Laboratory of Microbiology of the Hôpital Necker-Enfants Malades, Paris, France. A total of 49 strains from 16 patients were examined; 47 constituted 15 longitudinal series, as they were isolated from the sputa of patients seen on multiple occasions in the clinic. The longest interval between initial and final samples was 25 months, and the shortest was 2 months. Patients ranged in age from 6 months to 20 years (mean, 11.9 years; standard deviation [SD], 4.5 years). To protect confidentiality, letters were substituted for patient names. Longitudinal series obtained from patients I and M were age matched as a surrogate for infection duration (the patients were 13.5 and 14.9 years old, respectively, at the start of collection); as such, both series likely represent intermediate or late stages of adaptation to chronic infection.
Clinical isolates were obtained as previously described (21), colony purified from selective Pseudomonas isolation agar (PIA), and then frozen as −80°C glycerol stocks. Strains PAO1 and PA14 were provided by Samuel Miller, University of Washington.
Phenotypic characterization.
Archived samples were tested for susceptibility to amikacin (30 μg), tobramycin (10 μg), ciprofloxacin (5 μg), and ticarcillin (75 μg) by disc diffusion assay and scored for resistance, sensitivity, or intermediate resistance using guidelines established by CAFSM (Comité de l'Antibiogramme de la Société Française de Microbiologie). Strains found to have complete or intermediate resistance to three or more classes of drugs were defined as MAR (Table 1). Mucoidy was assayed by plating isolates on PIA agar and then scoring colony morphology after aerobic growth at 37°C for 15 h followed by anaerobic growth at 37°C for 12 h, as previously described (9).
Table 1.
Phenotypic variation in the Necker collection
| Patient | Strain | Isolation date (mo/day/yr) | Phenotype |
Frequency of spontaneous Rifr mutantsa | Resistanceb |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mucoid | Mutator | MAR | Ami | Cip | Tob | Tic | ||||
| A | A1 | 3/13/2002 | − | − | − | 3.60E−08 | R | S | S | R |
| A2 | 1/15/2003 | − | − | − | 4.15E−08 | S | S | S | S | |
| B | B1 | 2/25/2002 | − | − | − | 1.60E−08 | I | S | S | S |
| B2a | 2/10/2003 | + | − | − | 1.00E−08 | R | S | S | S | |
| B2b | − | − | − | 5.10E−09 | R | I | S | S | ||
| C | C1 | 3/5/2002 | + | − | − | 5.00E−09 | I | S | R | R |
| C2a | 8/22/2002 | − | − | − | 2.00E−09 | R | S | R | I | |
| C2b | − | − | − | 1.00E−08 | I | S | R | R | ||
| D | D1 | 10/15/2002 | − | − | − | 5.60E−08 | I | S | S | R |
| D2 | 9/5/2003 | − | − | − | 1.25E−07 | R | S | I | S | |
| E | E1 | 1/13/2002 | − | − | − | 1.93E−09 | R | S | I | S |
| E2 | 11/8/2002 | + | + | − | 1.46E−05 | R | S | S | S | |
| F | F1 | 12/10/2001 | + | − | − | 2.00E−09 | R | S | R | R |
| F2 | 1/29/2003 | − | − | − | 5.85E−09 | R | S | S | S | |
| G | G1 | 3/6/2002 | − | − | − | 3.85E−08 | S | S | S | S |
| G2 | 2/13/2003 | − | − | − | 7.70E−09 | S | S | S | S | |
| H | H1 | 1/25/2002 | − | + | + | 6.10E−06 | R | I | I | R |
| H2 | 5/30/2002 | − | + | − | 1.27E−05 | I | S | S | R | |
| I | I1 | 2/16/2002 | − | + | + | 1.30E−06 | R | I | S | R |
| I2 | 9/12/2002 | + | + | + | 1.29E−06 | R | R | S | R | |
| I3a | 9/25/2002 | + | − | − | 1.25E−07 | R | S | S | S | |
| I3b | + | − | + | 1.95E−07 | R | R | I | R | ||
| I4 | 3/14/2003 | − | − | + | 3.90E−08 | R | I | S | R | |
| J | J1 | 3/6/2002 | − | + | − | 7.26E−07 | S | I | S | S |
| J2a | 5/31/2002 | + | − | − | 1.43E−07 | S | S | S | S | |
| J2b | − | + | − | 4.00E−07 | R | S | S | S | ||
| K | K1 | 3/28/2002 | + | − | − | 8.75E−09 | R | S | S | S |
| K2a | 6/18/2002 | − | − | − | 5.42E−08 | R | S | I | S | |
| K2b | + | − | − | 1.80E−08 | R | S | R | S | ||
| K3a | 9/25/2002 | − | − | − | 1.27E−08 | R | S | R | S | |
| K3b | − | − | + | 5.50E−08 | R | I | R | R | ||
| K4a | 9/26/2002 | − | − | − | 1.60E−08 | R | S | S | S | |
| K4b | + | − | − | 1.50E−08 | R | S | I | I | ||
| K5a | 7/1/2003 | − | − | − | 7.51E−08 | R | S | R | S | |
| K5b | − | − | − | 8.00E−08 | R | S | I | S | ||
| L | L1 | 8/22/2001 | − | − | − | 0.00E−00 | R | S | I | S |
| L2 | 9/18/2003 | − | − | − | 2.50E−09 | R | I | I | S | |
| M | M1 | 10/12/2001 | + | − | − | 4.88E−09 | S | S | S | S |
| M2 | 8/16/2002 | − | − | − | 5.55E−09 | R | S | R | S | |
| M3a | 12/19/2002 | + | − | − | 9.50E−09 | S | S | S | S | |
| M3b | + | − | − | 3.00E−09 | R | S | S | S | ||
| N | N1a | 6/13/2003 | − | − | − | 2.88E−08 | I | S | S | R |
| N1b | − | + | − | 4.00E−07 | I | S | S | R | ||
| O | O1 | 2/26/2002 | − | − | − | 2.60E−09 | I | S | S | S |
| O2 | 4/28/2003 | − | − | − | 9.00E−09 | S | I | S | S | |
| P | P1 | 7/23/2001 | + | + | − | 6.50E−06 | I | S | S | S |
| P2a | 10/29/2001 | − | + | − | 7.60E−06 | S | R | S | S | |
| P2b | − | + | − | 3.54E−07 | S | S | S | S | ||
| P2c | − | + | + | 6.00E−06 | R | I | I | R | ||
Ami, amikacin; Cip, ciprofloxacin; Tob, tobramycin; Tic, ticarcillin; R, resistant; S, susceptible; I, intermediate.
Hypermutator strains having a Rifr frequency of >2.5 × 10−7 are shaded.
Mutator screening.
Mutation frequency was estimated as described in reference 21 by plating cells on Mueller-Hinton (MH) agar supplemented with 300 μg ml−1 of rifampin (Rif) (ICN, Fair Lawn, NJ). Briefly, glycerol stocks were streaked on PIA agar and grown overnight at 32°C. For each strain, three freshly grown colonies were used to inoculate 4-ml cultures of MH broth. Following overnight growth at 32°C, 100 μl of 10−6, 10−7, and 10−8 dilutions were plated in triplicate on MH agar, while 100 μl of 100, 10−1, and 10−2 dilutions were plated on MH-Rif. Plates were incubated at 32°C for 24 h and 48 h, respectively, and mutation frequencies estimated by dividing the total number of spontaneous Rifr mutants estimated from colonies on MH-Rif agar by the total number of all bacteria estimated from colony number on nonselective agar. “Mutators” were defined as clones where Rifr mutants appeared at a frequency 1 order of magnitude greater than the population median value of 2.5 × 10−8.
gDNA extractions.
Genomic DNA (gDNA) extractions were performed using a cetyltrimethylammonium bromide (CTAB) method of phenol-chloroform extraction (6) after overnight culture in Pseudomonas F broth, adjusting the number of extractions as necessary for mucoid strains. Genomic DNA was visualized, quantified, and verified for purity using gel electrophoresis and spectrophotometry.
rep-PCR.
Enterobacterial repetitive intergenic consensus sequence (ERIC) and BOXA1 (88) primers were independently used to produce repetitive-sequence PCR (rep-PCR) fingerprints. rep-PCR amplicons were electrophoresed at 4°C and 70V for 14.5 h in 250-ml 1.5% (wt/vol) agarose gels made up 1× Tris-acetate-EDTA (TAE) (40 mM Tris-acetate, 1 mM EDTA, pH 8.0), with continuous recirculation of 1× TAE buffer.
PFGE. (i) Plug preparation.
Pulsed-field gel electrophoresis (PFGE) plugs (100 μl) were prepared according to methods outlined in the CHEF-DR II instruction manual (no. 170-3612 through 170-3729) with modifications described below. P. aeruginosa cultures were grown overnight in 4 ml Luria-Bertani (LB) medium and diluted to an A600 of 0.8 to 1.0 using sterile LB, after which ∼1.0 ml of diluted culture was transferred to a 1.5-ml Eppendorf tube containing 180 μg chloramphenicol. A 100-μl aliquot of this culture was centrifuged for 3 min in a Sorvall MC12V centrifuge at 9,660 ×g and then resuspended in 50 μl of cell suspension buffer containing 5 mM EDTA.
(ii) Restriction digestion and electrophoresis.
To produce two PFGE fingerprints, duplicate plugs were prepared for each clone. Chromosomal DNA was restriction digested using either SpeI or DraI (New England BioLabs, Ipswich, MA) by first equilibrating plugs for 1 h in 0.5 ml of NEBuffer 2 or NEBuffer 4 and then digesting plugs for 4 h with 20 U restriction enzyme in 0.3 ml of the appropriate 1× buffer with 0.1 mg ml−1 bovine serum albumin (BSA). Following digestion, plugs were washed for 30 min in 1× wash buffer. Contour-clamped homogeneous electric field (CHEF) plugs were loaded onto 160-ml 1% (wt/vol) agarose gels and electrophoresed in 2 liters of 0.5× Tris-borate-EDTA (TBE) buffer, pH 8.3, at 12°C in a Bio-Rad CHEF-DR II apparatus (Bio-Rad, Hercules, CA). Each cycle was run at 6 V cm−1 at an angle of 120°, with pulse times of from 1 to 60 s for 31 h.
Image analysis.
rep-PCR and PFGE gel images were downloaded into Bionumerics v4.50. (Applied Maths, Sint-Martens-Latem, Belgium) and standardized using 1-kb ladders and Lambda ladder PFG markers (Promega, Madison, WI), and dendrograms were produced using Jaccard correlations, which account for band number and spacing. Images were analyzed as composite data sets so that each band could be assigned to a group specifying its gel position. Following band assignment, a Bootstrap analysis of 1,000 cycles was performed to assign confidence values to the dendrograms. Bootstrap values greater than or equal to 70% are reported (34).
MLST.
PCR amplification from genomic DNA and subsequent sequencing were performed using primers specific for acsA, aroE, guaA, mutL, nuoD, ppsA, and trpE (http://pubmlst.org/paeruginosa/info/primers.shtml) designed by Curran et al. (14). Amplification reaction mixtures were prepared in 50-μl volumes consisting of 40 ng of genomic DNA, 1× PCR buffer minus MgCl2 (Invitrogen, Carlsbad, CA), 1.5 mM MgCl2 (Invitrogen), 0.2 mM exACTGene PCR deoxynucleoside triphosphate (dNTP) mix (Fisher BioReagents, Pittsburgh, PA), 0.5 μM each primer (Sigma-Genosys, St. Louis, MO), and 1.25 U Taq DNA polymerase (Invitrogen) using standard P. aeruginosa multilocus sequence typing (MLST) thermal profiles (14). Amplification products were purified with the QIAquick PCR purification kit (Qiagen Sciences, Valencia, CA) or QIAquick gel extraction kit (Qiagen) for samples exhibiting nonspecific amplification. Internal fragments of ∼800- to 1,000-bp amplicons were sequenced on an ABI 3130 Genetic Analyzer. MLST allele assignments were obtained by BLAST similarity searches against the P. aeruginosa pubMLST database (http://pubmlst.org/paeruginosa/).
