Abstract
Severe malarial anemia (SMA) is a leading cause of morbidity and mortality in children residing in regions where Plasmodium falciparum transmission is holoendemic. Although largely unexplored in children with SMA, interleukin-18 (IL-18) is important for regulating innate and acquired immunity in inflammatory and infectious diseases. As such, we selected two functional single-nucleotide polymorphisms (SNPs) in the IL-18 promoter (−137G→C [rs187238] and −607C→A [rs1946518]) whose haplotypes encompass significant genetic variation due to the presence of strong linkage disequilibrium among these variants. The relationship between the genotypes/haplotypes, SMA (hemoglobin [Hb], <5.0 g/dl], and longitudinal clinical outcomes were then investigated in Kenyan children (n = 719). Multivariate logistic regression analyses controlling for age, gender, sickle cell trait, glucose-6-phosphate dehydrogenase (G6PD) deficiency, HIV-1, and bacteremia revealed that carriage of the −607AA genotype was associated with protection against SMA (odds ratio [OR] = 0.440 [95% confidence interval {CI} = 0.21 to 0.90], P = 0.031) in children with acute infection. In contrast, carriers of the −137G/−607C (GC) haplotype had increased susceptibility to SMA (OR = 2.050 [95% CI = 1.04 to 4.05], P = 0.039). Measurement of IL-18 gene expression in peripheral blood leukocytes demonstrated that elevated IL-18 transcripts were associated with reduced hemoglobin concentrations (ρ = −0.293, P = 0.010) and that carriers of the “susceptible” GC haplotype had elevated IL-18 transcripts (P = 0.026). Longitudinal investigation of clinical outcomes over a 3-year follow-up period revealed that carriers of the rare CC haplotype (∼1% frequency) had 5.76 times higher mortality than noncarriers (P = 0.001). Results presented here demonstrate that IL-18 promoter haplotypes that condition elevated IL-18 gene products during acute infection are associated with increased risk of SMA. Furthermore, carriage of the rare CC haplotype significantly increases the risk of childhood mortality.
INTRODUCTION
Malaria remains one of the most important parasitic infections in the world (49). Of the known apicomplexan Plasmodium parasites that can infect humans, P. falciparum, a species endemic to Africa, accounts for a vast majority of malaria-associated morbidity and mortality (10, 18, 49). Each year, malaria accounts for an estimated 247 million new cases, which result in approximately 881,000 deaths, 91% of which occur in Africa and 85% being in children under 5 years of age (50). In areas of high P. falciparum transmission, malaria infection manifests primarily as severe anemia, high-density parasitemia (HDP), respiratory distress, acute renal failure, and in rare cases, hypoglycemia and cerebral malaria (11, 25, 28, 41). The most common of these disease sequelae, severe malarial anemia (SMA), is responsible for the majority of the malaria-associated mortality in western Kenya (7, 32, 54). Based on historical presence of the disease, malaria has exerted a large impact on the human genome such that potentially harmful variants are preserved, largely because of the advantage offered in heterozygous individuals that are often protected from severe, complicated, and fatal malaria (14, 19, 21). Studies in our laboratory focused on variations in key cytokine genes have demonstrated associations between polymorphisms and SMA (4, 37, 38).
Interleukin-18 (IL-18) is a proinflammatory cytokine with diverse pleiotropic effects (30). Earlier studies designated IL-18 a gamma interferon (IFN-γ)-inducing factor due to its ability to induce production of IFN-γ from natural killer (NK) cells, T cells, and activated macrophages (31). IL-18 is synthesized as a precursor protein (proIL-18) and processed by an intracellular cysteine protease, caspase-1 (30). IL-18 is known to regulate both T helper 1 (Th1) and Th2 responses, depending on the cytokine milieu (31), and acts in vivo in synergy with IL-12 (33). Although IL-18 has a structure homologous to IL-1 and a significant functional homology to IL-12 in mediating Th1 responses and NK cell activity (30, 31), the mechanisms by which IL-18 induces IFN-γ seem to differ from those of IL-12 (46). Given its important role in the inflammatory process, IL-18 has extensively been studied in various disease pathologies, including digestive inflammatory diseases, human immunodeficiency virus (HIV) infection, diabetes, arthritis, asthma, tuberculosis, and cancer (27, 31).
A previous study in western Kenya investigating the relationship between IL-12 and IL-18 and also clinical malaria phenotypes in children (2 to 12 years of age) reported upregulation of IL-18 in uncomplicated malaria, which progressively declined in moderate malaria, and there was a further decrease in children with SMA (hemoglobin [Hb] <5.0 g/dl and any density parasitemia and fever) cases (8). These results parallel another study showing significantly elevated IL-12 and IL-18 in children (2 to 144 months of age) with mild malaria that decreased as disease severity progressed (24). In contrast, a study in adults (14 to 63 years of age), investigating the association between cytokine and antibody responses and uncomplicated, severe, and cerebral forms of malaria, demonstrated a close association between increased IL-18 levels and severe falciparum malaria (20, 29). These results are similar to an investigation showing elevated IL-18 in adult patients (mean age, 37.7 ± 5.9 years) with uncomplicated malaria, which decreased upon recovery of disease (44). Taken together, these results suggest that IL-18 plays an important role in conditioning severe malaria. However, no studies to date have reported the role of polymorphic variants of the IL-18 gene in modulating malaria.
