Abstract
Pathogenic biofilms have been associated with persistent infections due to their high resistance to antimicrobial agents, while commensal biofilms often fortify the host's immune system. Hence, controlling biofilm formation of both pathogenic bacteria and commensal bacteria is important in bacterium-related diseases. We investigated the effect of plant flavonoids on biofilm formation of enterohemorrhagic Escherichia coli O157:H7. The antioxidant phloretin, which is abundant in apples, markedly reduced E. coli O157:H7 biofilm formation without affecting the growth of planktonic cells, while phloretin did not harm commensal E. coli K-12 biofilms. Also, phloretin reduced E. coli O157:H7 attachment to human colon epithelial cells. Global transcriptome analyses revealed that phloretin repressed toxin genes (hlyE and stx2), autoinducer-2 importer genes (lsrACDBF), curli genes (csgA and csgB), and dozens of prophage genes in E. coli O157:H7 biofilm cells. Electron microscopy confirmed that phloretin reduced fimbria production in E. coli O157:H7. Also, phloretin suppressed the tumor necrosis factor alpha-induced inflammatory response in vitro using human colonic epithelial cells. Moreover, in the rat model of colitis induced by trinitrobenzene sulfonic acid (TNBS), phloretin significantly ameliorated colon inflammation and body weight loss. Taken together, our results suggest that the antioxidant phloretin also acts as an inhibitor of E. coli O157:H7 biofilm formation as well as an anti-inflammatory agent in inflammatory bowel diseases without harming beneficial commensal E. coli biofilms.
INTRODUCTION
Bacterial biofilms are sessile microbial communities attached to a surface by polysaccharides, proteins, and nucleic acids. Biofilms are ubiquitous in natural, medical, and engineering environments. Due to their increased resistance to antimicrobial treatment, biofilms formed by pathogenic bacteria pose serious problems to human health, such as cystic fibrosis, prostatitis, and periodontitis (7). In contrast, some commensal bacteria are crucial for nutrient assimilation and beneficial to the human immune system (16).
While most antibiotics that primarily aim to inhibit cell growth may result in bacterial drug resistance, biofilm inhibitors do not affect cell growth and there is less of a chance of resistance development (15, 27). Since biofilms play an important role in bacterial pathogenesis and drug resistance, biofilm inhibitors will help combat infectious diseases. Several nontoxic biofilm inhibitors effective against pathogenic bacteria have been found recently: for example, bacterium-origin compounds (d-amino acids [20], cis-2-decenoic acid [8], and indole derivatives [25]) and plant-origin compounds (furanones [15, 33], ursolic acid [34], corosolic acid and asiatic acid [12], and plant auxin 3-indolylacetonitrile [23]).
As antimicrobial compounds often eradicate intestinal commensal bacteria as well as pathogenic bacteria (16), potent biofilm inhibitors may unwantedly remove beneficial commensal biofilms. Hence, it is important to develop therapeutic strategies that can control the growth of pathogenic biofilms while leaving the beneficial biofilm intact (21).
Many flavonoids that are ubiquitous in the plant kingdom are biologically active in combating diseases due to their diverse beneficial roles in humans. Among numerous flavonoids, phloretin is a dihydrochalcone, a type of polyphenol mainly found in apples (about 5 mg/100 g of fresh apple) and strawberries. Phloretin has various biological functions, such as antioxidative (36), anticarcinogenic (50), and estrogenic (6) activities and inhibition of cardiovascular disease (10, 41). Recently, several other flavonoids showed the ability to inhibit the biofilm formation of Streptococcus mutans (22), Aeromonas hydrophila (18), and Escherichia coli O157:H7 (48). Additionally, a transcriptome analysis demonstrated that apple polyphenols, including phloretin, possessed anti-inflammatory effects against inflammatory bowel diseases (IBDs) in vitro (17).
IBD is a chronic inflammatory disorder of the gastrointestinal tract. The etiology of IBD includes multiple factors, such as genetic, immunologic, and environmental factors (11). It is generally accepted that commensal and pathogenic bacteria, particularly invasive bacteria, provide the constant antigenic stimulation that continuously activates pathogenic T cells to cause IBD (39, 40). Hence, it has long been suggested that both protecting the epithelial barrier from invasive bacteria and fortifying the mucosal immune system are useful in treating IBD (16, 39).
In this study, nine flavonoids were initially screened for nontoxic biofilm inhibitors against enterohemorrhagic E. coli O157:H7, and the effect of phloretin, one of the most effective inhibitors among the compounds tested, was further investigated with five commensal E. coli strains. In order to understand the molecular basis of the biofilm inhibition of phloretin, transcriptome analysis and electron microscopy were utilized. We also investigated the adhesion of E. coli O157:H7 cells or inflammatory cells to HT-29 human colonic epithelial cells, as this adhesion is an initial step in colon inflammation. Moreover, the rat model of colitis induced by trinitrobenzene sulfonic acid (TNBS), which is a well-established model for IBD (45), was used to study the anti-inflammatory effect of phloretin in vivo.
MATERIALS AND METHODS
Bacterial strains, materials, and growth rate measurements.
Enterohemorrhagic E. coli serotype O157:H7 (ATCC 43895, EDL933 strain [42]), four commensal E. coli K-12 strains, strains MG1655 (3), BW25113 (1), TG1 (38), and DH5α, and nonpathogenic E. coli ATCC 4157 were used. Luria-Bertani (LB) medium (38) was used as the medium for the growth of all E. coli strains. Chemicals, including nine flavonoids (6-aminoflavone [97%], 6-hydroxyflavone [98%], apigenin [99%], chrysin [97%], curcumin [94%], daidzein [98%], flavones [99%], genistein [98%], and phloretin [99%]), vitamin C and vitamin E, sodium phosphate, amyl alcohol, formaldehyde, glutaraldehyde, ethyl alcohol, hydrochloric acid, OsO4, p-dimethylamino-benzaldehyde, crystal violet, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, sodium pyruvate, 2,7-bis(2-carboxyethyl)-5 (6)-carboxyfluorescein acetoxymethyl ester (BCECF/AM), 5-aminosalicylic acid (5-ASA), and TNBS were purchased from Sigma-Aldrich Co. (St. Louis, MO). All flavonoids and vitamins were dissolved in dimethyl sulfoxide (DMSO). Ethanol and DMSO were purchased from Duksan Pure Chemical Co. (Ansan, South Korea). Each bacterial strain was initially streaked from −80°C glycerol stocks onto LB medium plates, and a fresh single colony was inoculated into LB medium (25 ml) in 250-ml flasks and routinely cultured at 250 rpm and 37°C unless otherwise indicated. Overnight cultures were diluted 1:100 using LB medium. For cell growth measurements, the optical density was measured at 600 nm (OD600) with a spectrophotometer (UV-160; Shimadzu, Japan). The specific growth rates were determined by measuring the OD600 and calculated using the linear portion of the natural logarithm of OD600 versus time.