Population genetic analyses by multilocus sequence analysis (MLSA).
Sequences were aligned using ClustalW with default algorithm parameters (85). For each MLST locus, DnaSP was used to determine the number of single-nucleotide polymorphisms (SNPs) and distinct alleles in the sample (72), estimates of the amount of genetic variation, and the minimum number of recombination events (40). Minimum spanning trees (50) were inferred for each locus from the matrix of pairwise nucleotide differences in Arlequin v2.0 (Genetics and Biometry, University of Geneva) (75). Subsequently, genealogical networks were manually derived from these trees such that the sum of the branch lengths separating two alleles corresponded exactly to the number of nucleotide differences between them. Genetic differentiation within and between patient series was estimated by analyses of molecular variance (AMOVA) models implemented in Arlequin using default settings.
mutL and mutS sequencing.
Primers were designed using FastPCR 6.0 (C.B.) and Primer3 (K.C.) and conserved sequences of mutL and mutS from PAO1, UCBPP-PA14, and LESB58, screened for predicted nonspecific amplification based on available whole-genome sequences (see Table S1 in the supplemental material), and then synthesized by Integrated DNA Technologies (Coralville, IA). PCRs were optimized for MgCl2 concentration and thermal profile, and betaine added to improve performance. PCR products were purified by Qiagen gel extraction following visualization and band excision and then Sanger sequenced at either the University of Washington or the University of Montana sequencing facilities. Consensus sequences for each strain were determined using DNA Baser 2.80.0 with manual editing based on trace quality scores. Alignments for mutS and mutL were constructed using ClustalW v2.0.12-Win. SNPs, nonsynonymous and synonymous nucleotide diversity (π), and DNA polymorphisms were identified using DnaSP v5. Minimum spanning networks under AMOVA were determined using Arlequin v3.11 and drawn in Network v4.5.1.6.
a-CGH.
Genome content was evaluated for the I and M series (series containing mutators and nonmutators, respectively) by hybridizing genomic DNA to Affymetrix GeneChip arrays. Array-based comparative genome hybridization (a-CGH) data were validated by Pathogen Functional Genomics Resource Center (PFGRC) arrays (http://pfgrc.jcvi.org/). For Affymetrix a-CGH, 10 mg P. aeruginosa genomic DNA (gDNA) was fragmented using DNase I for 5 to 10 min at 37°C, yielding ∼50- to 200-bp fragments. DNase I was inactivated at 99°C for 15 min. Fragmentation was analyzed using the Agilent Bioanalyzer. Fragmented gDNA was labeled using terminal deoxynucleotidyl transferase and Affymetrix GeneChip labeling reagent as specified by the manufacturer. Three milligrams of fragmented, labeled gDNA was loaded onto Affymetrix PAO1 GeneChips and incubated at 50°C for 16 h while shaking at 60 rpm. Microarrays were processed using a GeneChip450 fluidics station and scanned using a GeneChip 7G scanner, and data were analyzed using GCOS v1.4 and GeneSpring v7.3.a. PFGRC array experiments were performed according to protocols established by the Pathogen Functional Genomics Resource Center (http://pfgrc.jcvi.org/index.php/microarray/protocols.html). The dUTP/dTTP ratio was adjusted to 2:1 to accommodate the high P. aeruginosa G-C content (63%).
Statistical analyses of microarray data.
To determine a reliable cutoff value for calling genes present or absent, pairwise Pearson correlations were computed for Affymetrix and PFGRC microarray data between corresponding strains having nonzero values below a specified cutoff. The maximum average pairwise correlation occurred at −1.27, with an average correlation value of 0.73. To establish a confidence interval for this correlation, bootstrapping was performed at the maximum point for 10,000 repetitions to obtain an estimated standard deviation of 0.035. The range of cutoff values between −1.60 and −0.96 produced average correlation values within 2% of the maximum correlation of 0.73. The lower end of this interval, −1.60, was chosen to minimize the chance of false positives (see Fig. S2 in the supplemental material).
Nucleotide sequence accession numbers.
The sequences reported in this paper have been deposited in the GenBank database under accession numbers HQ605971 to HQ605981 (mutL) and HQ632813 to HQ632822 (mutS). MLST profiles have been submitted to the P. aeruginosa MLST database as isolates 696 to 736 and 755 to 762, sequence types 1007 to 1013 and 1066 to 1069, and novel trpE alleles trpE136 and trpE137. The CHP and CEL files generated from Affymetrix analyses, as well as normalized and log2 values, have been deposited in GEO (accession number GSE25481) and can be accessed at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=pfwntyiioqooqfq&acc=GSE25481.
RESULTS
The Necker P. aeruginosa collection exhibits widespread phenotypic polymorphism within and among patient series.
We investigated 49 clones isolated from 16 patients whose age at first P. aeruginosa isolation ranged from 0.6 to 16.3 years (mean, 12.2 years; SD, 4.4 years). Forty-seven of these isolates constituted 15 longitudinal series that ranged in duration of collection from 2 to 25 months (mean, 10.7 months; SD, 5.8 months) (Table 1). Thirty-four isolates were nonmucoid, 15 were mucoid, and 7 were multiply antibiotic resistant (MAR). The frequency with which Necker strains produced spontaneous rifampin-resistant mutants ranged from 1.29 × 10−5 to <1.9 × 10−9. Twelve clones exhibiting a Rifr frequency 1 order of magnitude greater than the population median (2.5 × 10−8) were designated mutators. Mutators were isolated from six patient series: E (10 months in duration), H (4 months), I (13 months), J (2.5 months), N (0 months), and P (3 months). By Fisher's two-tailed exact test we found no significant relationship between mutator status and mucoid status (P = 0.73); however, we did find a significant relationship between mutator and MAR status (P < 0.05). This finding is consistent with previous reports that mutators are more likely to be MAR than nonmutators at the Necker Hospital (21) and elsewhere (54).
Some degree of phenotypic polymorphism was evident in bacteria from all patients. In 8 of 16 patients, two or more strains isolated at the same time point differed with respect to either mucoidy (B, J, and K), antibiotic resistance profile (all [B, C, I, J, K, M, N, and P]), or mutator status (I, J, and N) (Table 1). As these phenotypes are vertically transmitted and, with the exception of mucoidy, stably maintained in the laboratory, we conclude they must have some heritable component. This finding, especially viewed in the light of the limited number of samples analyzed, supports the view that the CF lung contains genetically diverse P. aeruginosa populations, regardless of whether that diversity arises solely from mutation or from the combined forces of spontaneous mutation, horizontal gene transfer, and secondary infection.
Genome fingerprinting shows that clinical P. aeruginosa populations are founded by unique strains, evolve clonally, and undergo within-patient differentiation.
rep-PCR and PFGE are widely used to generate P. aeruginosa genome fingerprints that illuminate clone identity and population structure (15). The methods are mutually validating, as one is based on distribution of repetitive-sequence primer sites whereas the other is based on the distribution of restriction endonuclease recognition sites. To control for between-experiment variation, we performed rep-PCR amplifications on the entire collection simultaneously, using the same master mix. To control for variation in PFGE banding patterns, we ran the entire collection on the same gel with multiple lanes of the products from PAO1. rep-PCR and PFGE data were clustered using Bionumerics image analysis software.
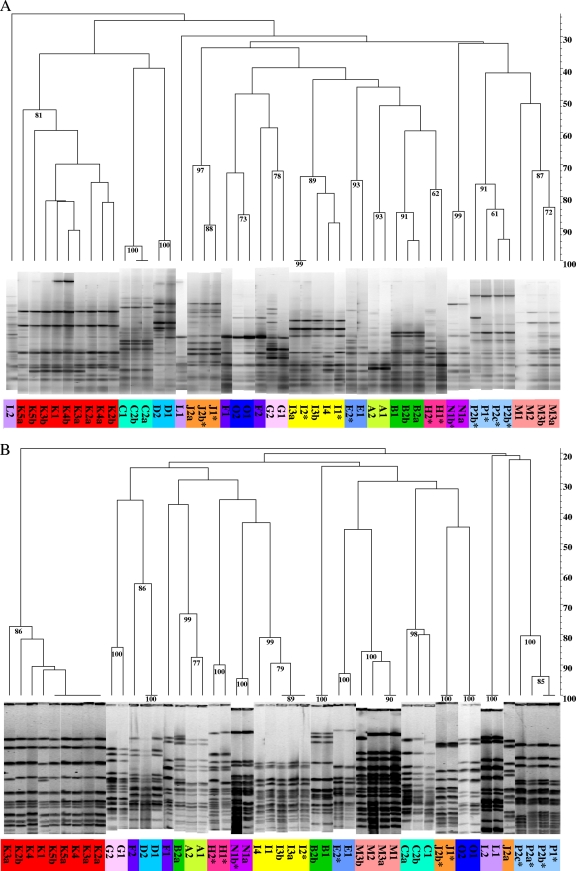
Genomic similarities within each lineagevaried, ranging from 52% to 98% for BOXA1, 78% to 100% for SpeI, 40% to 91% using ERIC, and 71% to 100% for DraI (Fig. 1; see Fig. S1 in the supplemental material). rep-PCR and PFGE data provided no evidence that certain fingerprints are more likely to be seen among initial isolates. Likewise, in no instance did we observe independent lineages to converge on particular fingerprints. These observations are consistent with previous reports that chronic P. aeruginosa infections arise from founders drawn from a genetically diverse environmental reservoir (71, 93) and that lineages evolve more or less independently within individual patients (82). Regardless of methodology, dendrograms show that isolates within patient series are much more similar to one another than they are to isolates from other patients. For example, the average within-patient similarity in SpeI PFGE dendrograms is 71.3%, whereas the among-patient similarity is only 34.2%. This overall trend holds whether fingerprints are complex (Fig. 1) or simple (see Fig. S1 in the supplemental material). The inference that within-patient similarity is greater than among-patient similarity is further supported by bootstrap estimates based on total band number and total number of distinguishing bands per group per experimental treatment. High bootstrap values have been shown to be a conservative estimate of confidence (34), with a value of 70 corresponding to ∼95% confidence.
Fig. 1.
Genomic fingerprinting. (A) rep-PCR using BOXA1 primers; (B) SpeI PFGE of SpeI-digested genomic DNA. Percent similarity is indicated on the y axis. *, mutator. Numbers at branch points indicate bootstrap values; only values of >70 are reported, as they provide 95% confidence in tree topology (34).
In certain instances, phenotypic differentiation was also manifest in different genome fingerprints. For example, J2a and J2b, which differed with respect to all three phenotypes assayed (Table 1), displayed distinct PFGE genotypes (Fig. 1B; see Fig. S1B in the supplemental material). In contrast, in the K series, where phenotypic polymorphism was evident at four of five time points (Table 1), we observed identical ERIC and DraI fingerprints (see Fig. S1 in the supplemental material) but well-differentiated BOXA1 and SpeI patterns (Fig. 1). In the only instance where a pairwise comparison between a non-mutator-containing series (M) and a mutator-containing series (I) of similar duration was possible, we found little correlation between the presence of mutators and within-lineage genetic differentiation resolvable by rep-PCR or PFGE. Interestingly, in the I series we noted that genome fingerprints of hypermutator I2 (Rifr = 1.29E−06) strongly resembled those of subsequent weak mutator I3a (Rifr = 1.25E−07) (e.g., similarity > 70% and bootstrap = 99% [Fig. 1A] and similarity > 95% and bootstrap = 89 [Fig. 1B]). This intriguing observation suggests that a pattern established by or carried within a mutator lineage can appear in a descendant clone having a lower mutation rate, an interpretation supported by our MLST data and by mutS sequence data for this series, as discussed below.