Two IL-18 single-nucleotide polymorphisms (SNPs; −137G→C [rs187238] and −607C→A [rs1946518]) have consistently been associated with altered IL-18 transcriptional activity (13, 23, 53). The G-to-C substitution at position −137 abolishes a histone 4 transcription factor-1 (H4TF-1) nuclear factor-binding site, while the C-to-A transversion at position −607 disrupts a cyclic AMP (cAMP)-responsive element protein-binding site (13). Consequently, lower promoter activity has been reported for the minor alleles −137C and −607A, respectively (3, 23, 53). Furthermore, haplotypes carrying these minor alleles also correlate with decreased IL-18 levels in peripheral blood mononuclear cells or plasma (13, 53). These haplotypes appear to capture the majority of genetic variation in IL-18, due to the presence of strong linkage disequilibrium among the variants (3). Although these SNPs have been implicated in various disorders such as type I diabetes, asthma, hepatitis C virus, and rheumatoid arthritis (2, 15, 43), no studies have examined the relationships between the variants and malaria disease outcomes. As such, we examined the impact of IL-18 promoter polymorphisms (−137G→C and −607C→A) on susceptibility to severe malarial anemia (Hb < 5.0 g/dl and any density parasitemia) and IL-18 transcriptional expression in children residing in a region where P. falciparum transmission is holoendemic in western Kenya.
(Portions of this work were presented at the 58th Annual Meeting of the American Society of Tropical Medicine and Hygiene held at Washington, DC, in 2009 [abstract 140].)
MATERIALS AND METHODS
Study site.
The present study was carried out among a pediatric population in Siaya District, western Kenya, an equatorial climate 1,140 m to 1,430 m above sea level with an annual rainfall between 800 mm to 2,000 mm. Comprehensive measurement of the Plasmodium falciparum entomological inoculation rate (EIR), more than a decade ago, was estimated at up to 300 infectious bites per person per year (5). Although recent data are not available for the EIR, the primary hospital within the region, Siaya District Hospital (SDH), has had increased pediatric malaria admissions from mid-2006 onward (34). Falciparum malaria prevalence is ∼83% in children <4 years of age, with severe disease manifesting as severe anemia and/or high-density parasitemia (HDP) (32, 35). The region is inhabited by the Luo ethnic tribe (>96%), a predominant homogenous population appropriate for genetic-based investigations. Details of the study site and clinical definitions of disease manifestations in the cohort are described in our previous report (35).
Study participants.
Children (n = 719) of both genders (3 to 36 months of age) were recruited at SDH and included children presenting for treatment of febrile illnesses as well as children visiting the Maternal and Child Health Clinic for childhood vaccinations. Enrollment was confined to those children presenting for their first “hospital contact.” Written informed consent was obtained from the parent/guardian of each child participating in the study. Parasite densities and Hb levels were determined, and the results were used to stratify parasitemic children into (i) non-SMA (Hb ≥ 5.0 g/dl) and (ii) SMA (Hb < 5.0 g/dl) groups, both with any density of parasitemia (48). In addition, analyses were carried out with children being categorized into either the non-SMA (Hb ≥ 6.0 g/dl and any density of parasitemia) or SMA (Hb < 6.0 g/dl and any density of parasitemia) group, based on the distribution of anemia determined by >14,000 longitudinal Hb measurements in age- and gender-matched children from the same geographic location (26). Children were excluded from the study for any one of the following reasons: (i) positive blood smears with non-P. falciparum species, (ii) previous hospitalization (for any reason), (iii) documented or reported use of antimalarial therapy 2 weeks prior to enrollment, and/or (iv) cerebral malaria diagnosis. Patients were treated according to the Ministry of Health (MOH)—Kenya guidelines, which included oral artemether-lumefantrine (Coartem) for uncomplicated malaria and intravenous quinine for severe malaria. The study was approved by the Ethics Committees of the Kenya Medical Research Institute and University of New Mexico Institutional Review Board.
Longitudinal follow-up.
Following enrollment (day 0), parents/guardians were asked to return with their child every 3 months throughout the 3-year follow-up period. If the parent/guardian had not returned to hospital by 1:00 pm on the day of the quarterly follow-up visit, our study staff visited the child's residence to check on his or her health status, including mortality. Since we determined the exact location of each child's residence with our geographic information system (GIS)/global position system (GPS) surveillance system, we could readily locate each child. In addition, since children experience multiple episodes of malaria and other pediatric infectious diseases in this region, parents/guardians were asked to return to hospital during their child's febrile episode(s). All laboratory tests required for proper clinical management of the patients were performed at each acute and quarterly visit, including complete hematological indices, malaria parasitemia measures, and evaluation of bacteremia (if clinically indicated). In addition, all-cause mortality data were collected throughout the 3-year follow-up. Although most children within this region die at home, visits by the study team confirmed the date of mortality. Mortality data, along with clinical and laboratory measures for multiple episodes of malaria, were used to evaluate the association between genotype (and haplotypic structure) and longitudinal outcomes of malaria and mortality.