Crystal violet biofilm assay.
A static biofilm formation assay was performed in 96-well polystyrene plates (Fisher Scientific) as previously reported (31). Briefly, overnight cultures were diluted to an OD600 of 0.05 in LB medium (300 μl) containing phloretin or other compounds at 0, 5, 10, 25, 50, and 100 μg/ml and incubated for 10 h without shaking at 37°C. After the cell density was measured (turbidity at 620 nm), the plates were washed three times with water to remove all planktonic cells, and then crystal violet (0.1% [vol/vol], 300 μl) was added to each well. After 20 min at room temperature, the microplate was emptied and washed three times with water. Next, ethanol (95%, 300 μl) was added to resolve the stained biofilm cells. Total biofilm cells (absorbance at 570 nm) were measured. Total biofilm (OD570) was normalized with cell growth (OD620).
Total RNA isolation.
For transcriptomic analyses, E. coli O157:H7 and E. coli K-12 BW25113 were inoculated in 250 ml of LB medium at 37°C in 1-liter shake flasks with overnight cultures (1:100 dilution). To form biofilms, 10 g of glass wool (Corning Glass Works, Corning, NY) was used (26) and cells were cultured for 7 h with shaking at 100 rpm. Phloretin (50 μg/ml) or DMSO alone was added at the beginning of culture. Glass wool was used to increase the surface area so that enough RNA for the DNA microarrays could be readily obtained. Before samples were taken, RNase inhibitor (RNAlater; Ambion, TX) was added and biofilm cells were immediately chilled for 30 s with dry ice and 95% ethanol (to prevent RNA degradation) before centrifugation at 13,000 × g for 2 min. The cell pellets were immediately frozen with dry ice and stored at −80°C. Total RNA was isolated using a Qiagen RNeasy minikit (Valencia, CA). To remove all DNA, the purified RNA was treated for 15 min with 30 units of DNase I. RNA quality was assessed using an Agilent 2100 bioanalyzer and an RNA 6000 nanochip (Agilent Technologies, Amstelveen, The Netherlands), and the quantity was determined using an ND-1000 spectrophotometer (NanoDrop Technologies, Inc., DE).
DNA microarray analysis.
The E. coli GeneChip Genome (version 2.0) array (P/N 900551; Affymetrix, Santa Clara, CA) was used to study the differential gene expression of the E. coli O157:H7 cells after the treatment with phloretin. DNA microarray analysis with one biological replicate was performed with an Affymetrix system as previously reported (23).
qRT-PCR.
To corroborate the DNA microarray data, quantitative real-time reverse transcription-PCR (qRT-PCR) was used to investigate the levels of transcription of csgA, csgB, stx2a, lsrA, lsrB, hlyE, and mtr in E. coli O157:H7 and E. coli K-12 BW25113 with and without phloretin treatment. Two independent RNA samples tested using identical DNA microarray conditions were used for these studies. Gene-specific primers were used for these genes, and the rrsG housekeeping gene was used as a control (Table 1). The expression level of the rrsG housekeeping gene was used to normalize the data on the expression of the genes of interest. The qRT-PCR method was adapted from a previous study (26). qRT-PCR was performed using a SYBR green master mix (Applied Biosystems, Foster City, CA) and an ABI StepOne real-time PCR system (Applied Biosystems) with two independent cultures.
Table 1.
Primer sequences for quantitative RT-PCR
| Gene | Direction | Primer |
|---|---|---|
| csgA | Forward | 5′-AGA TGT TGG TCA GGG CTC AG-3′ |
| Reverse | 5′-CGT TGT TAC CAA AGC CAA CC-3′ | |
| csgB | Forward | 5′-AAT CAG GCA GCC ATA ATT GG-3′ |
| Reverse | 5′-CCA TAA GCA CCT TGC GAA AT-3′ | |
| stx2a | Forward | 5′-GTT CCG GAA TGC AAA TCA GT-3′ |
| Reverse | 5′-CGG CGT CAT CGT ATA CAC AG-3′ | |
| lsrA | Forward | 5′-AAC ATC CTG TTT GGG CTG GCA A-3′ |
| Reverse | 5′-AAA CAA GCG TTC GGT TTC CGC A-3′ | |
| lsrB | Forward | 5′-AGC ATC CTG GCT GGG AAA TTG T-3′ |
| Reverse | 5′-AAA TTC TTT CAC CGT GCC GCG T-3′ | |
| hlyE | Forward | 5′-AAC CGC AGA TGG AGC ATT AG-3′ |
| Reverse | 5′-CAC TGT TTG GGT TGC TTC AA-3′ | |
| mtr | Forward | 5′-ACC AGC AGC AGA TCC AGA CT-3′ |
| Reverse | 5′-AAG ATC CGA AAA CCA TCG TG-3′ | |
| rrsG | Forward | 5′-TAT TGC ACA ATG GGC GCA AG-3′ |
| Reverse | 5′-ACT TAA CAA ACC GCC TGC GT-3′ |
Fimbria analysis using SEM.
To examine fimbria production, scanning electron microscopy (SEM) following our previous protocol was used (23). Briefly, E. coli O157:H7 cells were grown on a nylon filter (0.5 by 0.5 mm square) to form biofilm cells. The initial turbidity was 0.05 at 600 nm with 300 μl of cells. The cells and the nylon filter were incubated together to form biofilm cells at 37°C for 24 h without shaking. After E. coli O157:H7 cells were cultured with and without phloretin (50 μg/ml), 2.5% glutaraldehyde and 2% formaldehyde were added to prefix the cells and the cells were incubated overnight at 4°C. The specimens were examined using an S-4100 SEM (Hitachi, Japan) at a voltage of 15 kV and magnifications ranging from ×2,000 to ×25,000.
Human cell line culture.
The HT-29 human colon cancer cell line and U937 human leukemia cell line were purchased from the American Type Culture Collection (ATCC; Manassas, VA), and the cells were grown in RPMI 1640 medium (Invitrogen Life Technologies, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS), 100 U/ml, penicillin, 100 μg/ml streptomycin, 1 mM sodium pyruvate, and 2 mM glutamine. The culture medium was replaced every other day.
Assay of monocyte or E. coli strain adhesion to human colonic epithelial cells.