MLSA confirms clonal population structure but also provides evidence for secondary infection in some patients.
For each strain, we sequenced seven loci for multilocus sequence analysis (MLSA). Necker collection isolates were genetically diverse, with a total of 151 single-nucleotide polymorphisms (SNPs) contributing to between 6 and 12 alleles at each locus (Table 2). With two exceptions, the trpE136 and trpE137 alleles observed in patients H and I, MLST alleles in the Necker collection had been previously reported in the pubMLST database (http://pubmlst.org/paeruginosa/) (Table 3). Strain allele combinations resulted in 19 distinct sequence types (STs), 11 of which were novel (Table 3). We estimated the amount of genetic differentiation between strains from different series with the statistic FST (90), which takes on values of between 0 and 1, with higher values indicating increasing differentiation between groups. Though some STs were isolated from multiple series (D and F, L and O, and E and N), we generally observed extreme genetic differences in strains between patient series for all loci (Table 2), with FST values ranging between 0.78 and 0.97. In many patients (A, C, D, E, G, K, L, M, and N), the within-patient STs were identical at different sampling times. Therefore, MLST data are consistent with rep-PCR and PFGE data indicating that patients acquire strains from a genetically diverse environmental reservoir and that once established, bacteria evolve clonally.
Table 2.
Population genetic sequence diversity of the Necker collection
| Locus | Sequence length (nt) | No. of: |
πa | Rmb | FSTc | ||
|---|---|---|---|---|---|---|---|
| Sequences sampled | Alleles | SNPs | |||||
| acsA | 810 | 49 | 12 | 21 | 0.0082 | 3 | 0.88 |
| aroE | 944 | 48 | 9 | 41 | 0.0136 | 5 | 0.97 |
| guaA | 832 | 49 | 11 | 13 | 0.0039 | 1 | 0.84 |
| mutL | 905 | 49 | 10 | 21 | 0.0073 | 4 | 0.90 |
| nuoD | 804 | 49 | 6 | 11 | 0.0037 | 0 | 0.97 |
| ppsA | 808 | 49 | 10 | 24 | 0.0076 | 3 | 0.78 |
| trpE | 575 | 48 | 9 | 20 | 0.0080 | 1 | 0.87 |
π, nucleotide diversity per nucleotide site.
Rm, minimum number of recombination events.
From AMOVA.
Table 3.
Necker collection multilocus sequence typesa
| Patient | Strain | Alleleb |
pubMLST typec | ||||||
|---|---|---|---|---|---|---|---|---|---|
| acsA | aroE | guaA | mutL | nuoD | ppsA | trpE | |||
| A | A1 | 6 | 68 | 20 | 11 | 4 | 4 | 7 | ST439 |
| A2 | 6 | 68 | 20 | 11 | 4 | 4 | 7 | ST439 | |
| B | B1 | 1 | 5 | 5 | 7 | 4 | 6 | 7 | ST1007 |
| B2a | 1 | 5 | 5 | 7 | 4 | 6 | 7 | ST1007 | |
| B2b | 125 | 5 | 6 | 3 | 4 | 13 | 7 | ST1066 | |
| C | C1 | 4 | 4 | 16 | 12 | 4 | 6 | 3 | ST828 |
| C2a | 4 | 4 | 16 | 12 | 4 | 6 | 3 | ST828 | |
| C2b | 4 | 4 | 16 | 12 | 4 | 6 | 3 | ST828 | |
| D | D1 | 125 | 5 | 6 | 3 | 4 | 13 | 23 | ST633 |
| D2 | 125 | 5 | 6 | 3 | 4 | 13 | 23 | ST633 | |
| E | E1 | 23 | 5 | 11 | 7 | 1 | 12 | 7 | ST274 |
| E2 | 23 | 5 | 11 | 7 | 1 | 12 | 7 | ST274 | |
| F | F1 | 6 | 84 | 11 | 3 | 4 | 76 | 91 | ST1008 |
| F2 | 125 | 5 | 6 | 3 | 4 | 13 | 23 | ST633 | |
| G | G1 | 6 | 5 | 11 | 7 | 3 | 12 | 19 | ST282 |
| G2 | 6 | 5 | 11 | 7 | 3 | 12 | 19 | ST282 | |
| H | H1 | 39 | 4 | 1 | 11 | 4 | 12 | 2 | ST1009 |
| H2 | 39 | 4 | 1 | 11 | 4 | 12 | 136 | ST1067 | |
| I | I1 | 23 | 5 | 11 | 7 | 1 | 12 | 137 | ST1068 |
| I2 | 23 | 5 | 11 | 7 | 1 | 12 | 137 | ST1068 | |
| I3a | 23 | 5 | 11 | 7 | 1 | 12 | 137 | ST1068 | |
| I3b | 23 | 5 | 11 | 7 | 1 | 12 | 137 | ST1068 | |
| I4 | 23 | 5 | 11 | 7 | 1 | 12 | 137 | ST1068 | |
| J | J1 | 28 | 5 | 36 | 3 | 3 | 13 | 7 | ST155 |
| J2a | 28 | 5 | 36 | 3 | 3 | 13 | 7 | ST155 | |
| J2b | 28 | 5 | 36 | 3 | 3 | 1 | 7 | ST1010 | |
| K | K1 | 89 | 30 | 64 | 26 | 48 | 24 | 32 | ST195 |
| K2a | 89 | 30 | 64 | 26 | 48 | 24 | 32 | ST195 | |
| K2b | 89 | 30 | 64 | 26 | 48 | 24 | 32 | ST195 | |
| K3a | 89 | 30 | 64 | 26 | 48 | 24 | 32 | ST195 | |
| K3b | 89 | 30 | 64 | 26 | 48 | 24 | 32 | ST195 | |
| K4a | 89 | 30 | 64 | 26 | 48 | 24 | 32 | ST195 | |
| K4b | 89 | 30 | 64 | 26 | 48 | 24 | 32 | ST195 | |
| K5a | 89 | 30 | 64 | 26 | 48 | 24 | 32 | ST195 | |
| K5 | 89 | 30 | 64 | 26 | 48 | 24 | 32 | ST195 | |
| L | L1 | 40 | 84 | 11 | 3 | 4 | 76 | 91 | ST675 |
| L2 | 40 | 84 | 11 | 3 | 4 | 76 | 91 | ST675 | |
| M | M1 | 11 | 3 | 11 | 3 | 4 | 15 | 2 | ST1011 |
| M2 | 11 | 3 | 11 | 3 | 4 | 15 | 2 | ST1011 | |
| M3a | 11 | 3 | 11 | 3 | 4 | 15 | 2 | ST1011 | |
| M3b | 11 | 3 | 11 | 3 | 4 | 15 | 2 | ST1011 | |
| N | N1a | 23 | 5 | 11 | 7 | 1 | 12 | 7 | ST274 |
| N1b | 23 | 5 | 11 | 7 | 1 | 12 | 7 | ST274 | |
| O | O1 | 40 | 84 | 11 | 3 | 4 | 76 | 91 | ST675 |
| O2 | 7 | 5 | 12 | 3 | 4 | 1 | 7 | ST549 | |
| P | P1 | 40 | 68 | 20 | 11 | 29 | 4 | 7 | ST1069 |
| P2a | 121 | 20 | 26 | 13 | 3 | 64 | 7 | ST1012 | |
| P2b | 121 | 20 | 26 | 13 | 3 | 64 | 7 | ST1012 | |
| P2c | 121 | 20 | 26 | 13 | 3 | 13 | 7 | ST1013 | |
Shaded rows indicate instances of possible co-/cross-infection.
MLST alleles were assigned based on the following nucleotide positions of the sequence data set: acsA, nt 95 to 484; aroE, nt 233 to 730; guaA, nt 273 to 645; mutL, nt 453 to 894; nuoD, nt 86 to 451; ppsA, nt 1 to 370; trpE, nt 1 to 443. trpE136 and trpE137 (boldface) are novel alleles not previously reported in the global MLST database.
Boldface indicates novel pubMLST types reported here for the first time.
The FST values suggest that the probability of strain migration between patients is low. However, MLSA reveals instances of secondary infection, manifest as multiple genotypes infecting the same individual and one of the genotypes infecting multiple individuals. Three clones (D1, D2, and F2) from two patients exhibit identical STs, sharing sequence identity over 5,678 nucleotides (nt) distributed across seven genes (shaded areas in Table 3). Likewise, both strains from patient L (L1 and L2) and one strain from patient O (O1) exhibit identical STs but for a different set of alleles than those infecting patients D and F. CLONDIAG (Clondiag Chip Technologies, Germany) biochip analysis of 13 SNPs in conserved genomic regions, including oriC, citS, ampC, oprI, fliC, oprL, and alkB2 (49), also supports the inference of co-/cross-infection by these strains (data not shown). Significantly, all patients in this study overlapped in their treatment schedules (see Fig. S5 in the supplemental material), and patients D, F, and L were all homozygous for the F508del cystic fibrosis transmembrane conductance regulator (CFTR) allele that appears to predispose patients to invasion by transmissible P. aeruginosa (45).
Mutator series appear to be no more variable in their MLST haplotypes than nonmutator series.
Whether populations with mutators are more genetically variable than those without is a question that has far-reaching implications for understanding P. aeruginosa evolutionary dynamics in the CF lung. As all but two MLST alleles in the Necker collection have been previously reported (Table 3), we conclude that most within-series MLST variation arose prior to the onset of infection. However, the limited number of samples available for any time point compromises the accuracy with which we can estimate within-series genetic variation. While our MLST data do not support the inference that mutator series are more genetically variable than nonmutators, it is noteworthy that the only novel alleles found in this study were from patients with mutator strains (H and I) (Table 3).
MLSA provides no evidence for between-strain recombination during chronic infection.
Secondary infection provides opportunities for recombination between strains to create novel genomic diversity of potential clinical importance. To test for recombination, we used Rm (40), a conservative estimator of the minimum number of recombination events in a sample of nucleotide data that conform to the infinite-site mutation model (i.e., where recurrent mutation has not occurred and a maximum of two nucleotide identity states are therefore observed at a position). For data that conform to this model, the observation of all four possible genotypes between a pair of polymorphic sites can be explained only by a recombination event between the sites. With only a single exception (in aroE), all SNPs in the MLSA data set conform to the infinite-site model. Recombination was detected for six of the seven loci (Table 2).
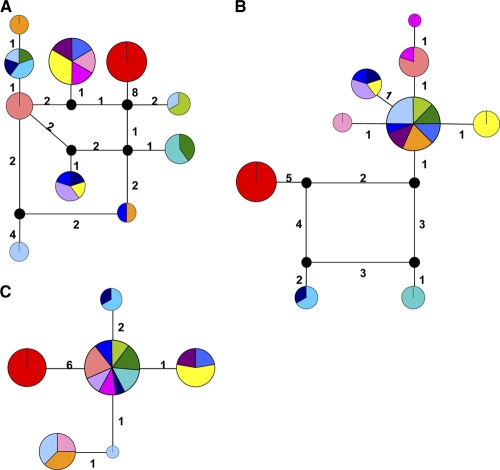
Recombination events are also evident in the genealogical histories of the loci (Fig. 2). Genealogical networks accommodate recombination, as well as other scenarios that traditional bifurcating phylogenies fail to accurately describe (66), such as ancestor-descendant relationships among extant alleles (with ancestors at internal nodes in the network) and multifurcations. Loci impacted by recombination are graphically represented as cycles (i.e., loops) in these networks to relate the four genotypes arising from only two mutations (Fig. 2). Though the respective evolutionary histories of these loci have been shaped by recombination, these events predate the population dynamics of the series under study, as the alleles connected via recombination cycles have been previously reported (http://pubmlst.org/paeruginosa/) (Table 3). We therefore conclude that recombination at these loci has not impacted the population genetics of individual series during the period of chronic infection for which we have records, including series for which there is evidence of secondary infection.