Clinical and laboratory evaluation.
Hb levels and complete blood counts were determined using the Beckman Coulter AcT diff2 (Beckman Coulter Corporation, Miami, FL). To determine parasitemia, thick blood smears stained with Giemsa stain were prepared and examined under high-power magnification. P. falciparum parasites per 300 white blood cells (WBCs) were determined, and parasitemia (per μl) was estimated using the total WBC count. In order to delineate severe anemia caused by malaria compared to that caused by additional anemia-promoting infections, human immunodeficiency virus (HIV)-1, bacteremia, sickle cell status, and glucose-6-phosphate dehydrogenase (G6PD) deficiency were determined. HIV-1 status was determined using two serological methods followed by proviral DNA PCR, as described previously (36). Parents/guardians of the study participants received pre- and posttest HIV/AIDS counseling and provided informed consent. At the time of enrollment, none of the children had been started on antiretroviral therapy. Bacteremia was determined by microbial cultivation according to our standard methods (47). Sickle cell status was determined by alkaline cellulose acetate electrophoresis on Titan III plates (Helena BioSciences, Sunderland, United Kingdom) according to the manufacturer's instructions. G6PD deficiency was determined by a fluorescent spot test using the manufacturer's methods (Trinity Biotech Plc., Bray, Ireland).
Genetic analyses.
Blood spots were collected on FTA Classic cards (Whatman Inc., Clifton, NJ), air dried, and stored at room temperature until use. DNA was extracted using the Gentra system (Gentra Systems, Inc., Minneapolis, MN) according to the manufacturer's recommendations. To obtain sufficient quantities for genetic analyses, genomic DNA was amplified using the GenomiPhi system (GE Healthcare, Piscataway, NJ). The IL-18 −137G→C promoter polymorphism was genotyped by using the high-throughput TaqMan 5′ allelic discrimination Assay-By-Design method according to the manufacturer's instructions (assay identification number C_2408543_10; Applied Biosystems, Foster City, CA). Samples were genotyped for the −607C→A polymorphism by using the PCR-restriction fragment length polymorphism (RFLP) technique previously described (22).
Extraction of RNA from WBC pellets.
Total RNA was extracted from frozen RNase-stored white blood cells (WBCs) by using the guanidium isothiocyanate (GITC)-isopropanol method as previously described (9). Briefly, WBCs were lysed for 5 min at room temperature in GITC solution containing 25 mM sodium acetate and 10% Sarkosyl. To completely dissociate the nucleoproteins, a layer of phenol was added, followed by chloroform-isoamyl alcohol mix, vortexed thoroughly, and incubated for 30 min on ice. The aqueous phase was recovered after centrifugation and the extracted RNA precipitated overnight at −20°C in cold isopropanol. The precipitated RNA was thereafter pelleted by centrifugation, washed in 70% ethanol, and solubilized in RNase-free water. RNA was stored at −80°C until use.
Reverse transcription-PCR and semiquantitative analysis of IL-18 gene expression.
To quantify the extracted RNA, absorbance (A260/A280) was measured using GeneQuant pro (GE Healthcare, Piscataway, NJ) for each sample, and concentrations were calculated based on optical densities. cDNA was synthesized using the TaqMan reverse transcription reagent kit (Applied Biosystems, Foster City, CA). Briefly, 1 μg of RNA was reverse transcribed in a 20-μl reaction volume mix containing, as final concentrations, 5 mM MgCl2, 1 mM deoxynucleoside triphosphates (dNTPs), 1.5 μM random hexamers, 1 U RNase inhibitor, 1.5 U reverse transcriptase enzyme, and 1× PCR buffer. Incubation was carried out at 48°C for 45 min in a GeneAmp PCR system 9700 (Applied Biosystems, Foster City, CA), followed by inactivation of the reverse transcriptase enzyme at 95°C for 5 min and cooling at 4°C.