The adhesion of monocytes or E. coli strains to colonic epithelial cells was evaluated using a coculture system in which HT-29 colonic epithelial cells were cocultured with either U937 monocytic cells or E. coli strains as previously described (19). U937 cells were prelabeled with BCECF/AM (10 μg/ml) for 1 h at 37°C. The cells were cocultured in the presence or absence of tumor necrosis factor alpha (TNF-α) for 3 h at 37°C. For the bacterial adhesion assay, both E. coli O157:H7/pCM18 cells and E. coli K-12 BW25113/pCM18 cells were grown for 16 h at 37°C in LB medium supplemented with 300 μg/ml of erythromycin to maintain plasmid pCM18, encoding green fluorescent protein (14). E. coli cells were harvested by centrifugation at 13,000 rpm for 5 min at 4°C, washed once with phosphate-buffered saline (PBS), and resuspended in PBS. In the case of the phloretin experiments, HT-29 cells were pretreated with phloretin (200 μM, corresponding to 54.8 μg/ml) prior to TNF-α or E. coli exposure. Then, the HT-29 cells were washed twice with the PBS to remove nonadhering cells. Cells representative of each group were taken and imaged by fluorescence microscopy with a fluorescence microscope connected to a digital camera (TE2000-U; Nikon, Japan). To quantify green fluorescence in the coculture of HT-29 cells with either U937 monocytic cells or E. coli O157:H7/pCM18, the mixtures were lysed with 0.1% Triton X-100 in 0.1 M Tris, and the BCECF/AM fluorescence was analyzed using a Fluostar Optima microplate reader (BMG Labtech GmbH, Offenburg, Germany) with excitation at 485 nm and emission at 520 nm. In the quantification of E. coli O157:H7/pCM18 adhesion to HT-29 cells, a negative-control group that contained E. coli O157:H7/pCM18 only was set, and the amount of fluorescence produced by this sample was subtracted from that produced by the other experimental groups. In a separate experiment, the number of E. coli O157:H7/pCM18 cells attached to HT-29 cells was counted by two observers in a double-blind manner and analyzed with an image analysis system (ImageInside, version 2.32).
Model of TNBS-induced rat colitis.
The method used to establish the model of TNBS-induced rat colitis was adapted from that described previously (46). Sprague-Dawley rats (age, 7 to 8 weeks) were purchased from Samtaco bio Korea (Osan, South Korea). Rats were fasted for 24 h prior to TNBS injection. Then, they were lightly anesthetized using diethyl ether. Using a polyethylene catheter fitted onto a 1-ml syringe, rats were slowly injected with 0.8 ml of 5% TNBS in 50% (vol/vol) ethanol into the lumen of the colon (8 cm proximal to the anus through the rectum), and they were kept in a vertical position for 60 s before being returned to their cages. Rats in the control group were handled similarly, but 50% (vol/vol) ethanol alone was administered instead. To investigate the effect of phloretin, rats were administered phloretin (20 mg/kg of body weight/day) via the oral route for 5 days after TNBS administration. As a positive control, 5-ASA (at 100 mg/kg/day), an active metabolite of sulfasalazine, which is a drug commonly used for treatment of IBD, was used. All rats were killed on day 7 after the TNBS administration. The macroscopic ulceration and severity of colitis were evaluated by two independent investigators who were blind to the treatment. The colon tissues from 5 to 7 cm proximal to the rectum were cut out and used for myeloperoxidase (MPO) activity measurements. Animal experiments were performed according to the institutional guidelines for the care and use of laboratory animals of the National Institutes of Health, Yeungnam University.
MPO activity for tissue neutrophil infiltration.
The method used to determine MPO activity for tissue neutrophil infiltration was adapted from a method described previously (46). MPO activity serves as a marker for leukocyte infiltration into tissues. For the measurement of MPO activity, we used an MPO assay kit (ASA-001; Cytostore, Canada). A segment of colon 1 cm in length was dissected, washed in cold PBS (pH 7.4), weighed, suspended in ice-cold sample buffer (0.5 ml/50 mg of tissue), and homogenized for 30 s using a tissue homogenizer at 4°C (Biospec Products Inc., Switzerland). The homogenate was mixed with an additional sample buffer to give a final concentration of 50 mg/ml and centrifuged twice at 10,000 rpm for 5 min. Each 20 μl of sample solution was mixed with 200 μl of development reagent (12.5 μl of H2O2 containing chromogen powder in 2.5 ml phosphate buffer), and then the level of MPO activity in the tissue was detected by measuring the absorbance at 450 nm immediately after the solution was mixed and at 1-min intervals using a spectrophotometer (Versa MAX; Molecular Devices, CA). One unit of MPO activity was defined by Bradley et al. (5) to be the activity required for converting 1 μM H2O2 at 25°C for 7 min and expressed as units/g of tissue.
Microarray data accession number.
The full microarray data are available in the Gene Expression Omnibus database under accession number GSE28144.
RESULTS
Phloretin inhibits E. coli O157:H7 biofilm formation without inhibiting the growth of planktonic cells.
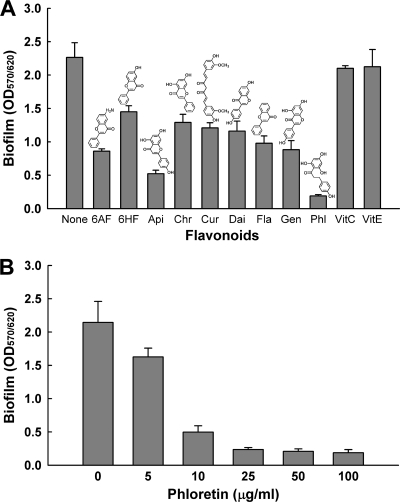
Nine flavonoids were screened for their ability to inhibit the biofilm formation of E. coli O157:H7 in LB medium in polystyrene 96-well plates. While most flavonoids showed inhibitory effects on E. coli O157:H7 biofilm formation, two well-known antioxidant compounds (vitamin C and vitamin E) did not affect E. coli O157:H7 biofilm formation (Fig. 1). This result indicated that biofilm reduction by flavonoids was due to their antibiofilm activity and not due to their antioxidant activity alone. Among nine flavonoids, phloretin led to the most significant reduction in E. coli O157:H7 biofilm formation in a dose-dependent manner (Fig. 1A and B). Specifically, 25 and 50 μg/ml of phloretin reduced E. coli O157:H7 biofilm formation by 89% and 93%, respectively (Fig. 1B).
Fig. 1.