Fig. 2.
Genealogical relationships among alleles recovered from the Necker collection for representative loci: ppsA (A), trpE (B), and nuoD (C). The area of the node is proportional to the relative frequency of the allele in the sample, and the wedge color corresponds to the series color depicted in Fig. 1 and in Fig. S1 in ths supplemental material. The sum of the branch lengths between two alleles corresponds exactly to the number of nucleotide sequence differences observed between them. Note the presence of cycles for loci impacted by recombination. Black nodes represent alleles not observed in the sample.
Comparative analysis of a mutator series and a nonmutator series.
Patients I and M were roughly age matched (13.5 and 14.9 years old, respectively) as a surrogate for infection duration. Because neither series from these patients showed evidence of P. aeruginosa co-/cross-infection by MLSA and because mutators were isolated from patient I but not from patient M (Table 1), we chose to study genetic variation in these two series comparatively and in greater detail. As a first step toward discovering the mechanistic basis for the I-series mutator phenotype and to test for the interesting possibility that the later-arising I clones displayed lower mutation frequencies due to compensatory mutations, we completely sequenced mismatch repair genes mutL and mutS, the loci most frequently associated with the mutator phenotype (62).
Complete sequences for mutL and mutS reveal the basis for mutator phenotypes in the I series.
An alignment (see Fig. S4 in the supplemental material) of 2,568 bp of mutS from the I and M series and four outgroups (Necker isolate K1, a Liverpool epidemic strain [LESB58], UCBPP-PA14, and PAO1) revealed 59 polymorphic sites (see Table S3 in the supplemental material) with only one nonsynonymous change in K1M (nt 841; S → G) and a novel 8-bp deletion at nt 1194 to 1201 (Table 4). The latter results in a frameshift mutation and a premature stop codon in I1, I2, I3a, and I3b. I3a and I3b exhibit spontaneous mutation to Rifr frequencies that are an order of magnitude lower than those of I1 and I2, falling just below the cutoff to be classified as mutators (Table 1). Thus, I3a and I3b may contain extragenic suppressors that partly compensate for the loss of MutS activity.
Table 4.
Summary of mutS polymorphism
| Isolate(s) | Base at nta: |
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 336 | 342 | 357 | 594 | 804 | 951 | 996 | 1047 | 1120 | 1194–1201 | 1245 | 1506 | 1539 | 1575 | 1599 | 1650 | 1668 | 1902 | 1926 | 1998 | 2052 | 2181 | 2187 | |
| I1, I3a | C | C | G | G | C | G | T | T | T | Deletion | G | A | A | G | C | G | C | T | G | C | T | C | G |
| I2 | C | C | G | G | C | G | T | T | T | Deletion | G | A | A | G | T | G | C | T | G | C | T | C | G |
| I3b | C | C | G | G | C | G | T | T | T | Deletion | G | A | A | A | C | G | C | T | G | C | T | C | G |
| I4 | C | C | G | G | C | G | T | T | T | CTATCCCG | G | A | A | G | C | G | C | T | G | C | T | C | G |
| M1, M3a | T | T | A | A | T | A | C | C | C | CTATCCCG | A | G | G | G | T | A | T | C | A | C | C | T | A |
| M2 | T | T | A | A | T | A | C | C | C | CTATCCCG | A | G | G | G | C | A | T | C | A | G | C | T | A |
| M3b | T | T | A | A | T | A | C | C | C | CTATCCCG | A | G | G | G | C | A | T | C | A | C | C | T | A |
| LESB58 | C | C | G | A | T | A | C | C | C | CTATCCCG | A | G | G | G | C | A | T | C | A | C | C | T | A |
| PA14 | C | C | G | G | C | G | T | T | T | CTATCCCG | G | G | G | A | T | G | C | T | G | C | T | C | G |
| PAO1 | C | C | G | A | T | A | C | C | C | CTATCCCG | A | G | G | G | C | A | T | C | A | C | C | T | A |
Shaded changes are unique to a series and do not represent underlying polymorphism observed in reference strains.
The novel 8-bp deletion is not detected in I4, a bona fide nonmutator (3.90E−08) and the final isolate of the I series, whose PFGE profiles and MLST haplotype match those of the earlier strains. Together, these observations suggest that I-series hypermutators and nonmutators all originate from an I4-like ancestor. The location of I4 at an internal node of a minimum spanning network analysis supports this conclusion (see see Fig. S3A in the supplemental material). Unlike diversity indices such as π, AMOVA calculations can consider the presence or absence of nucleotides as a fifth state; therefore, divergence due to deletion is reflected in the resulting distance matrix and network diagram. The mutS network coincides with other data placing the I series in closer proximity to PA14 and the M series nearer PAO1; K1 is a distant outlier, as in other analyses.
Restricting comparisons to the mutator-containing I series and the nonmutator M series, there are only 23 mutS polymorphisms (counting the 8-bp deletion in I-series mutators as a single mutational event); 16 polymorphisms simply reflect the underlying diversity of reference strains (Table 4; see Table S3 in the supplemental material). Seemingly contrary to a hypothesis that hypermutator status should generate more molecular diversity, four of the remaining polymorphisms are unique to the M series, while only two are unique to the I series; however, caution should be exercised when drawing genome-wide conclusions about diversity from just a few loci. The I series does exhibit within-series diversity at two sites (nt 1575 and 1599), in addition to the 8-bp deletion. The M series also exhibits polymorphism at nt 1599, suggesting that this site may be prone to mutation.
In contrast to the case for the mutS locus, analysis of 1,902-bp mutL sequences (see Fig. S4B in the supplemental material) indicates homogeneity in the I and M series (Table 5). Of 54 polymorphic sites (see Table S3 in the supplemental material), K1 exhibited the only nonsynonymous change (nt 5; S → N) in this subset of Necker strains. Haplotypic differences between the two patient series, outgroup isolate K1, and reference strains allowed construction of an alternate single-locus minimum spanning network. The structure of the mutL minimum spanning network reflects haplotypic similarity of the I series to the Liverpool epidemic strain LESB58 (see Fig. S3B in the supplemental material). M-series isolates appear to be more closely related to lab strain PAO1, a result that is consistent with MLST, a-CGH, and mutS network data.
Table 5.
Summary of mutL polymorphism
| Isolate(s) | Base at nta: |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 103 | 321 | 396 | 498 | 531 | 558 | 726 | 750 | 1035 | 1308 | 1365 | 1470 | 1689 | 1704 | 1717 | 1743 | 1771 | 1815 | 1857 | 1858 | |
| I series | C | T | T | C | A | T | T | T | G | C | C | T | C | G | T | T | C | G | A | C |
| M series | T | C | C | T | G | C | C | C | C | T | T | G | T | G | C | C | T | A | G | T |
| LESB58 | C | T | T | T | G | C | C | C | G | C | T | T | C | G | T | T | C | A | G | T |
| PA14 | C | C | C | C | A | T | C | C | C | C | C | T | C | A | T | C | C | A | G | T |
| PAO1 | C | C | C | T | G | C | C | C | C | T | T | T | T | G | C | C | T | A | G | T |
Shaded changes are unique to a series and do not represent underlying polymorphism observed in reference strains.
Comparative genome hybridization shows that mutator and nonmutator longitudinal series lack a common set of genes found in P. aeruginosa PAO1.
Analyses of genome fingerprints, MLST haplotypes, and mutL and mutS sequences provided no clear evidence that patient series from which we isolated mutators were genetically more variable than series from which we did not. Because mismatch repair (MMR)-deficient strains are more prone to homologous recombination (63, 68, 18) and thus susceptible to genome rearrangement, we also determined whether mutator and nonmutator populations exhibited differences in genome content using Affymetrix PAO1 microarrays and validated the results using PFGRC arrays.
Microarray analyses indicated that 126 PAO1 open reading frames (ORFs) could not be detected in multiple members of the mutator and nonmutator series (Table 6); this represents ∼142.4 kb or 2.3% of the ∼6.3-Mbp PAO1 reference genome. These genes fell into 49 contiguous blocks and consisted of 89 transcriptional units, 27 of which were polycistronic. A majority (70/126) of these ORFs were in the PseudoCAP function category “hypothetical, unclassified, unknown” (Table 6; see Table S4A in the supplemental material). Of the remainder, 7 were related to “phage, transposon, or plasmid” and included genes having homology to phage Pf1 (PA0717 and PA0724). Fifty-eight fell into one or more of functional categories: “motility and attachment” (4), “cell wall/lipopolysaccharide (LPS)” (15), “membrane proteins” (9), “transport of small molecules” (9), and “putative enzymes” (21). Most of these 58 ORFs have homology to proteins that carry out activities at the cell surface, e.g., a block of 20 known virulence factors (PA3142 to PA3160) dedicated to LPS O-antigen biosynthesis and export, as well as pilin genes pilA and pilC and fimbrial chaperone genes cupC1 and cupC2. Five PAO1 transcription factors were also absent, two having the lysR family signature (PA0207 and PA2220) and one, vqsM (PA2226), with 47% similarity to the essential quorum-sensing regulator OruR. As a group, therefore, all nine strains isolated from these two patients share attributes seen as characteristic of CRI in the cystic fibrosis lung (20, 22, 80): loss of certain genes whose products, by virtue of being secreted from or situated at the cell surface, offer targets for the immune system and loss of others whose products help mediate quorum sensing.
Table 6.
PAO1 genes not detected in either I or M series
| Boundary | Size (bp) | Presence in straina: |
Function | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| I series |
M series |
||||||||||
| I1 | I2 | I3a | I3b | I4 | M1 | M2 | M3a | M3b | |||
| PA0202-PA0207 | 6,568 | Putative enzymes | |||||||||
| Transport of small molecules | |||||||||||
| Membrane protein | |||||||||||
| Transcriptional regulator | |||||||||||
| PA0442 | 116 | Unknown | |||||||||
| PA0445 | 1,016 | Related to phage, transposon, or plasmid | |||||||||
| PA0642 | 788 | Unknown | |||||||||
| PA0715-PA0717 | 3,387 | Related to phage, transposon, or plasmid | |||||||||
| PA0724 | 1,262 | Related to phage, transposon, or plasmid | |||||||||
| PA0820 | 812 | Unknown | |||||||||
| PA0977 | 323 | Unknown | |||||||||
| PA0980-PA0981 | 943 | Unknown | |||||||||
| PA0983-PA0985 | 2,679 | Membrane proteins, secreted factors | |||||||||
| PA0992-PA0993 | 1,388 | Motility and attachment | |||||||||
| Motility and attachment | |||||||||||
| PA1133 | 350 | Unknown | |||||||||
| PA1152 | 344 | Unknown | |||||||||
| PA1204 | 557 | Unknown | |||||||||
| PA1239-PA1241 | 2,463 | Putative enzymes | |||||||||
| Transcriptional regulators | |||||||||||
| PA1368-PA1372 | 7,761 | Unknown | |||||||||
| PA1385 | 1,124 | Cell wall/LPS/capsule | |||||||||
| PA1394 | 254 | Unknown | |||||||||
| PA1509-PA1510 | 2,848 | Unknown | |||||||||
| Unknown | |||||||||||
| PA1887-PA1888 | 2,061 | Unknown | |||||||||
| PA1935-PA1936 | 1,067 | Unknown | |||||||||
| PA1939 | 1,997 | Unknown | |||||||||
| PA2037 | 1,460 | Unknown | |||||||||
| PA2046 | 407 | Unknown | |||||||||
| PA2218-PA2228 | 11,615 | Membrane proteins | |||||||||
| Transcriptional regulator | |||||||||||
| PA2372 | 572 | Unknown | |||||||||
| PA2427 | 467 | Unknown | |||||||||
| PA2456 | 341 | Unknown | |||||||||
| PA2459-PA2462 | 19,322 | Unknown | |||||||||
| PA2730-PA2736 | 10,319 | Putative enzymes | |||||||||
| PA2791 | 290 | Unknown | |||||||||
| PA2818-PA2819 | 2,125 | Unknown | |||||||||
| PA3142_i-PA3160_wzz | 19,498 | Cell wall/LPS/capsule, putative enzymes, amino acid biosynthesis and metabolism, membrane proteins, transport of small molecules | |||||||||
| PA3292 | 857 | Unknown | |||||||||
| PA3298 | 302 | Unknown | |||||||||
| PA3486-PA3488 | 6,856 | Fatty acid and phospholipid metabolism | |||||||||
| PA3497-PA3514 | 16,113 | Putative enzymes | |||||||||
| Transcriptional regulator | |||||||||||
| Membrane proteins | |||||||||||
| Transport of small molecules | |||||||||||
| PA3577_i | 113 | Unknown | |||||||||
| PA3866-PA3869 | 5,339 | Adaptation, protection; secreted factors | |||||||||
| DNA replication, recombination, modification, and repair | |||||||||||
| PA4255_rpmC | 191 | Translation, posttranslational modification, degradation | |||||||||
| PA4306 | 218 | Unknown | |||||||||
| PA4485 | 377 | Unknown | |||||||||
| PA4503 | 1,010 | Membrane proteins, transport of small molecules | |||||||||
| PA4525_pilA | 449 | Motility and attachment | |||||||||
| PA4527_pilC | 1,124 | Motility and attachment | |||||||||
| PA4999 | 1,205 | Unknown | |||||||||
| PA5086-PA5087 | 1,747 | Unknown | |||||||||
| Unknown | |||||||||||
| Total size | 142.4 kb | ||||||||||
Shading indicates that the gene is present in the genome.