Semiquantitative IL-18 gene expression experiments were performed using 1 μg of cDNA in a 25-μl PCR mix containing 0.3 μM (each) oligonucleotides with a sense sequence 5′-CCC TTT GCT CCC CTG GCG AC-3′ and antisense 5′-AGA CTG CAG CAG GTG GCA GC-3′ to generate a 301-bp fragment. To normalize the amount of cDNA loaded per reaction, an internal control, the cyclophilin A (CYC-A) housekeeping gene was amplified in a 25-μl reaction, containing final concentrations of 0.3 μM each CYC-A sense oligonucleotide 5′-GTC TCC TTT GAG CTG TTT GC-3′ and antisense oligonucleotide 5′-AAG CAG GAA CCC TTA TAA CC-3′. Amplification was carried out in a 96-well format GeneAmp PCR system 9700 (Applied Biosystems, Foster City, CA), in reaction mixes containing final concentrations of 1.5 mM MgCl2, 200 nM dNTPs, 1.0 U GoTaq polymerase enzyme (Promega Corporation, Madison, WI), and 1× PCR buffer supplied by the manufacturer. Cycling conditions were as follows: initial denaturation at 94°C for 2 min, followed by 30 cycles each of denaturation at 94°C for 30 s, annealing at 63°C for 30 s, and 72°C for 45 s. A final extension of 72°C for 5 min was included, before cooling the products to 4°C. The resulting products were resolved on a 2% agarose gel stained with 0.5 μg/ml ethidium bromide (Sigma-Aldrich, St. Louis, MO) and visualized on an UV transilluminator (Spectroline Corporation, Westbury, NY). Digital gel photos were taken using a Canon PowerShot A640 camera (Canon Inc. Lake Success, NY) and band images were processed using ImageJ software (1). The integrated mean band intensities are reported as arbitrary units (AU) and were determined as ratios of the IL-18 gene band's mean intensity value divided by the corresponding housekeeping gene mean intensity to normalize the values. All HIV-1-positive and bacteremia-positive children were excluded from this analysis.
Data analyses.
SPSS statistical software package version 15.0 (IBM SPSS Inc., Chicago, IL) was used for all statistical analyses. Demographic, clinical, and parasitological data between groups were compared using chi-square (χ2), Mann-Whitney U, and Student's t tests. Genotype, allele, and haplotypic frequencies of IL-18 were compared between non-SMA and SMA groups by using the χ2 test. Pairwise comparisons of variables (genotypes and haplotypes) between non-SMA and SMA were conducted using the Mann-Whitney U test. IL-18 mRNA levels were normalized by expressing their mean arbitrary units (AU) as ratios of the CYC-A (housekeeping) gene. Parametric analyses between and within groups were performed using Student's t test (with Welch's correction) and analysis of variance (ANOVA) for bivariate and multivariate comparisons, respectively, both with 95% confidence intervals (CI). To assess the effect conferred by a particular genotype or haplotype (cross-sectionally), multivariate logistic regression analysis was used to calculate the odds ratio (OR) and 95% CI in a model controlling for the potential confounding effects of age, gender, G6PD deficiency, sickle cell trait, HIV-1 status, and presence of bacterial infections. The Hardy-Weinberg equilibrium (HWE) was tested using a χ2 goodness-of-fit test. Linkage disequilibrium (LD) between polymorphisms was quantified using Multiallelic Interallelic Disequilibrium Analysis (MIDAS) software version 1.0 (12). Haplotypes were constructed using the HPlus software program (Fred Hutchinson Cancer Research Center, Seattle, WA) and their frequencies estimated based on a Bayesian algorithm. In addition, hierarchical logistic regression was used to investigate the association between haplotypes and longitudinal outcomes of repeated SMA episodes and mortality. Under this model, the covariates (i.e., age, gender, G6PD deficiency, sickle cell trait, HIV-1, and bacteremia status) were entered as independent confounding effects with haplotype contrast (carrier versus noncarrier) in predicting outcomes (SMA and mortality). All P values of <0.100 were further analyzed by using Cox regression/survival analysis, and differences in the distributions of hazard rate functions (i.e., the probabilities of experiencing the event) between carriers and noncarriers were examined using Mann-Whitney U tests. Statistical significance was defined by a P value of ≤0.050 for all analyses.
RESULTS
Characteristics of malaria-infected study participants upon enrollment.
Since one of the primary aims of the study was to determine the genetic variants that condition susceptibility to SMA in children with falciparum malaria, we first conducted cross-sectional analyses. As such, children with P. falciparum parasitemia (3 to 36 months; n = 523) were grouped according to the WHO definition of SMA, with non-SMA patients having Hb of ≥5.0 g/dl (n = 400) and SMA patients having Hb of <5.0 g/dl (n = 123) (48). A summary of demographic, clinical, and parasitological characteristics of the parasitemic study participants (upon enrollment) is shown in Table 1 . The gender ratios in the clinical groups were comparable (P = 0.098). However, age differed across the groups, with the SMA group being significantly younger (P = 0.017). Consistent with the a priori stratification, Hb levels differed between the groups (P < 0.001). Total white blood cell (WBC) counts were significantly higher in children with SMA (P < 0.001). The admission temperature was, however, comparable between the groups (P = 0.364). Mean parasite densities (MPS/μl), geometric mean parasitemias (per μl), and the proportions of children with high-density parasitemia (HDP; MPS ≥ 10,000/μl) were not significantly different between the groups (P values of 0.340, 0.984, and 0.209, respectively). Similarly, the distributions of carriers of sickle cell traits (Hb AS, carriers [heterozygous] of the sickle cell trait) and glucose 6 phosphate (G6PD) deficiency were comparable between non-SMA and SMA groups (P = 0.110 and 0.210, respectively).