Effect of flavonoids on E. coli O157:H7 biofilm formation in LB medium at 37°C after 10 h in 96-well plates. Total biofilm formation (OD570) was normalized by bacterial growth (OD620). (A) 6-Aminoflavone (6AF), 6-hydroxyflavone (6HF), apigenin (Api), chrysin (Chr), curcumin (Cur), daidzein (Dai), flavone (Fla), genistein (Gen), phloretin (Phl), vitamin C (VitC), and vitamin E (VitE) at 50 μg/ml were used. (B) Dose-dependent response of phloretin to E. coli O157:H7 biofilm formation. All of the compounds were dissolved in DMSO. DMSO was used as a control. The structures of 6-aminoflavone, 6-hydroxyflavone, apigenin, chrysin, curcumin, daidzein, flavone, genistein, and phloretin are shown. The experiment was done in triplicate, and data represent the means of three independent experiments.
A potential biofilm inhibitor is preferred not to have toxicity that may lead to bacterial drug resistance (32). Thus, the toxicity of phloretin was investigated by measuring the specific rate of growth of planktonic E. coli O157:H7 cells. The growth rates with and without phloretin were almost identical: 1.32 ± 0.01/h in the absence of phloretin and 1.31 ± 0.03/h with 50 μg/ml of phloretin. Therefore, a dosage of 50 μg/ml of phloretin was not toxic to planktonic cells of E. coli O157:H7 and was seen to reduce its biofilm formation.
Phloretin does not harm commensal E. coli K-12 biofilm formation.
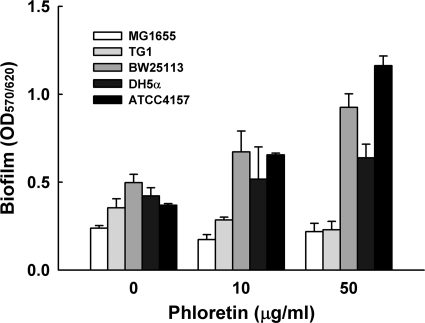
It is important to develop therapeutic compounds that reduce pathogenic biofilms while leaving the beneficial commensal biofilm unharmed (21). Thus, the effect of phloretin was also investigated with five commensal E. coli strains: MG1655, TG1, BW25113, DH5α, and ATCC 4157. Interestingly, there was no inhibitory effect by phloretin on biofilm formation in four E. coli K-12 strains and a nonpathogenic E. coli strain (Fig. 2). In the case of E. coli K-12 BW25113 and nonpathogenic E. coli ATCC 4157, phloretin even enhanced their biofilm formation in a dose-dependent manner.
Fig. 2.
Effect of phloretin on the biofilm formation of E. coli K-12 BW25113, MG1655, TG1, and DH5α and nonpathogenic E. coli ATCC 4157 in LB medium at 37°C after 10 h in 96-well plates. Total biofilm formation (OD570) was normalized by bacterial growth (OD620). The experiment was done in triplicate, and data represent the means of three independent experiments.
Differential gene expression of E. coli O157:H7 cells with phloretin.
To investigate the molecular mechanism of biofilm reduction by phloretin, DNA microarrays were used to determine differential gene expression for E. coli O157:H7 biofilm cells with and without phloretin. It was found that 156 genes were significantly regulated (more than 2.5-fold); 70 genes were repressed and 86 genes were induced by phloretin (see Tables S1 and S2 in the supplemental material). Most noticeably, two toxin genes (hemolysin hlyE and Shiga toxin 2 stx2), autoinducer-2 (AI-2) importer genes (lsrACDBF), and dozens of prophage genes were the most repressed (2.5- to 3.8-fold), while purin metabolism-related genes, transport-related genes, and stress-related genes were the most induced (2.5- to 4.7-fold) (see Tables S1 and S2 in the supplemental material).
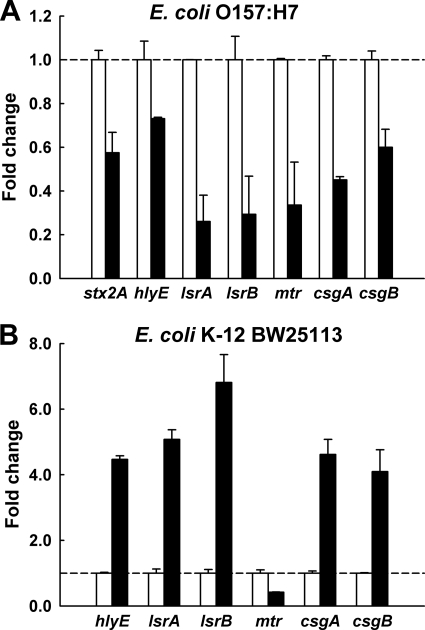
qRT-PCR was used to corroborate the DNA microarray data. Using independent cultures from a DNA microarray sample, qRT-PCR showed differential changes in expression comparable to those shown by the DNA microarray data for E. coli O157:H7 biofilm cells with and without phloretin treatment, as phloretin clearly repressed two toxin genes (stx2 and hlyE), two autoinducer-2 importer genes (lsrA and lsrB), an indole importer gene (mtr), and a curlin major subunit (csgA) and a curlin minor subunit (csgB) (Fig. 3A).
Fig. 3.
Transcriptional profiles of biofilm gene expression of E. coli O157:H7 biofilm cells (A) and E. coli K-12 BW25113 biofilm cells (B) in the absence (white bars) and presence (black bars) of phloretin (50 μg/ml). Transcriptional profiles were measured by qRT-PCR. Relative fold expression represents the change (n-fold) in transcription compared to the data in the absence of phloretin (white bars, value of 1.0).
Divergent gene expression of E. coli K-12 BW25113 cells with phloretin.
Since phloretin inhibited E. coli O157:H7 biofilm formation while it enhanced E. coli K-12 BW25113 biofilm formation (Fig. 1 and 2), qRT-PCR was also used to investigate the gene expression of E. coli K-12 BW25113 biofilm cells. Interestingly, the genes that were repressed in E. coli O157:H7 biofilm cells (hlyE, lsrA, lsrB, csgA, and csgB) were highly induced in E. coli K-12 BW25113 biofilm cells (Fig. 3B), while stx2 (Shiga toxin 2 gene) was absent in the E. coli K-12 strain and the expression of the mtr gene was repressed in both strains. The results suggest the importance of the expression of the hlyE, lsrA, lsrB, csgA, and csgB genes in controlling the biofilm formation of E. coli O157:H7 and E. coli K-12 BW25113.
Phloretin reduced fimbria formation in E. coli O157:H7.
Since fimbriae, including curli and pili, are important factors for E. coli O157:H7 biofilm formation (35, 37, 47) and since phloretin repressed the expression of the curli genes csgA and csgB in E. coli O157:H7 biofilm cells (Fig. 3A), fimbria production was investigated with SEM. Corroborating the transcriptome analysis, phloretin significantly decreased the fimbria production of E. coli O157:H7 (Fig. 4). Therefore, it appeared that the biofilm inhibition by phloretin was partially caused by the reduction of fimbriae.