Comparative genome hybridization also illuminates differences in genome content between a mutator and a nonmutator series.
Of particular relevance to our study was the distribution of PAO1 genes whose presence or absence distinguished the nonmutator M series from the mutator I series. All strains in the M series lacked 14 ORFs present in both PAO1 and every member of the I series (Table 7; see Table S4B in the supplemental material). These genes were located in 8 contiguous stretches of DNA and consisted of 14 transcriptional units, 4 of which were polycistronic; most (12/14) were in the PseudoCAP function class “hypothetical, unclassified, unknown.” A conspicuous genetic polymorphism was evident at the last sample collection, as one of two strains isolated (M3b) had an additional 86-gene, ∼105-kb deletion (PA2425 to PA2510) as well as a deletion in PA3904. The inference that this block of genes was lost during chronic infection is supported by the facts that, aside from this one large deletion, M-series isolates exhibit identical MLST haplotypes (Table 3) and similar PFGE fingerprints (Fig. 1B; see Fig. S1B in the supplemental material) (but note the diminished size of second largest SpeI band in M3b [Fig. 1B]). The large M3b deletion contains 53 transcriptional units, of which 21 are polycistronic. Of 86 PAO1 genes that are absent, almost half (40/86) are classified as “hypothetical, unclassified, unknown,” while 23 fall into the functional categories “adaptation and protection” (2), “membrane protein” (13), and “transport of small molecules” (8), activities that likely occur at the cell surface. The deleted segment includes 3 known virulence factors (PA2425 to PA2427) involved in pyoverdine gene expression. Also deleted are multiple activities (sdaA, glyA2, gcvP2, and gcvH2) related to serine and glycine metabolism as well as those encoded by the multidrug efflux transporter genes mexE and mexF and their regulator, mexT. Indeed, a remarkable aspect of this ∼105-kb deletion is that 13 of the 86 PAO1 ORFs lost are known or putative transcriptional regulators, including foxI, foxR, and the LysR substrate gene bexR, in addition to mexT. While the ultimate evolutionary fate of M3b is unknown, either the loss of these genes, so many of which regulate gene expression, does not impose a fitness cost or that cost is offset by fitness gains arising from loss of virulence factors.
Table 7.
PAO1 genes not detected in M series only
| Boundary | Size (bp) | Presence in straina: |
Function | |||
|---|---|---|---|---|---|---|
| M1 | M2 | M3a | M3b | |||
| PA0053 | 254 | Unknown | ||||
| PA0643 | 1,091 | Unknown | ||||
| PA0982 | 548 | Unknown | ||||
| PA1366 | 770 | Unknown | ||||
| PA2073-PA2074 | 2,242 | Transport of small molecules | ||||
| Unknown | ||||||
| PA2425-PA2510_catR | 107,346 | Putative enzymes | ||||
| Transcriptional regulator | ||||||
| Membrane proteins | ||||||
| Transport of small molecules | ||||||
| Amino acid biosynthesis and metabolism | ||||||
| Central intermediary metabolism | ||||||
| Carbon compound catabolism | ||||||
| Adaptation, protection | ||||||
| Chaperones and heat shock proteins | ||||||
| Translation, posttranslational modification, degradation | ||||||
| Two-component regulatory systems | ||||||
| Energy metabolism | ||||||
| Antibiotic resistance and susceptibility | ||||||
| Carbon compound catabolism | ||||||
| PA2794 | 1,316 | Unknown | ||||
| PA3291 | 554 | Unknown | ||||
| PA3904_i | 395 | Unknown | ||||
| Total | 114.5 kb | |||||
Shading indicates that the gene is present in the genome.
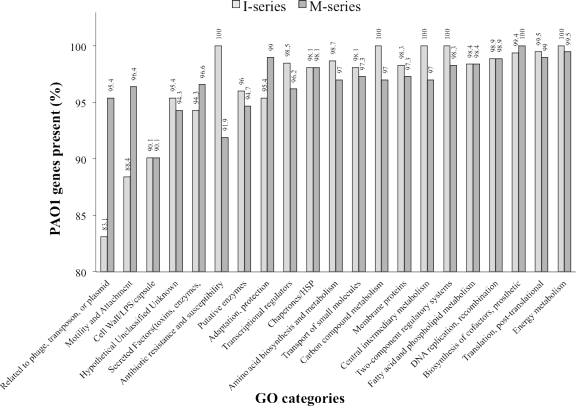
In contrast to 14 PAO1 ORFs that were absent from all nonmutators (Table 7), microarray analysis uncovered 61 ORFs that were present in nonmutators but absent from the I series (Table 8; see Table S4C in the supplemental material). These genes fell into 20 contiguous blocks, and consisted of 33 transcriptional units, of which 16 are polycistronic. Remarkably, unlike the set of genes absent from both series or those absent from the M series only, few (15/61) were classified as “hypothetical, unclassified, or unknown.” Rather, a clear majority (35/61) fell into three PseudoCAP functional categories: 18 ORFs were “related to phage, transposon, or plasmids,” including 14 clustered genes (PA0615 to PA0628) having homology to P. aeruginosa phage φCTX and bacteriophage P2, and another 17 ORFs were functionally related to “motility and attachment” (9) and “adaptation and protection” (8). All of the former are cotranscribed at operon 932 (PA4549 to PA4556) and comprise genes encoding type IV fimbrial biogenesis proteins. Among the latter category are a contiguous set of siderophore loci (PA2396 to PA2401) that constitute virulence factors, as well as pyocin S2 killer protein and immunity genes (PA1150 to PA1151). I-series strains therefore recapitulate the theme of loss of function in potential host immune system targets seen in cystic fibrosis CRI. Further, of 15 ORFs having no known function, 6 have computationally predicted export signals (see Table S4C in the supplemental material). Overall, and even including the M3b deletion strain in our analyses, the high-mutation-frequency I series is distinguished from the low-mutation-frequency M series by the absence of more PAO1 genes functionally related to mobile elements, motility and attachment, and adaptation and protection, while retaining a nearly complete complement of PAO1 genes involved in carbon and central intermediary metabolism and in antibiotic resistance (Fig. 3). Regarding this last observation, it is noteworthy that while no M-series strain was multiply antibiotic resistant, all but one of the I-series strains were (Table 1).
Table 8.
PAO1 genes not detected in I series only
| Boundary | Size (bp) | Presence in straina: |
Function | ||||
|---|---|---|---|---|---|---|---|
| I1 | I2 | I3a | I3b | I4 | |||
| PA0098-PA0099 | 2,197 | Unknown | |||||
| PA0100 | 920 | Unknown | |||||
| PA0497-PA0499 | 2,803 | Unknown | |||||
| Motility and attachment, chaperones and heat shock proteins | |||||||
| PA0615-PA6028 | 12,085 | Related to phage, transposon, or plasmid | |||||
| PA0646-PA0648 | 1,581 | Unknown | |||||
| PA0719 | 377 | Related to phage, transposon, or plasmid | |||||
| PA0723_coaB | 248 | Related to phage, transposon, or plasmid | |||||
| PA0729 | 347 | Unknown | |||||
| PA0821-PA0824 | 2,321 | Unknown | |||||
| PA1150_pys2-PA1151_imm2 | 2,334 | Adaptation, protection | |||||
| Secreted factors (toxins, enzymes, alginate) | |||||||
| PA1664 | 140 | Unknown | |||||
| PA1914 | 1,226 | Unknown | |||||
| PA2036 | 521 | Unknown | |||||
| PA2100-PA2106 | 6,801 | Transcriptional regulator | |||||
| Biosynthesis of cofactors, prosthetic groups, and carriers | |||||||
| Amino acid biosynthesis and metabolism | |||||||
| Putative enzymes | |||||||
| PA2386_pvdA-PA2387 | 1,943 | Adaptation, protection | |||||
| Transcriptional regulator | |||||||
| PA2392 | 1,634 | Unknown | |||||
| PA2396-PA2401 | 19,398 | Unknown | |||||
| Adaptation, protection | |||||||
| Transport of small molecules | |||||||
| Secreted factors (toxins, enzymes, alginate) | |||||||
| Membrane proteins | |||||||
| PA2403 | 1,211 | Unknown | |||||
| PA2816 | 380 | Unknown | |||||
| PA4514 | 2,261 | Transport of small molecules | |||||
| PA4549_fimT-PA4556_pilE | 7,342 | Motility and attachment | |||||
| PA5191 | 356 | Unknown | |||||
| Total | 68.4 kb | ||||||
Shading indicates that the gene is present in the genome.
Fig. 3.
Percentages of PAO1 genes detected in the I and M series by functional category.
DISCUSSION
P. aeruginosa population genetic structure in nature and in the clinic.
P. aeruginosa is a ubiquitous microbe that can be isolated from soils, waters, and plant surfaces, as well as from infection sites of immunocompromised individuals (53). Although environmental P. aeruginosa strains differ widely in genome content (47, 51), evidence suggests that most retain infectious potential (93). The conserved P. aeruginosa core genome contains recombinogenic regions where genomic content is modulated by entry and exit of plasmids (46), integrons (23), and other active mobile genetic elements (69). Incorporation of certain accessory elements into the core genome may in some cases exclude others, resulting in different regulatory networks and antibiotic resistance profiles (51, 93). For example, Kiewitz et al. reported two reversible entry sites for a 106-kbp plasmid among sequential P. aeruginosa isolates (46). Strains occupying the same habitat may therefore possess identical accessory elements but regulate them differently according to where they are integrated.
Because the P. aeruginosa genome is malleable, the total population genetic diversity observed locally can approach that observed globally (92). Still, regionally dominant clones can arise and become disproportionately represented in a particular hospital or in closely associated groups of patients and family members (3, 41, 86). Although it is widely accepted that CF patients generally acquire infections from environmental reservoirs (71), cross-infection has been reported between patients harboring strains especially adept at colonization (44). Typically, however, such instances of cross-infection occur between CF patients having prolonged contact, such as siblings, or between uninfected and infected CF patients at clinics where patients are not segregated. Also, a recent study reported an apparent link between host CFTR genotype homozygous F508del and susceptibility to transmissible strains (45). Transmissible-strain infections are generally associated with worse clinical outcomes (1).