Table 1.
Demographic, clinical, and laboratory characteristics of the study participants
| Characteristica | Non-SMA patientsb | SMA patientsb | P |
|---|---|---|---|
| No. of participants | 400 | 123 | |
| No. (%) of: | |||
| Males | 213 (53.9) | 55 (44.7) | 0.098c |
| Females | 187 (46.8) | 68 (55.3) | |
| Age (mo) | 10.0 (10.0) | 8.0 (7.0) | 0.017d |
| Hemoglobin (g/dl) | 7.1 (2.8) | 4.2 (1.1) | <0.001d |
| WBCs (×103/μl) | 11.2 (5.8) | 15.2 (9.6) | <0.001d |
| Temperature (οC) | 37.5 (2.0) | 37.5 (2.0) | 0.364d |
| Parasite density (MPS/μl) | 18,983 (39,222) | 14,952 (44,241) | 0.340d |
| Geometric mean parasitemia/μl | 13,849 | 10,667 | 0.984e |
| No. (%) of patients with: | |||
| HDP (≥10,000 parasites/μl) | 256 (64.0) | 71 (57.7) | 0.209c |
| Sickle cell trait | 62 (15.5) | 12 (9.8) | 0.110c |
| G6PD deficiency | 45 (11.3) | 9 (7.3) | 0.210c |
WBCs, white blood cells; MPS, malaria parasites; HDP, high-density parasitemia; G6PD deficiency, glucose-6-phosphate dehydrogenase deficiency.
Data are presented as medians (interquartile ranges [IQRs]) unless otherwise noted. SMA was categorized according to the WHO definition (i.e., Hb < 5.0 g/dl, with any density of parasitemia) (48).
Statistical significance determined by the chi-square analysis.
Statistical significance determined by Mann-Whitney U test. (Significant differences are in bold.)
Statistical significance determined by Student's t test.
Association between IL-18 gene expression and anemia in children with P. falciparum infections.
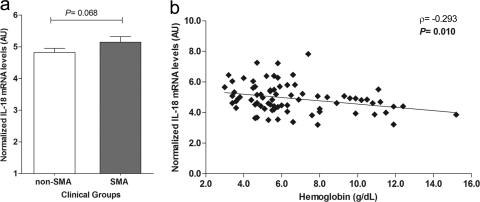
Prior to embarking on the genetic analyses, gene expression profiles of IL-18 were measured in the non-SMA (n = 53) and SMA (n = 24) groups upon enrollment by use of semiquantitative mRNA analyses, normalized to the housekeeping gene cyclophilin A. Although there were higher IL-18 mRNA expression levels in children with SMA, the difference between the groups did not reach statistical significance (P = 0.068; Fig. 1 a). An identical pattern was observed when children were categorized according to the definition using an Hb value of <6.0 g/dl as the cutoff criterion for SMA (P = 0.053; data not shown). Additional analysis of the relationship between IL-18 mRNA expression and Hb levels revealed that higher IL-18 transcripts were associated with lower Hb concentrations (ρ = −0.293; P = 0.010) (Fig. 1b).
Fig. 1.
Relationship between IL-18 mRNA expression and anemia. IL-18 gene expression was measured in peripheral blood leukocytes collected from children (n = 77) with malaria upon enrollment in the study (day 0). (a) Bivariate analysis of normalized IL-18 gene expression (arbitrary units [AU]) in the non-SMA (Hb ≥ 5.0 g/dl; n = 53) and SMA (Hb < 5.0 g/dl; n = 24) groups was performed using Student's t test. (b) Association between normalized IL-18 mRNA levels (n = 77), expressed as AU, and hemoglobin levels (Hb; g/dl) at admission was determined by the Spearman rank correlation test.
Distribution of IL-18 promoter genotypes and alleles in children with acute malaria.