Fig. 4.
Effect of phloretin on fimbria production in E. coli O157:H7 biofilm cells. Fimbria production was observed by SEM for biofilm cells grown with phloretin (50 μg/ml) and without phloretin. For SEM analysis, biofilm cells were cultured on a nylon membrane in a 96-well plate at 37°C for 24 h.
Phloretin inhibits E. coli O157:H7 adhesion to HT-29 human colonic epithelial cells.
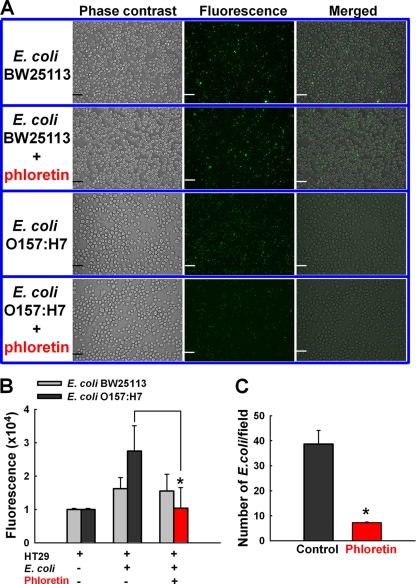
As phloretin was seen to reduce E. coli O157:H7 biofilm formation on abiotic surfaces (Fig. 1), we evaluated the ability of E. coli O157:H7 to adhere to epithelial cells, an initial step that leads to infection and virulence. As expected, more E. coli O157:H7 cells than commensal E. coli K-12 BW25113 cells were attached to HT-29 human colonic epithelial cells, indicating that E. coli O157:H7 was more adhesive than E. coli K-12 BW25113 cells (Fig. 5A). This result is consistent with the result of biofilm formation for both the E. coli O157:H7 and E. coli K-12 BW25113 strains in 96-well plates (Fig. 1 and 2).
Fig. 5.
Inhibitory effects of phloretin on E. coli cell adhesion to HT-29 human colonic epithelial cells. (A) Fluorescence microscopy appearance of E. coli O157:H7 cell or E. coli K-12 BW25113 cell attachment to HT-29 cells. HT-29 cells were cocultured with E. coli O157:H7 cells or E. coli K-12 BW25113 cells. Both E. coli O157:H7/pCM18 cells and E. coli K-12 BW25113/pCM18 cells were tagged with green fluorescent protein. Bars, 50 μm. (B) Quantitative fluorescence. (C) Number of E. coli O157:H7 cells attached to HT-29 cells with phloretin (200 μM, corresponding to 54.8 μg/ml) and without phloretin. The green fluorescence of the cells was captured using a fluorescence microscope, and the green fluorescence represents E. coli cells attached to HT-29 cells. Green fluorescence was also quantified using a fluorescent plate reader, and the number of E. coli O157:H7/pCM18 cells attached to HT-29 cells was analyzed by an image analysis system (ImageInside, version 2.32). *, P < 0.05 compared to the control group (no treatment with phloretin). The experiment was done in duplicate, and data represent the means of two independent experiments.
More importantly, the treatment with phloretin (200 μM, corresponding to 54.8 μg/ml) apparently reduced the attachment of E. coli O157:H7 cells to HT-29 epithelial cells, while phloretin did not influence the attachment of E. coli K-12 BW25113 cells (Fig. 5A). A quantitative analysis of green fluorescence from E. coli O157:H7 cells also clearly indicated the ability of phloretin to inhibit O157:H7 adhesion in HT-29 epithelial cells (Fig. 5B). Similarly, the number of E. coli O157:H7 cells attached to HT-29 epithelial cells was significantly decreased by phloretin treatment (Fig. 5C). Therefore, phloretin reduced the biofilm formation of E. coli O157:H7 cells on human colonic epithelial cells (Fig. 5) as well as on abiotic surfaces (Fig. 1).
Phloretin inhibits TNF-α-induced monocyte adhesion to HT-29 human colonic epithelial cells.
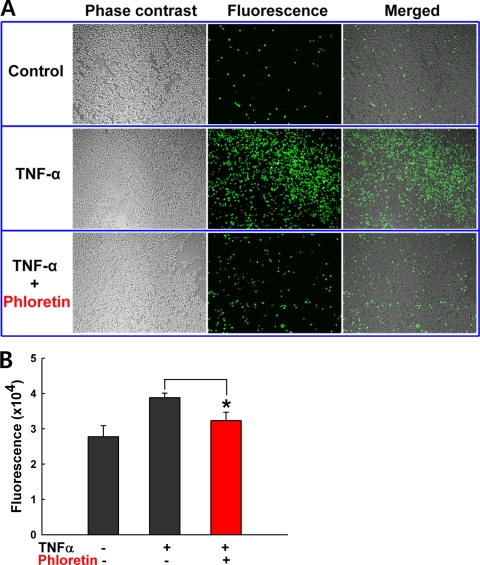
Since a previous transcriptome analysis showed that phloretin inhibited proinflammatory gene expression in vitro (17), we examined the ability of phloretin to prevent intestinal inflammation by using an in vitro model of colitis. Since TNF-α is one of the major inflammatory cytokines involved in IBD, TNF-α-induced adhesion of monocytes to HT-29 human colonic epithelial cells represents an in vitro model of intestinal inflammation (46). While TNF-α induced U937 premonocytic cell adhesion to HT-29 cells, as expected, phloretin significantly inhibited TNF-α-induced U937 premonocytic cell adhesion to HT-29 cells (Fig. 6A and B). This result supported well the potential of phloretin as an anti-inflammatory agent in IBD.
Fig. 6.
Inhibitory effects of phloretin on the TNF-α-induced adhesion of U937 cells to HT-29 human colonic epithelial cells. Fluorescence microscopy appearance (A) and quantitative fluorescence (B) of U937 cells attached to HT-29 cells with phloretin (200 μM, corresponding to 54.8 μg/ml) and without phloretin. U937 cells were prelabeled with BCECF/AM (10 μg/ml) for 1 h and then stimulated by 10 ng/ml of TNF-α. After 3 h of incubation, HT-29 cells were cocultured with U937 cells which had been prelabeled with BCECF/AM (10 μg/ml). The adhesion of BCECF/AM fluorescence-labeled U937 cells to HT-29 colon epithelial cells was captured using a fluorescence microscope. *, P < 0.05 compared to no treatment with phloretin. The control was no TNF-α treatment.
Anti-inflammatory effect of phloretin on TNBS-induced rat colitis.