Clonality and secondary infection in the Necker collection.
The population genetic structure of the Necker isolates conforms to the view that local diversity can approach that seen globally (92). Genome fingerprint and MLST data indicate that the first strain isolated from each longitudinal series is genetically distinct from the first strain isolated from other series. Necker strains cluster within individual patients as clonal groups, with no evidence for interclonal recombination over periods of up to 25 months. Significantly, every patient in our study group entered the Necker CF clinic for treatment and sampling during 2002 (Table 1; see Fig. S5 in the supplemental material). In most cases genome fingerprint and MLST data provide no evidence for co-/cross-infection among patients. However, three clones isolated from patients D (D1 and D2) and F (F2) exhibit identical 7-locus MLST genotypes, and three clones isolated from patients L (L1 and L2) and O (O1) share a different 7-locus MLST genotype. In both groups, among-strain MLST identity is reflected in similar PFGE fingerprints (Fig. 1B; see Fig. S1B in the supplemental material). Further, strains isolated from patients D and F have identical CLONDIAG SNP-chip profiles that differ from identical SNP-chip profiles shared by strains from patients L and O (N. Cramer and B. Tümmler, unpublished data). Although postisolation cross-contamination is formally possible, these observations, coupled with the facts that patients overlapped in their treatment schedules and were homozygous for the F508del CFTR mutation, more likely indicate secondary infection.
Establishment of chronic P. aeruginosa infections in the CF lung: successful colonists may not make successful residents.
Evidence now suggests that following multiple acute episodes early in life, CF patients are colonized by one or a few persistent P. aeruginosa strains that establish a chronic infection (44, 61). Transition from the acute to chronic phase is often marked by the appearance of mucA mutants that produce copious exopolysaccharide (EPS) (19), as well as by differential regulation of the GAC histidine sensor-kinase complex (26). We observed both mucoid and nonmucoid phenotypes within lineages, sometimes isolated from the same patient at the same time (Table 1, patients B, J, and K). This observation may reflect heterogeneity in the lung environment and/or clonal differentiation as strains adaptively radiate to anaerobic niches within the lung (94) or to aerobic regions in the upper respiratory tract (39, 44). We also observe instances where a mucoid strain fingerprint is later observed in a nonmucoid strain (Fig. 1; see Fig. S1 in the supplemental material [patient K]). These findings, considered in light of the fact that mucoid reversion to the nonmucoid phenotype can occur by a single point mutation in algT (19), raise the possibility that mucoid polymorphism may be an example of “social cheating” (74). Overproduction of EPS promotes bacterial colony formation and attachment to epithelial surfaces, and biofilms consolidated with EPS appear to be more resistant to antibodies and antibiotics (31, 55). However, because EPS overproduction carries a cost and because reversion can occur at high frequency, nonmucoid strains may arise that enjoy the benefits of a “public good,” namely, EPS, without having to bear the burden of its synthesis.
P. aeruginosa strains residing in the CF lung are polymorphic but experience limited gene flow with P. aeruginosa strains living in the external environment.
Traits required for P. aeruginosa to successfully invade and colonize the CF lung appear to differ from those required for it to persist (76). Motility, type III secretion, and O-antigen biosynthesis, important factors in the onset of an acute infection, are commonly lost during the course of chronic infection (28, 43, 52). Over time, progressive loss of these traits should diminish the likelihood of transmission between CF hosts. Moreover, because the number of cells in an established population is vastly greater than the number of any new colonist cells, established P. aeruginosa populations should resist invasion by novel environmental strains. Thus, while horizontal gene transfer can shape genome architecture in the external environment (67), most evidence suggests that evolutionary dynamics of chronic P. aeruginosa infections in the CF lung are driven by vertical gene transfer (77, 83). Successive clones within patients should therefore more closely resemble one another than those in other patients.
Analysis of the Necker collection generally upholds this prediction: genome fingerprints (Fig. 1; see Fig. S1 in the supplemental material) and MLST haplotypes (Table 3) cluster within series. Nevertheless, Necker isolates exhibit extensive phenotypic variation, evident as polymorphism in antibiotic sensitivities, mucoidy, and spontaneous mutation to rifampin resistance within all 16 patient series. MLST analysis reveals genetic polymorphism in 6 of 16 series. Of these six, two (F and O) appear to be polymorphic by virtue of secondary infection involving strains shared with patients D and L, respectively. Significantly, the MLSA-based inference of clonal relatedness of both D isolates to F2 and relatedness of both L isolates to O1 is supported by PFGE data (Fig. 1B; see Fig. S1B in the supplemental material). Of the other patient series that 7-locus MLSA reveals to be polymorphic, four (H, I, J, and P) contain mutators, and clonal relatedness is discernible in three: J is polymorphic only at pps, and H and I are polymorphic only at trpE. Overall, MLST polymorphism is greatest in the all-mutator P series.
Population genetic structure in the Necker collection thus appears to arise from each patient being successfully colonized by an environmental strain, possibly including those originating from other patients, followed by chronic infections wherein relatedness is evident among successively isolated clones. Superimposed on this underlying structure is widespread phenotypic and genotypic polymorphism, in several instances arising from likely co-/cross-infection and in others presumably from the interaction of selection, drift, and mutation, the last of which is augmented in MMR-deficient mutators.
Considering the limited number of strains isolated from each patient and the limited amount of sequence data obtained for any strain, the amount of phenotypic and genetic diversity that we report almost certainly underestimates total P. aeruginosa diversity within and among patients (24). Unfortunately, sampling limitations complicate inference of ancestor-descendant relationships, making it difficult to distinguish alternative evolutionary models of clonal succession and clonal interference. Distinguishing between these alternatives will require a much more extensive assessment of within-patient clonal diversity, involving scores rather than pairs of strains isolated at regular time intervals, regardless of whether or not the patient is suffering from acute exacerbation of symptoms.
Parallel evolution in P. aeruginosa strains chronically infecting the CF lung.
In addition to selective pressures arising from interactions with other microbes (48, 77), P. aeruginosa evolves in the CF lung under host-specific pressures that include immune system surveillance, intermittent antibiotic therapy, low iron availability, and physicochemical factors arising from defects in the CFTR gene, notably copious secretions abundant in branched-chain amino acids and low O2 tension (29, 94). Because these same selective pressures occur to various degrees in all CF patients, it is perhaps not surprising to find parallel adaptive evolution in P. aeruginosa, specifically loss-of-function mutations in motility, type III secretion, O-antigen biosynthesis, and production of virulence factors such as exotoxins, phenazine, and proteases (78), as well as diminished expression of type IV fimbrial biogenesis genes (42). Indeed, many of the functions required to invade and successfully colonize the CF lung are dispensed with as P. aeruginosa transitions from acute to chronic infection, presumably because the secreted molecules associated with these functions provide inviting targets for the host immune system (60). Other parallel changes include regulatory mutations in mucA that may enhance cells' propensity to form biofilms, mutations in mexZ that increase expression of multidrug efflux pumps (2, 78), and loss-of-function mutations in lasR, a central regulator of intercellular quorum sensing via the synthesis and recognition of acyl-homoserine lactones. Relative to wild-type strains, lasR mutants exhibit multiple phenotypes that are likely to increase fitness in the CF lung; these include improved growth on amino acids that are abundant in CF lung secretions (7) and metabolic shifts that diminish O2 consumption and increase nitrate utilization (37), which indirectly enhance tobramycin and ciprofloxacin resistance (37).
Comparative genome hybridization suggests parallel evolution in longitudinal series that contain or lack mutators.
Microarray analysis shows that P. aeruginosa strains in two patient series lack ∼70 genes categorized as “hypothetical, unclassified, unknown.” However, both series also lack nearly as many genes that fall into categories where loss of function has been associated with CRI. Most of the latter have homology to cell surface protein genes and include virulence determinants associated with LPS O-antigen biosynthesis and export, as well as pilins and fimbrial chaperones. Multiple transcription factors were also absent, two having the lysR family signature and one (vqsM) having similarity to the quorum-sensing regulator OruR. Of the PAO1 genes whose absence distinguishes the nonmutator M series from the mutator I series, most (12/14) have no known function. In contrast, many (18/61) of the PAO1 loci whose absence distinguishes the mutator-containing I series are related to phages, transposons, or plasmids, including a cluster of 14 genes homologous to P. aeruginosa phage φCTX and bacteriophage P2. Another 17 ORFs were functionally related to motility and attachment and to adaptation and protection. Many of the former are cotranscribed and encode type IV fimbrial biogenesis proteins, whereas several of the latter encode siderophore proteins, which are recognized virulence factor loci where diversity has previously been observed (79).
The 105-kb deletion that we uncovered in nonmutator M3b also fits the pattern of parallel phenotypic evolution during chronic infection. This single massive deletion results in loss of function in activities related to serine and glycine metabolism, regulation of mex multidrug efflux transporters, and multiple virulence factors related to pyoverdine gene expression. Deletions of similar size involving pyoverdine loci have been observed in strains from other collections (20), indicating that such deletants may have an adaptive advantage in the CF lung environment.
Evolution of mutators: another example of parallel evolution.
In additional to loss-of-function and regulatory mutations, P. aeruginosa isolates from chronic but not from acute CF infections show a high incidence of defects in DNA repair and error avoidance pathways (63), resulting in heritable mutators that should increase population genetic variation. First reported by Oliver et al. (61), the unusually high incidence of P. aeruginosa mutators in CF patients has since been associated with hallmark features of chronic infection: antibiotic resistance (this study and references 21, 38, and 63), cytotoxicity to bronchial epithelial cells (38), increased anaerobic and microaerobic respiration (35), and attenuation of virulence (38, 58). Ferroni et al. (21) recently reported diminished host lung function as a function of increased incidence of MMR-deficient strains, though they concede that this observation could be an artifact of increased mutator incidence in late-stage infections.
Examining genome sequences from the endpoints of an 8-year infection, Smith et al. (78) confirmed that many of the characteristic phenotypes of chronic CF isolates could be attributed to mutations in mexZ, mexA, lasR, and vfr and further that there was a strong bias toward nonsynonymous changes, as would be expected if those mutations were under positive selection. Using the complete set of genes in which mutations were seen to occur, those authors screened intervening strains in the 8-year patient series, as well as series in 29 other patients, and found additional evidence to support the inferences of parallel evolution and positive selection.
Do mutators catalyze evolutionary change? (i) Conflicting evidence from recent studies.
The late-stage (96-month) isolate for which Smith et al. (78) collected whole-genome sequence data was a mutator, bearing a single nonsynonymous change in mutS. Mena et al. (58) screened the entire collection and found that while only 17% of strains exhibited a mutator phenotype, a disproportionate number of all mutations in the collection were carried in mutator genomes (e.g., 87% of clones had ≥7 mutations). Indeed, sequential mutator lineages accumulated >3 mutations per year, while nonmutators accumulated 0.25 mutation per year. Strikingly, Mena et al. uncovered an instance where two independent MMR-inactivating mutations occurred in the same lineage (lesions in mutL and mutS), providing even stronger evidence for positive selection for the mutator trait.