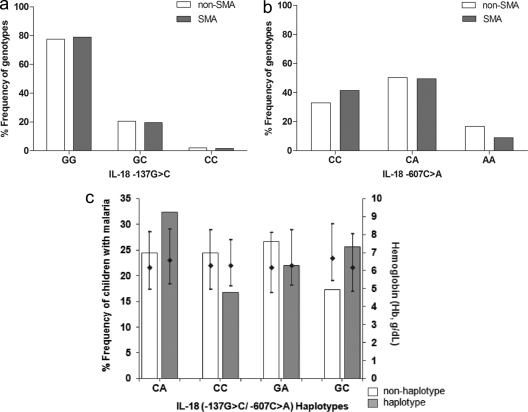
To investigate the association between IL-18 promoter variants (−137G→C and −607C→A) and cross-sectional susceptibility to SMA, genotype and allele frequencies were determined in non-SMA and SMA groups (Fig. 2 a and b). Genotypic frequencies for the polymorphic variant at −137 were 0.78 (GG), 0.20 (GC), and 0.02 (CC) in the non-SMA group and 0.79 (GG), 0.19 (GC), and 0.02 (CC) in the SMA group, respectively (Fig. 2a). Both frequencies were consistent with HWE (non-SMA, χ2 = 0.863 [P = 0.353]; SMA, χ2 = 0.132 [P = 0.716]). Overall genotype distributions in the two groups were 0.78 (GG), 0.20 (GC), and 0.02 (CC) and were in equilibrium (χ2 = 0.990; P = 0.320). The genotypic proportions between non-SMA and SMA groups were not significantly different (P = 0.934; Fig. 2a). Frequencies of the −137G and C alleles were 0.88 and 0.12 for non-SMA cases and 0.89 and 0.11 for the SMA group, respectively. The overall proportions were 0.88 and 0.12 for G and C alleles, respectively, and did not significantly differ between the groups (P = 0.853).
Fig. 2.
Distribution of IL-18 genotypes and haplotypes in parasitemic children upon enrollment. Parasitemic children (n = 523) upon enrollment were categorized into the non-SMA (n = 400; Hb ≥ 5.0 g/dl) or SMA (n = 123; Hb < 5.0 g/dl) group. Frequency distributions of the individual IL-18 genotypes are presented as proportions (percentages) for the −137G→C (a) and −607C→A (b) polymorphisms. (c) Proportion of children with SMA stratified according to IL-18 haplotypes, along with hemoglobin (Hb; g/dl) levels presented as medians (25th and 75th percentiles). The diamonds and error bars show the Hb median and 25th/75th percentiles, respectively, for children with the indicated haplotypes relative to those without these haplotypes. Comparison of proportions was determined by the chi-square analysis. The frequency distributions of IL-18 genotypes between non-SMA and SMA groups were comparable for −137 (P = 0.934; panel a) and −607 (P = 0.056; panel b). Analysis showed that there were more GC haplotypes in the SMA category than non-GC haplotypes (P = 0.034). Additionally, analysis of Hb levels of non-GC and GC haplotypes revealed a significant difference (P = 0.029). All the other haplotypes were comparable between carriers (haplotypes present) and noncarriers (haplotypes absent) (panel c).
Genotypic distributions for the variants at −607 were 0.33 (CC), 0.50 (CA), and 0.17 (AA) in the non-SMA group and 0.42 (CC), 0.50 (CA), and 0.08 (AA) in the SMA group (Fig. 2b). Distributions in both groups were in equilibrium (non-SMA, χ2 = 0.416 [P = 0.519]; SMA, χ2 = 1.466 [P = 0.226]). Overall frequency distributions were 0.35 (CC), 0.50 (CA), and 0.15 (AA) and were consistent with HWE (χ2 = 1.102; P = 0.314). The proportions of genotypes in the non-SMA and SMA groups were marginally different (P = 0.056; Fig. 2b). Distribution of alleles C and A of the −607 variant were 0.58 and 0.42 for the non-SMA group and 0.66 and 0.34 for SMA, respectively. Overall proportions were 0.60 and 0.40 for C and A alleles, respectively, and did not significantly differ between the non-SMA and SMA groups (P = 0.121).
Distribution of IL-18 promoter haplotypes in children with acute malaria.
Construction of haplotypes for the two polymorphic loci yielded the following overall frequencies: 0.11 for −137C/−607A (CA), 0.01 for CC, 0.29 for GA, and 0.59 for GC. Distribution of haplotypes in children with SMA (n = 123) were 0.32 for CA, 0.17 for CC, 0.22 for GA, and 0.26 for GC (Fig. 2c). Comparison of SMA percentages among haplotypic carriers and noncarriers revealed that the group with the GC haplotype had a higher proportion of SMA patients (P = 0.034) and lower Hb concentrations (P = 0.029) than the group with the non-GC haplotype (Fig. 2c). The proportions of children with SMA did not significantly differ for any of the other two haplotypes (non-CA versus CA, P = 0.826; non-CC versus CC, P = 0.691) but marginally differed between non-GA and GA (P = 0.074) haplotypes. Consistent with this distribution for the three haplotypes, Hb levels were comparable between haplotypic carriers and noncarriers (non-CA versus CA, P = 0.381; non-CC versus CC, P = 0.857; non-GA versus GA, P = 0.255). Additional analyses demonstrated that the two loci were in linkage disequilibrium (LD; ∣D′∣ = 0.889).
Relationship between IL-18 polymorphisms and SMA.