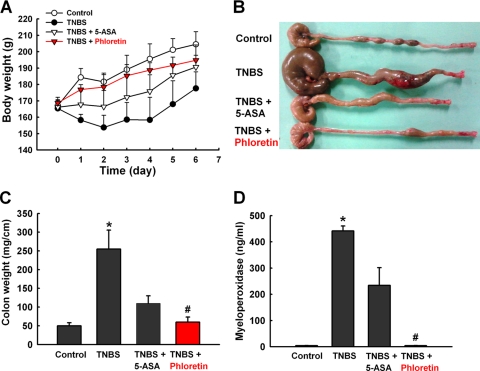
In order to demonstrate the efficacy of phloretin as an anti-inflammatory agent, the TNBS-induced rat colitis model (45) was also used. As expected, TNBS as a control prohibited body weight increase, in contrast to the weight gain in the vehicle-treated control group (Fig. 7A). The TNBS-treated rats developed significant signs of colitis, such as bloody diarrhea and wasting conditions, revealed by remarkable changes in body weight (Fig. 7A and B). The weight of colon tissue (1 cm near the TNBS injection site) was significantly increased by TNBS (Fig. 7C), whereas the colon length distal to the cecum was shortened by TNBS (Fig. 7B). Most importantly, the rats administered phloretin (20 mg/kg/day) recovered their body and colon weights as significantly as those administered a commonly used drug, 5-ASA (100 mg/kg/day), did (Fig. 7A and C). We also measured the colonic MPO activity, which serves as a marker for tissue neutrophil infiltration. The MPO activity was highly elevated in the TNBS-treated colon, and this increase in MPO activity was significantly suppressed by phloretin treatment almost to the level of the vehicle-treated control group (Fig. 7D). It is noteworthy that the effect of phloretin (20 mg/kg/day) was more prominent than that of the conventional IBD drug 5-ASA (100 mg/kg/day) in every aspect of the inflammatory response: body weight, colon weight, and MPO activity. These results suggest that phloretin can be a potent therapeutic agent for IBD.
Fig. 7.
Phloretin ameliorates TNBS-induced colitis in rats. Colitis was induced by rectal administration of TNBS. (A) Body weight was recorded daily from day 1 to day 6. The macroscopic appearance of the large intestine (B), the wet weight of the distal 5-cm portion of the colon (C), and the MPO activity of colon tissue (D) were analyzed. The control group received 50% ethanol as a vehicle. To investigate the effect of phloretin, rats were administered phloretin (20 mg/kg/day) via the oral route for 5 days after TNBS administration. 5-ASA (100 mg/kg/day), a drug commonly used for treatment of IBD, was used as a positive control. The data represents the means ± standard errors of the means for 5 rats per group. *, P < 0.05 compared to the vehicle-treated control group; #, P < 0.05 compared to the TNBS-treated group.
DISCUSSION
The intestinal tract is colonized by approximately 1013 to 1014 commensal bacteria consisting of thousands of species, including E. coli (13). E. coli O157:H7 is a common human pathogen responsible for outbreaks of hemorrhagic colitis causing bloody diarrhea that can lead to life-threatening hemolytic-uremic syndrome (4). The hallmark of E. coli O157:H7 infection is attaching and effacing, and the first step of infection involves adhesion of bacteria to host cells and the formation of microcolonies (30). However, to date, no effective therapy has been found. Antibiotics, antimotility agents, narcotics, and nonsteroidal anti-inflammatory drugs are not usually provided, as they increase the risk of developing hemolytic-uremic syndrome, a major cause of acute renal failure (44).
Flavonoids are a diverse group of plant natural products which play important roles in plant growth and in plant defense against microorganisms and pests. It has recently been reported that several flavonoids, such as naringenin, kaempferol, quercetin, and apigenin, suppressed autoinducer-2 activity in Vibrio harveyi, and these flavonoids also reduced biofilm formation in V. harveyi and E. coli O157:H7 (48). In this study, we investigated different kinds of flavonoids for their ability to control biofilms of E. coli O157:H7 as well as five commensal E. coli strains and found that phloretin specifically reduced E. coli O157:H7 biofilm formation. DNA microarray and qRT-PCR studies showed that phloretin suppressed autoinducer-2 importer genes (lsrACDBF) of E. coli O157:H7 biofilm cells. This result supported the previous finding (48) that flavonoids could interfere with bacterial quorum-sensing (AI-2) signaling and reduced E. coli O157:H7 biofilm formation.
In addition to AI-2 signaling, recent advances in elucidating the diverse molecular mechanisms underlying E. coli biofilm formation have been made (2, 49), for example, mechanisms involving fimbriae (curli and pili), adhesion proteins, cyclic di-GMP, indole, and even phage genes. In particular, curli possess characteristics of amyloid fibers and tend to link bacterial cells, and curli are considered to be an important factor for E. coli O157:H7 biofilm formation and also considered a virulence attribute (35, 37, 47). In this study, phloretin reduced the expression of the curli genes (csgA and csgB) (Fig. 3) and fimbria production (Fig. 4). Also, we previously reported that plant auxin 3-indolylacetonitrile inhibited E. coli O157:H7 biofilm formation by reducing fimbria formation (23). Therefore, this study also supports the idea that the inhibition of fimbria formation could be a practical way to decrease the biofilm formation of E. coli O157:H7. Since E. coli O157:H7 contains at least 16 putative fimbrial operons, including the csgBAC curli operon and several pilus operons (28), and since curli and pili have similar structures of fibers that influence bacterial adhesion in epithelial cells (35), a further experiment is required to clearly characterize if the fibers in SEM images (Fig. 4) are curli or pili.
Additional qRT-PCR results for two E. coli strains (Fig. 3) again support the mechanism in which AI-2 signal transport and fimbria production are important in the action of phloretin since phloretin treatment resulted in divergent biofilm formation (Fig. 1 and 2) and divergent expression of biofilm-controlling genes between E. coli O157:H7 biofilm cells and E. coli K-12 BW25113 biofilm cells (Fig. 3). However, it is still unclear which protein(s), such as a phloretin-binding protein(s) or a phloretin-sensing protein(s), is directly involved in the action of phloretin in E. coli; this should be studied further.
Although another extracellular signal, indole, inhibited E. coli O157:H7 biofilm formation (24, 25), the addition of phloretin did not change the accumulation of extracellular indole in either E. coli O157:H7 or E. coli K-12 BW25113 (data not shown), and phloretin repressed the indole importer gene (mtr) in both E. coli O157:H7 and E. coli K-12 BW25113. Therefore, it appeared that the indole signal may not be involved in the action of phloretin for control of E. coli biofilms.