While mutators show the capacity to accelerate mutation accumulation in the CF lung, they are not a necessary prerequisite to characteristic adaptive changes such as the switch to mucoidy and the loss of quorum sensing. Examining 70 strains isolated as longitudinal series from 10 patients, Ciofu et al. (11) reported that mucA and lasR mutations usually preceded mutations in antimutator genes. They also saw pervasive polymorphism in their collection, manifest as long-term coexistence of clones that were strong, weak, and nonmutators, with hypermutators being amplified in later stages by hitchhiking along with strongly selected antibiotic-resistant variants (11). Thus, overall, the picture of P. aeruginosa evolutionary dynamics emerging from these studies is one of periodic selection on an ancestral clone manifest as intermittent clonal expansion, with multiple, related genotypes coexisting over extended periods of time. Mutators appear to have accelerated rates of mutation accumulation, hitchhiking along with late-stage adaptive mutations, especially those that confer drug resistance.
(ii) Evidence from the Necker collection.
If most P. aeruginosa genes are vertically transmitted in the CF lung, then evolution of genetic novelty in this environment depends on the rate of mutation supply. Theory suggests that relative to small populations in constant environments, large populations that undergo strong and/or variable selection are supplied with many more beneficial mutations (8). However, under these circumstances, the number of favorable adaptations that attain high frequency may be limited by competition among clones with different but equally beneficial mutations (33, 17). Should clonal interference delay successive sweeps of the fittest genotypes, population genetic variation should increase over time. However, because we have imperfect knowledge of P. aeruginosa deme size and population genetic structure in the CF lung, it is unclear how often conditions favor clonal interference.
Genetic novelty may also originate from mutators, which are a conspicuous feature of P. aeruginosa evolving in the CF lung (61, 64). Theory and anecdotal evidence suggest that within the CF lung strong and variable selective pressures intermittently favor mutators, enabling mutator alleles to hitchhike along with the adaptive mutations they help generate (83). Although previous studies (16, 54, 59) as well as our own have shown positive correlation between mutator status and multiple antibiotic resistance, it is not yet certain how much mutators contribute to adaptive genetic variation. Because the evolutionary disadvantages associated with an elevated mutation rate limit the long-term evolutionary success of a mutator lineage (25), favorable traits generated by mutators should persist only within descendant nonmutator lineages. In our collection, the I patient series shows genomic architectures in early mutator isolates that appear later in nonmutators. These observations may reflect the persistence of a selectively favored architecture established by or passaged through a mutator lineage. Under the latter scenario, an adaptive architecture arose within a nonmutator lineage in which the I1 mutS allele appeared and transiently hitchhiked. Nonmutator I4, the last isolate in this series, does not contain the 8-bp mutS I1 deletion and exhibits a background frequency of mutation to rifampin resistance. I3a and I3b do contain the 8-bp mutS deletion and exhibit mutation frequencies that are below the mutator cutoff but still severalfold higher than the population median (Table 1). This suggests that in I3a and I3b compensatory mutations rather than reversions may have diminished the cost of an elevated mutation rate, enabling these genotypes to retain adaptive changes generated via hypermutation. How often and by what mechanism(s) this occurs remain to be discovered.
Concluding remarks.
The goals of this study were to assay phenotypic and genotypic variability among P. aeruginosa strains evolving in CF patients and to assess the relative contributions of secondary infection, recombination, and mutator activity to that variability. Our results support the view that chronic infections are established independently by one or a few founder clones from environmental reservoirs (10) and that their evolutionary trajectories are rarely influenced by recombination and horizontal gene transfer, even though co-/cross-infection is not uncommon. The mutator incidence in the Necker collection was comparable to those in previous reports (reference 61 and others), and while mutators were more likely to be antibiotic resistant, their presence in longitudinal series was not associated with significant differences in genetic variability assessed by genome fingerprinting and MLST profiling. In two age-matched series with or without mutators, microarray assay revealed a widespread absence of genes whose loss of function has been associated with P. aeruginosa adaptation during chronic infection, with the mutator series further distinguished by the absence of more such genes than the nonmutator series. However, mutator status was not required to generate adaptive macromutations, as a large deletion discovered in the nonmutator series resulted in loss of multiple virulence factors. Much more intensive sampling of longitudinal isolates is needed to gain deeper insights into the population diversity and evolutionary dynamics of P. aeruginosa in the CF lung, as well as the special role, if any, that mutators may play in disease progression.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant NIAID R15-AI079708 to F.R. and S.R.M., The University of Montana's Office of Research and Sponsored Programs, NCRR P20 RR016455, and the Functional Genomics Core at Montana State University, a Nakamura Graduate Fellowship to C.B.C., Cystic Fibrosis Foundation undergraduate traineeships (WARREN07H0 and WARREN08H0) to A.E.W., and Howard Hughes undergraduate fellowships to A.E.W. and Meghan Zimmerman.
Nick McClure and Leonid Kalachev performed analyses that established a-CGH cutoff values; clinical data were represented by Meghan Zimmerman. The manuscript was improved by the comments of Luke Hoffman, Michael Minnick, Mark Pershouse, Scott Samuels, and Gavin Sherlock.
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
Published ahead of print on 19 September 2011.
REFERENCES
- 1. Aaron S. D., et al. 2010. Infection with transmissible strains of Pseudomonas aeruginosa and clinical outcomes in adults with cystic fibrosis. JAMA 304:2145–2153 [DOI] [PubMed] [Google Scholar]
- 2. Alguel Y., Lu D., Quade N., Sauter S., Zhang X. 2010. Crystal structure of MexZ, a key repressor responsible for antibiotic resistance in Pseudomonas aeruginosa. J. Struct. Biol. 172:305–310 [DOI] [PubMed] [Google Scholar]
- 3. Anthony M., et al. 2002. Genetic analysis of Pseudomonas aeruginosa isolates from the sputa of Australian adult cystic fibrosis patients. J. Clin. Microbiol. 40:2772–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arber W. 2000. Genetic variation: molecular mechanisms and impact on microbial evolution. FEMS Microbiol. Rev. 24:1–7 [DOI] [PubMed] [Google Scholar]
- 5. Arjan J. A., et al. 1999. Diminishing returns from mutation supply rate in asexual populations. Science 283:404–406 [DOI] [PubMed] [Google Scholar]
- 6. Ausubel F., et al. 2002. Short protocols in molecular biology, 5th ed. Wiley, New York, NY [Google Scholar]
- 7. Barth A. L., Pitt T. L. 1996. The high amino-acid content of sputum from cystic fibrosis patients promotes growth of auxotrophic Pseudomonas aeruginosa. J. Med. Microbiol. 45:110–119 [DOI] [PubMed] [Google Scholar]
- 8. Bollback J. P., Huelsenbeck J. P. 2007. Clonal interference is alleviated by high mutation rates in large populations. Mol. Biol. Evol. 24:1397–1406 [DOI] [PubMed] [Google Scholar]
- 9. Bragonzi A., et al. 2005. Nonmucoid Pseudomonas aeruginosa expresses alginate in the lungs of patients with cystic fibrosis and in a mouse model.J. Infect. Dis. 192:410–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burns J. L., et al. 2001. Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis.J. Infect. Dis. 183:444–452 [DOI] [PubMed] [Google Scholar]
- 11. Ciofu O., Mandsberg L. F., Bjarnsholt T., Wassermann T., Høiby N. 2010. Genetic adaptation of Pseudomonas aeruginosa during chronic lung infection of patients with cystic fibrosis: strong and weak mutators with heterogeneous genetic backgrounds emerge in mucA and/or lasR mutants. Microbiology 156:1108–1119 [DOI] [PubMed] [Google Scholar]
- 12. Cirz R. T., O'Neill B. M., Hammond J. A., Head S. R., Romesberg F. E. 2006. Defining the Pseudomonas aeruginosa SOS response and its role in the global response to the antibiotic ciprofloxacin. J. Bacteriol. 188:7101–7110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Conibear T. C. R., Collins S. L., Webb J. S. 2009. Role of mutation in Pseudomonas aeruginosa biofilm development. PLoS One 4:e6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Curran B., Jonas D., Grundmann H., Pitt T., Dowson C. G. 2004. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J. Clin. Microbiol. 42:5644–5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dawson S. L., Fry J. C., Dancer B. N. 2002. A comparative evaluation of five typing techniques for determining the diversity of fluorescent pseudomonads. J. Microbiol. Methods 50:9–22 [DOI] [PubMed] [Google Scholar]
- 16. Denamur E., et al. 2005. Intermediate mutation frequencies favor evolution of multidrug resistance in Escherichia coli. Genetics 171:825–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de Visser J. A. G. M., Rozen D. E. 2005. Limits to adaptation in asexual populations. J. Evol. Biol. 18:779–788 [DOI] [PubMed] [Google Scholar]
- 18. de Visser J. A. G. M. 2002. The fate of microbial mutators. Microbiology 148:1247–1252 [DOI] [PubMed] [Google Scholar]
- 19. DeVries C. A., Ohman D. E. 1994. Mucoid-to-nonmucoid conversion in alginate-producing Pseudomonas aeruginosa often results from spontaneous mutations in algT, encoding a putative alternate sigma factor, and shows evidence for autoregulation. J. Bacteriol. 176:6677–6687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ernst R. K., et al. 2003. Genome mosaicism is conserved but not unique in Pseudomonas aeruginosa isolates from the airways of young children with cystic fibrosis. Environ. Microbiol. 5:1341–1349 [DOI] [PubMed] [Google Scholar]
- 21. Ferroni A., et al. 2009. Effect of mutator P. aeruginosa on antibiotic resistance acquisition and respiratory function in cystic fibrosis. Pediatr. Pulmonol. 44:820–825 [DOI] [PubMed] [Google Scholar]
- 22. Finnan S., Morrissey J. P., O'Gara F., Boyd E. F. 2004. Genome diversity of Pseudomonas aeruginosa isolates from cystic fibrosis patients and the hospital environment. J. Clin. Microbiol. 42:5783–5792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fonseca E. L., Vieira V. V., Cipriano R., Vicente A. C. P. 2005. Class 1 integrons in Pseudomonas aeruginosa isolates from clinical settings in Amazon region, Brazil. FEMS Immunol. Med. Microbiol. 44:303–309 [DOI] [PubMed] [Google Scholar]
- 24. Foweraker J. E., Laughton C. R., Brown D. F. J., Bilton D. 2005. Phenotypic variability of Pseudomonas aeruginosa in sputa from patients with acute infective exacerbation of cystic fibrosis and its impact on the validity of antimicrobial susceptibility testing. J. Antimicrob. Chemother. 55:921–927 [DOI] [PubMed] [Google Scholar]
- 25. Giraud A., et al. 2001. Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science 291:2606–2608 [DOI] [PubMed] [Google Scholar]
- 26. Goodman A. L., et al. 2004. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 7:745–754 [DOI] [PubMed] [Google Scholar]
- 27. Grundmann H., Schneider C., Hartung D., Daschner F. D., Pitt T. L. 1995. Discriminatory power of three DNA-based typing techniques for Pseudomonas aeruginosa. J. Clin. Microbiol. 33:528–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hancock R. E., et al. 1983. Pseudomonas aeruginosa isolates from patients with cystic fibrosis: a class of serum-sensitive, nontypable strains deficient in lipopolysaccharide O side chains. Infect. Immun. 42:170–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hassett D. J., et al. 2009. Pseudomonas aeruginosa hypoxic or anaerobic biofilm infections within cystic fibrosis airways. Trends Microbiol. 17:130–138 [DOI] [PubMed] [Google Scholar]