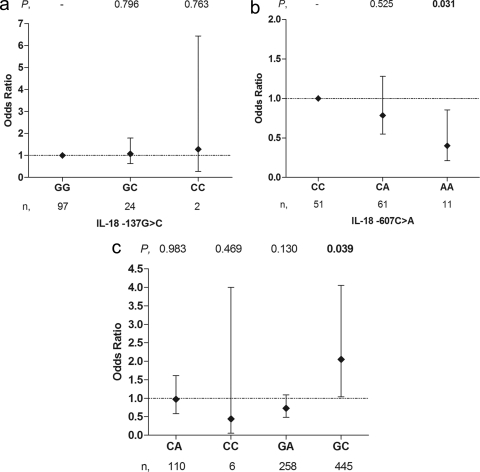
To investigate the relationship between the IL-18 polymorphisms and susceptibility to SMA in children with acute disease, we performed multivariate logistic regression modeling, controlling for covariates. Analysis of the −137 variants showed no association between SMA and the homozygous mutant (CC, OR = 1.282 [95% CI = 0.255 to 6.443], P = 0.763) or the heterozygote variant (GC, OR = 1.071 [95% CI = 0.639 to 1.794], P = 0.796), relative to wild-type genotype GG (Fig. 3 a).
Fig. 3.
Relationship between IL-18 genotypes/haplotypes and SMA. The cross-sectional associations between genotypes/haplotypes and SMA (Hb < 5.0 g/dl) were determined in children with malaria (n = 523). Odds ratios (ORs) and 95% confidence intervals (CIs) were determined using multivariate logistic regression analyses, controlling for age, gender, G6PD deficiency, HIV-1 and sickle cell trait status, and presence of bacteremia. To determine the impact of each genotype on susceptibility to SMA, individuals with the homozygous wild-type variant (GG for −137 [a] and CC for −607 [b]) were used as the reference groups. (c) Similarly, to determine the association between haplotypes and susceptibility to SMA, individuals without the haplotype were used as the reference group. The diamonds and error bars represent the ORs and 95% CIs associated with susceptibility to SMA. Numbers at the top represent significance (P values), while the n values at the bottom represent the numbers of children with the genotype/haplotype.
Investigation of the −607 SNP showed that, relative to the CC (wild-type) group, children carrying the homozygous AA (mutant) had a 56% reduced risk of developing SMA (OR = 0.440 [95% CI = 0.212 to 0.856], P = 0.031) (Fig. 3b). Heterozygosity (CA) at −607 showed a similar pattern and was associated with a 22% decrease in susceptibility to SMA (OR = 0.786 [95% CI = 0.741 to 1.280], P = 0.525). Additional multivariate logistic regression analyses using an Hb value of <6.0 g/dl as the definition of SMA showed identical results for both the −137 and −607 variants (data not shown).
Association between IL-18 haplotypes and SMA.
Multivariate modeling of the haplotypes, controlling for covariates, showed that there was no cross-sectional association between susceptibility to SMA and carriage of the −137C/−607A (CA; OR = 0.966 [95% CI = 0.581 to 1.605], P = 0.983) or CC (OR = 0.441 [95% CI = 0.048 to 4.040], P = 0.469) haplotype. However, the GA haplotype showed a trend toward protection (OR = 0.725 [95% CI = 0.478 to 1.099], P = 0.130) against SMA (Fig. 3c). Furthermore, children with the GC haplotype demonstrated a risk of developing SMA 2-fold higher than that for children without the haplotype (OR = 2.050 [95% CI, 1.037 to 4.054], P = 0.039). Consistent with these analyses, when <6.0 g/dl Hb was used to define SMA, there was no association between susceptibility to SMA and the CA (OR = 0.911 [95% CI, 0.588 to 1.412], P = 0.668), CC (OR = 0.449 [95% CI, 0.077 to 2.630], P = 0.374), or GA (OR = 0.944 [95% CI, 0.659 to 1.351], P = 0.752) haplotype, while carriers of the GC haplotype showed a tendency toward enhanced risk of SMA (OR = 1.552 [95% CI, 0.925 to 2.607], P = 0.096).
Levels of IL-18 transcripts in the genotypic groups.
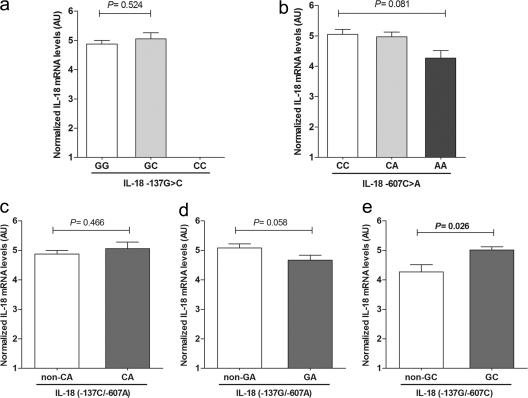
IL-18 transcript levels were quantified and compared across the genotypic groups (Fig. 4). Due to a lower prevalence of homozygous C individuals in the population, we were unable to obtain mRNA levels for this genotypic group. As such, we carried out a bivariate analysis to compare the mRNA levels in the two available genotypes (GG [n = 58] and GC [n = 19]). Results revealed that the transcriptional levels were comparable between the genotypes (P = 0.524; Student's t test) (Fig. 4a).