DNA microarray data (see Table S1 in the supplemental material) and qRT-PCR data (Fig. 3) showed that phloretin repressed the expression of the genes for the toxins hemolysin (hlyE) and Shiga toxin 2 (stx2). The results indicate that phloretin may alleviate the virulence of E. coli O157:H7. In contrast, phloretin induced several stress resistance genes, such as marRAB and hcsBA. Thus, it is possible that phloretin could positively influence antibiotic resistance, which should be investigated further.
Growing evidence regarding the role of the intestinal microflora in IBD exists (39). Although all commensal bacteria are probably not responsible for IBD, it has been demonstrated that patients with IBD have high concentrations of mucosal bacteria and that the concentrations increase progressively with the severity of IBD (43). In particular, enteroaggregative E. coli (EAEC) colonization can occur in the mucosa of both the small and large bowels, which can lead to inflammation in the colon (29, 30), and the several fimbriae are involved in EAEC adherence to the surface of HEp-2 cells (9). However, there is not yet a clear relationship between E. coli O157:H7 biofilm formation and IBD. This study found for the first time that apple phloretin reduced the biofilm formation of E. coli O157:H7, partially via fimbria production, and also reduced bacterial adhesion to colonic epithelial cells (Fig. 5), while phloretin did not reduce commensal E. coli biofilms or adherence to epithelial cells (Fig. 2 and 5). Moreover, the previous anti-inflammatory effect of phloretin (17) has been confirmed in the TNBS-induced rat colitis model (Fig. 7). Therefore, it will be interesting to further investigate the effects of phloretin on IBD-associated E. coli strains such as EAEC. Although this study investigated the effects of phloretin with five commensal E. coli strains and one E. coli O157:H7 strain, more independent isolates of both pathogenic and commensal E. coli strains should be further investigated to understand the reciprocal impact of phloretin on E. coli biofilm formation.
Phloretin is part of the human daily diet, since it is abundantly found in apples and strawberries. Phloretin accounts in part for the antioxidant capacity of apples, and phloretin has also been revealed to have anticarcinogenic activities (50) and in vitro anti-inflammatory activity on colon epithelial cells (17) and on vein endothelial cells (41). In a previous study, it was shown that phloretin downregulated the mRNA levels of most tested proinflammatory genes (NF-κB, TNF-α, interleukin-8, CXCL3, CXCL10, signal transducer and activator of transcription 1, and interferon gamma-induced protein 10) in human colon epithelial cells in vitro (17).
This study demonstrated for the first time that phloretin, a natural flavonoid, is a nontoxic inhibitor of enterohemorrhagic E. coli O157:H7 biofilms but does not harm commensal E. coli K-12 biofilms. Also, importantly, our results confirmed that phloretin shows anti-inflammatory properties in both the in vitro and in vivo inflammatory colitis models. The effect of phloretin was noticeably more pronounced than that of the conventional IBD drug 5-ASA. Taken together, our results show that there exists a triple biological activity of phloretin that may make it useful as a lead compound for biofilm inhibitors of E. coli O157:H7 as well as an antioxidant compound and may have beneficial effects on IBD.
Supplementary Material
ACKNOWLEDGMENT
This research was supported by the International Research & Development Program of the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science and Technology (MEST) of the Republic of Korea.
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
Published ahead of print on 19 September 2011.
REFERENCES
- 1. Baba T., et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006 0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beloin C., Roux A., Ghigo J. M. 2008. Escherichia coli biofilms. Curr. Top. Microbiol. Immunol. 322:249–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blattner F. R., et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1474 [DOI] [PubMed] [Google Scholar]
- 4. Boyce T. G., Swerdlow D. L., Griffin P. M. 1995. Escherichia coli O157:H7 and the hemolytic-uremic syndrome. N. Engl. J. Med. 333:364–368 [DOI] [PubMed] [Google Scholar]
- 5. Bradley P. P., Priebat D. A., Christensen R. D., Rothstein G. 1982. Measurement of cutaneous inflammation: estimation of neutrophil content with an enzyme marker. J. Investig. Dermatol. 78:206–209 [DOI] [PubMed] [Google Scholar]
- 6. Calliste C. A., et al. 2001. Chalcones: structural requirements for antioxidant, estrogenic and antiproliferative activities. Anticancer Res. 21:3949–3956 [PubMed] [Google Scholar]
- 7. Costerton J. W., Stewart P. S., Greenberg E. P. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322 [DOI] [PubMed] [Google Scholar]
- 8. Davies D. G., Marques C. N. 2009. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol. 191:1393–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Farfan M. J., Inman K. G., Nataro J. P. 2008. The major pilin subunit of the AAF/II fimbriae from enteroaggregative Escherichia coli mediates binding to extracellular matrix proteins. Infect. Immun. 76:4378–4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Figtree G. A., et al. 2000. Plant-derived estrogens relax coronary arteries in vitro by a calcium antagonistic mechanism. J. Am. Coll. Cardiol. 35:1977–1985 [DOI] [PubMed] [Google Scholar]
- 11. Fiocchi C. 1998. Inflammatory bowel disease: etiology and pathogenesis. Gastroenterology 115:182–205 [DOI] [PubMed] [Google Scholar]
- 12. Garo E., et al. 2007. Asiatic acid and corosolic acid enhance the susceptibility of Pseudomonas aeruginosa biofilms to tobramycin. Antimicrob. Agents Chemother. 51:1813–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gill S. R., et al. 2006. Metagenomic analysis of the human distal gut microbiome. Science 312:1355–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hansen M. C., Palmer R. J., Jr., Udsen C., White D. C., Molin S. 2001. Assessment of GFP fluorescence in cells of Streptococcus gordonii under conditions of low pH and low oxygen concentration. Microbiology 147:1383–1391 [DOI] [PubMed] [Google Scholar]
- 15. Hentzer M., et al. 2002. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 148:87–102 [DOI] [PubMed] [Google Scholar]
- 16. Hooper L. V., Gordon J. I. 2001. Commensal host-bacterial relationships in the gut. Science 292:1115–1118 [DOI] [PubMed] [Google Scholar]