- 30. Henrichfreise B., Wiegand I., Pfister W., Wiedemann B. 2007. Resistance mechanisms of multiresistant Pseudomonas aeruginosa strains from Germany and correlation with hypermutation. Antimicrob. Agents Chemother. 51:4062–4070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hentzer M., et al. 2001. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 183:5395–5401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Heo M., Shakhnovich E. I. 2010. Interplay between pleiotropy and secondary selection determines rise and fall of mutators in stress response. PLoS Comput. Biol. 6:e1000710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hill W. G., Robertson A. 1966. The effect of linkage on limits to artificial selection. Genet. Res. 8:269–294 [PubMed] [Google Scholar]
- 34. Hillis D. M., Bull J. J. 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 42:182–192 [Google Scholar]
- 35. Hoboth C., et al. 2009. Dynamics of adaptive microevolution of hypermutable Pseudomonas aeruginosa during chronic pulmonary infection in patients with cystic fibrosis.J. Infect. Dis. 200:118–130 [DOI] [PubMed] [Google Scholar]
- 36. Hoffman L. R., et al. 2006. Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 103:19890–19895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hoffman L. R., et al. 2010. Nutrient availability as a mechanism for selection of antibiotic tolerant Pseudomonas aeruginosa within the CF airway. PLoS Pathog. 6:e1000712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hogardt M., et al. 2007. Stage-specific adaptation of hypermutable Pseudomonas aeruginosa isolates during chronic pulmonary infection in patients with cystic fibrosis.J. Infect. Dis. 195:70–80 [DOI] [PubMed] [Google Scholar]
- 39. Høiby N. 2004. New insight into the pathogenesis and epidemiology of Pseudomonas aeruginosa infection in cystic fibrosis, p. 11–25 In Struelens M. J. (ed.), Proceedings of the 4th Elzenveld Workshop on Infectious Diseases Bristol-Myers Squibb, Antwerpen, Belgium [Google Scholar]
- 40. Hudson R. R., Kaplan N. L. 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111:147–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hunfeld K. P., et al. 2000. Risk of Pseudomonas aeruginosa cross-colonisation in patients with cystic fibrosis within a holiday camp—a molecular-epidemiological study. Wien. Klin. Wochenschr. 112:329–333 [PubMed] [Google Scholar]
- 42. Huse H. K., et al. 2010. Parallel evolution in Pseudomonas aeruginosa over 39,000 generations in vivo. mBio 1:e00199–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jain M., et al. 2004. Type III secretion phenotypes of Pseudomonas aeruginosa strains change during infection of individuals with cystic fibrosis. J. Clin. Microbiol. 42:5229–5237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jelsbak L., et al. 2007. Molecular epidemiology and dynamics of Pseudomonas aeruginosa populations in lungs of cystic fibrosis patients. Infect. Immun. 75:2214–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jones A. M., et al. 2010. Clinical outcome for cystic fibrosis patients infected with transmissible pseudomonas aeruginosa: an 8-year prospective study. Chest 137:1405–1409 [DOI] [PubMed] [Google Scholar]
- 46. Kiewitz C., Larbig K., Klockgether J., Weinel C., Tümmler B. 2000. Monitoring genome evolution ex vivo: reversible chromosomal integration of a 106 kb plasmid at two tRNA(Lys) gene loci in sequential Pseudomonas aeruginosa airway isolates. Microbiology 146:2365–2373 [DOI] [PubMed] [Google Scholar]
- 47. Kiewitz C., Tümmler B. 2000. Sequence diversity of Pseudomonas aeruginosa: impact on population structure and genome evolution. J. Bacteriol. 182:3125–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Klepac-Ceraj V., et al. 2010. Relationship between cystic fibrosis respiratory tract bacterial communities and age, genotype, antibiotics and Pseudomonas aeruginosa. Environ. Microbiol. 12:1293–1303 [DOI] [PubMed] [Google Scholar]
- 49. Köhler T., Buckling A., van Delden C. 2009. Cooperation and virulence of clinical Pseudomonas aeruginosa populations. Proc. Natl. Acad. Sci. U. S. A. 106:6339–6344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Krustal J. B., Jr. 1956. On the shortest spanning subtree of a graph and the traveling salesman problem. Am. Math. Soc. 7:48–50 [Google Scholar]
- 51. Liang X., Pham X. Q., Olson M. V., Lory S. 2001. Identification of a genomic island present in the majority of pathogenic isolates of Pseudomonas aeruginosa. J. Bacteriol. 183:843–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Luzar M. A., Thomassen M. J., Montie T. C. 1985. Flagella and motility alterations in Pseudomonas aeruginosa strains from patients with cystic fibrosis: relationship to patient clinical condition. Infect. Immun. 50:577–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lyczak J. B., Cannon C. L., Pier G. B. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2:1051–1060 [DOI] [PubMed] [Google Scholar]
- 54. Maciá M. D., et al. 2005. Hypermutation is a key factor in development of multiple-antimicrobial resistance in Pseudomonas aeruginosa strains causing chronic lung infections. Antimicrob. Agents Chemother. 49:3382–3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mah T.-F., et al. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306–310 [DOI] [PubMed] [Google Scholar]
- 56. McAlester G., O'Gara F., Morrissey J. P. 2008. Signal-mediated interactions between Pseudomonas aeruginosa and Candida albicans. J. Med. Microbiol. 57:563–569 [DOI] [PubMed] [Google Scholar]
- 57. McCallum S. J., et al. 2001. Superinfection with a transmissible strain of Pseudomonas aeruginosa in adults with cystic fibrosis chronically colonised by P aeruginosa. Lancet 358:558–560 [DOI] [PubMed] [Google Scholar]
- 58. Mena A., et al. 2008. Genetic adaptation of Pseudomonas aeruginosa to the airways of cystic fibrosis patients is catalyzed by hypermutation. J. Bacteriol. 190:7910–7917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Miller K., O'Neill A. J., Chopra I. 2004. Escherichia coli mutators present an enhanced risk for emergence of antibiotic resistance during urinary tract infections. Antimicrob. Agents Chemother. 48:23–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nguyen D., Singh P. K. 2006. Evolving stealth: genetic adaptation of Pseudomonas aeruginosa during cystic fibrosis infections. Proc. Natl. Acad. Sci. U. S. A. 103:8305–8306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Oliver A., Cantón R., Campo P., Baquero F., Blázquez J. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251–1254 [DOI] [PubMed] [Google Scholar]
- 62. Oliver A., Mena A. 2010. Bacterial hypermutation in cystic fibrosis, not only for antibiotic resistance. Clin. Microbiol. Infect. 16:798–808 [DOI] [PubMed] [Google Scholar]
- 63. Oliver A. 2010. Mutators in cystic fibrosis chronic lung infection: prevalence, mechanisms, and consequences for antimicrobial therapy. Int. J. Med. Microbiol. 300:563–572 [DOI] [PubMed] [Google Scholar]
- 64. Oliver A., Baquero F., Blázquez J. 2002. The mismatch repair system (mutS, mutL and uvrD genes) in Pseudomonas aeruginosa: molecular characterization of naturally occurring mutants. Mol. Microbiol. 43:1641–1650 [DOI] [PubMed] [Google Scholar]
- 65. Perron G. G., Hall A. R., Buckling A. 2010. Hypermutability and compensatory adaptation in antibiotic-resistant bacteria. Am. Nat. 176:303–311 [DOI] [PubMed] [Google Scholar]
- 66. Posada D., Crandall K. A. 2001. Selecting the best-fit model of nucleotide substitution. Syst. Biol. 50:580–601 [PubMed] [Google Scholar]
- 67. Qiu X., Kulasekara B. R., Lory S. 2009. Role of horizontal gene transfer in the evolution of Pseudomonas aeruginosa virulence. Genome Dyn. 6:126–139 [DOI] [PubMed] [Google Scholar]
- 68. Rayssiguier C., Thaler D. S., Radman M. 1989. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature 342:396–401 [DOI] [PubMed] [Google Scholar]
- 69. Römling U., Schmidt K. D., Tümmler B. 1997. Large chromosomal inversions occur in Pseudomonas aeruginosa clone C strains isolated from cystic fibrosis patients. FEMS Microbiol. Lett. 150:149–156 [DOI] [PubMed] [Google Scholar]
- 70. Römling U., Schmidt K. D., Tümmler B. 1997. Large genome rearrangements discovered by the detailed analysis of 21 Pseudomonas aeruginosa clone C isolates found in environment and disease habitats. J. Mol. Biol. 271:386–404 [DOI] [PubMed] [Google Scholar]
- 71. Römling U., Wingender J., Müller H., Tümmler B. 1994. A major Pseudomonas aeruginosa clone common to patients and aquatic habitats. Appl. Environ. Microbiol. 60:1734–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rozas J., Sánchez-DelBarrio J. C., Messeguer X., Rozas R. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497 [DOI] [PubMed] [Google Scholar]
- 73. Salinas D., et al. 2005. Submucosal gland dysfunction as a primary defect in cystic fibrosis. FASEB J. 19:431–433 [DOI] [PubMed] [Google Scholar]
- 74. Sandoz K. M., Mitzimberg S. M., Schuster M. 2007. Social cheating in Pseudomonas aeruginosa quorum sensing. Proc. Natl. Acad. Sci. U. S. A. 104:15876–15881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Schneider S., Roessli D., Excoffier L. 2000. Arlequin: a software for population genetics data analysis. User manual version 2.00. Genetics and Biometry Lab, Department of Anthropology, University of Geneva, Geneva, Switzerland [Google Scholar]
- 76. Shen K., et al. 2006. Extensive genomic plasticity in Pseudomonas aeruginosa revealed by identification and distribution studies of novel genes among clinical isolates. Infect. Immun. 74:5272–5283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sibley C. D., et al. 2008. Discerning the complexity of community interactions using a Drosophila model of polymicrobial infections. PLoS Pathog. 4:e1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Smith E. E., et al. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. U. S. A. 103:8487–8492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Smith E. E., Sims E. H., Spencer D. H., Kaul R., Olson M. V. 2005. Evidence for diversifying selection at the pyoverdine locus of Pseudomonas aeruginosa. J. Bacteriol. 187:2138–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Spencer D. H., et al. 2003. Whole-genome sequence variation among multiple isolates of Pseudomonas aeruginosa. J. Bacteriol. 185:1316–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Stover C. K., et al. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964 [DOI] [PubMed] [Google Scholar]
- 82. Struelens M. J., Schwam V., Deplano A., Baran D. 1993. Genome macrorestriction analysis of diversity and variability of Pseudomonas aeruginosa strains infecting cystic fibrosis patients. J. Clin. Microbiol. 31:2320–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tanaka M. M., Bergstrom C. T., Levin B. R. 2003. The evolution of mutator genes in bacterial populations: the roles of environmental change and timing. Genetics 164:843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tenover F. C., et al. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Tümmler B., et al. 1991. Nosocomial acquisition of Pseudomonas aeruginosa by cystic fibrosis patients. J. Clin. Microbiol. 29:1265–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Tümmler B. 2006. Clonal variations in Pseudomonas aeruginosa, p. 35–68 In Ramos J. L., Levesque R. C. (ed.), Pseudomonas, vol. 4. Molecular biology of emerging issues. Springer, New York, NY [Google Scholar]
- 88. Versalovic J., Koeuth T., Lupski J. R. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823–6831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Waine D. J., Honeybourne D., Smith E. G., Whitehouse J. L., Dowson C. G. 2008. Association between hypermutator phenotype, clinical variables, mucoid phenotype, and antimicrobial resistance in Pseudomonas aeruginosa. J. Clin. Microbiol. 46:3491–3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Weir B. S., Cockerham C. C. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370 [DOI] [PubMed] [Google Scholar]
- 91. Welsh M. J., Boat T. F., Beaudet A. L. 1995. Cystic Fibrosis, p. 5121–5188 In Scriver C. R., Beaudet A. L., Sly W. S., Valle D. (ed.), The metabolic and molecular bases of inherited disease, 7th ed. McGraw-Hill, New York, NY [Google Scholar]
- 92. Wiehlmann L., et al. 2007. Population structure of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 104:8101–8106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wolfgang M. C., et al. 2003. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 100:8484–8489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Worlitzsch D., et al. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Invest. 109:317–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Zeng A.-P., Kim E.-J. 2004. Iron availability, oxygen limitation, Pseudomonas aeruginosa and cystic fibrosis. Microbiology 150:516–518 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.