- 17. Jung M., Triebel S., Anke T., Richling E., Erkel G. 2009. Influence of apple polyphenols on inflammatory gene expression. Mol. Nutr. Food Res. 53:1263–1280 [DOI] [PubMed] [Google Scholar]
- 18. Kappachery S., Paul D., Yoon J., Kweon J. H. 2010. Vanillin, a potential agent to prevent biofouling of reverse osmosis membrane. Biofouling 26:667–672 [DOI] [PubMed] [Google Scholar]
- 19. Kobayashi N., Ikesue A., Majumdar S., Siahaan T. J. 2006. Inhibition of e-cadherin-mediated homotypic adhesion of Caco-2 cells: a novel evaluation assay for peptide activities in modulating cell-cell adhesion. J. Pharmacol. Exp. Ther. 317:309–316 [DOI] [PubMed] [Google Scholar]
- 20. Kolodkin-Gal I., et al. 2010. d-Amino acids trigger biofilm disassembly. Science 328:627–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kolter R., Greenberg E. P. 2006. Microbial sciences: the superficial life of microbes. Nature 441:300–302 [DOI] [PubMed] [Google Scholar]
- 22. Koo H., et al. 2003. Inhibition of Streptococcus mutans biofilm accumulation and polysaccharide production by apigenin and tt-farnesol. J. Antimicrob. Chemother. 52:782–789 [DOI] [PubMed] [Google Scholar]
- 23. Lee J.-H., Cho M. H., Lee J. 2011. 3-Indolylacetonitrile decreases Escherichia coli O157:H7 biofilm formation and Pseudomonas aeruginosa virulence. Environ. Microbiol. 13:62–73 [DOI] [PubMed] [Google Scholar]
- 24. Lee J.-H., Lee J. 2010. Indole as an intercellular signal in microbial community. FEMS Microbiol. Rev. 34:426–444 [DOI] [PubMed] [Google Scholar]
- 25. Lee J., Bansal T., Jayaraman A., Bentley W. E., Wood T. K. 2007. Enterohemorrhagic Escherichia coli biofilms are inhibited by 7-hydroxyindole and stimulated by isatin. Appl. Environ. Microbiol. 73:4100–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee J., et al. 2008. Indole cell signaling occurs primarily at low temperatures in Escherichia coli. ISME J. 2:1007–1023 [DOI] [PubMed] [Google Scholar]
- 27. Lesic B., et al. 2007. Inhibitors of pathogen intercellular signals as selective anti-infective compounds. PLoS Pathog. 3:1229–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Low A. S., et al. 2006. Analysis of fimbrial gene clusters and their expression in enterohaemorrhagic Escherichia coli O157:H7. Environ. Microbiol. 8:1033–1047 [DOI] [PubMed] [Google Scholar]
- 29. Nataro J. P. 2005. Enteroaggregative Escherichia coli pathogenesis. Curr. Opin. Gastroenterol. 21:4–8 [PubMed] [Google Scholar]
- 30. Nataro J. P., Kaper J. B. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pratt L. A., Kolter R. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285–293 [DOI] [PubMed] [Google Scholar]
- 32. Rasmussen T. B., Givskov M. 2006. Quorum-sensing inhibitors as anti-pathogenic drugs. Int. J. Med. Microbiol. 296:149–161 [DOI] [PubMed] [Google Scholar]
- 33. Ren D., Sims J. J., Wood T. K. 2001. Inhibition of biofilm formation and swarming of Escherichia coli by (5Z)-4-bromo-5-(bromomethylene)-3-butyl-2(5H)-furanone. Environ. Microbiol. 3:731–736 [DOI] [PubMed] [Google Scholar]
- 34. Ren D., et al. 2005. Differential gene expression for investigation of Escherichia coli biofilm inhibition by plant extract ursolic acid. Appl. Environ. Microbiol. 71:4022–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rendón M. A., et al. 2007. Commensal and pathogenic Escherichia coli use a common pilus adherence factor for epithelial cell colonization. Proc. Natl. Acad. Sci. U. S. A. 104:10637–10642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rezk B. M., Haenen G. R., van der Vijgh W. J., Bast A. 2002. The antioxidant activity of phloretin: the disclosure of a new antioxidant pharmacophore in flavonoids. Biochem. Biophys. Res. Commun. 295:9–13 [DOI] [PubMed] [Google Scholar]
- 37. Ryu J. H., Beuchat L. R. 2005. Biofilm formation by Escherichia coli O157:H7 on stainless steel: effect of exopolysaccharide and curli production on its resistance to chlorine. Appl. Environ. Microbiol. 71:247–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 39. Sartor R. B. 2008. Microbial influences in inflammatory bowel diseases. Gastroenterology 134:577–594 [DOI] [PubMed] [Google Scholar]
- 40. Sasaki M., et al. 2007. Invasive Escherichia coli are a feature of Crohn's disease. Lab. Invest. 87:1042–1054 [DOI] [PubMed] [Google Scholar]
- 41. Stangl V., et al. 2005. The flavonoid phloretin suppresses stimulated expression of endothelial adhesion molecules and reduces activation of human platelets. J. Nutr. 135:172–178 [DOI] [PubMed] [Google Scholar]
- 42. Strockbine N. A., et al. 1986. Two toxin-converting phages from Escherichia coli O157:H7 strain 933 encode antigenically distinct toxins with similar biologic activities. Infect. Immun. 53:135–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Swidsinski A., et al. 2002. Mucosal flora in inflammatory bowel disease. Gastroenterology 122:44–54 [DOI] [PubMed] [Google Scholar]
- 44. Tarr P. I., Gordon C. A., Chandler W. L. 2005. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 365:1073–1086 [DOI] [PubMed] [Google Scholar]
- 45. Ten Hove T., et al. 2001. Blockade of endogenous IL-18 ameliorates TNBS-induced colitis by decreasing local TNF-α production in mice. Gastroenterology 121:1372–1379 [DOI] [PubMed] [Google Scholar]
- 46. Thapa D., et al. 2008. Clotrimazole ameliorates intestinal inflammation and abnormal angiogenesis by inhibiting interleukin-8 expression through a nuclear factor-κB-dependent manner. J. Pharmacol. Exp. Ther. 327:353–364 [DOI] [PubMed] [Google Scholar]
- 47. Uhlich G. A., Cooke P. H., Solomon E. B. 2006. Analyses of the red-dry-rough phenotype of an Escherichia coli O157:H7 strain and its role in biofilm formation and resistance to antibacterial agents. Appl. Environ. Microbiol. 72:2564–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vikram A., Jayaprakasha G. K., Jesudhasan P. R., Pillai S. D., Patil B. S. 2010. Suppression of bacterial cell-cell signalling, biofilm formation and type III secretion system by citrus flavonoids. J. Appl. Microbiol. 109:515–527 [DOI] [PubMed] [Google Scholar]
- 49. Wood T. K. 2009. Insights on Escherichia coli biofilm formation and inhibition from whole-transcriptome profiling. Environ. Microbiol. 11:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yang C. S., Landau J. M., Huang M. T., Newmark H. L. 2001. Inhibition of carcinogenesis by dietary polyphenolic compounds. Annu. Rev. Nutr. 21:381–406 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.