Fig. 4.
IL-18 transcript levels stratified according to genotype/haplotype. Analyses of IL-18 gene expression levels among children (n = 77) stratified according to the IL-18 −137G→C (a) and −607C→A (b) genotypes. (c to e) IL-18 mRNA transcript levels stratified according to haplotypes. Statistical significance across the genotypes was determined by ANOVA, while bivariate comparisons were performed using two-tailed unpaired Student's t test with Welch's correction at a 95% confidence interval. Error bars represents standard error of means (SEM).
IL-18 mRNA levels across the −607 genotypes were marginally different (P = 0.081; ANOVA) (Fig. 4b). Additional bivariate analyses demonstrated comparable expression levels for the CA (n = 37, P = 0.751; Student's t test) genotype relative to wild-type (CC, n = 31) genotype, as well as significantly different levels between AA (n = 9, P = 0.025; Student's t test) and the CC genotype and also between the CA and AA (P = 0.047) groups.
Levels of IL-18 transcripts in the haplotypic groups.
Stratification of IL-18 mRNA expression according to haplotypes showed that IL-18 transcripts were similar between carriers and noncarriers of the CA (n = 18; P = 0.466) (Fig. 4c) haplotypes. IL-18 levels could not be determined in the rare −137C/−607C (CC) haplotype, since no peripheral blood samples were available for this group. Carriers of the GA haplotype had marginally lower IL-18 transcripts (n = 30; P = 0.058) (Fig. 4d) than those with the non-GA haplotypes. IL-18 mRNA levels in carriers of the GC haplotype were observed to be significantly higher than those in the non-GC group (n = 68; P = 0.026) (Fig. 4e).
Association between haplotypes and longitudinal outcomes (SMA and mortality).
After we determined the cross-sectional relationship between genotypes/haplotypes and susceptibility to SMA, hierarchical logistic regression was used to investigate the relationship between carriage of the different haplotypes and longitudinal outcomes (i.e., repeated episodes of SMA and mortality). Haplotypic distributions for the overall cohort (n = 719) were 0.11 (CA), 0.01 (CC), 0.29 (GA), and 0.59 (GC), consistent with those documented cross-sectionally. In addition, as with the cross-sectional analyses, the two loci were in linkage disequilibrium (∣D′∣ = 0.837) for the entire cohort. Longitudinal analyses failed to identify any significant relationships between repeated episodes of SMA and the CA (β = 0.130, P = 0.530), CC (β = 0.202, P = 0.788), GA (β = −0.103, P = 0.543), and GC (β = −0.199, P = 0.410) haplotypes. Over the 3-year follow-up period, there was an 8.2% mortality rate (59/719). Longitudinal modeling via hierarchical logistic regression revealed that there was a 33.3% mortality rate (4/12) in carriers of the CC haplotype and 7.8% (55/707) in noncarriers (β = −1.727, P = 0.066). Consistent with these results, Cox regression modeling, controlling for the same confounding variables, revealed mean hazard rates (the probability of dying over time) of 37.2% and 6.5% for carriers and noncarriers of the CC haplotype, respectively (P = 0.001). Thus, there was a 5.72 higher risk of all-cause mortality in carriers of the CC haplotypes. The longitudinal mortality did not differ between carriers and noncarriers of the CA (β = 0.499, P = 0.320), GA (β = −0.128, P = 0.710), and GC (β = −0.992, P = 0.184) haplotypes.
DISCUSSION
While uncovering genes that condition susceptibility to malaria is clearly important for an improved understanding of the disease, our laboratory has taken a different strategy. For example, since nearly all children in regions where transmission is holoendemic have repeated malarial infections, the question is not “What protects against acquisition of malaria?” but rather “What genes/gene pathways are associated with susceptibility to SMA?” with SMA being the single severe disease manifestation in regions of holoendemicity. Utilizing such an approach, focusing specifically on innate immune response genes in parasitized children, we have identified a number of genes that have significant relationships with both the risk of developing SMA and functional changes in their respective gene products (4, 37, 38). To extend these results, we investigated two functional promoter polymorphisms in IL-18 at positions −137 and −607 in children with and without SMA.
Prior to embarking on discerning the relationship between genotypes/haplotypes and susceptibility to SMA, we first determined if IL-18 gene expression differed between nonsevere and severe infections. Contrary to previous observations (8, 24), our results demonstrated that children with SMA had significantly higher levels of IL-18 transcripts. However, consistent with other studies (20, 29), we demonstrate that levels of IL-18 mRNA were inversely associated with Hb concentrations, suggesting that IL-18 may play an important role in conditioning anemia outcomes in children with falciparum malaria. Differences between our findings and previous ones (8, 24) are likely due to differences in study design and populations. For example, a previous study investigated responders and nonresponders to IL-18 among adult populations (8), while the current study evaluated IL-18 gene expression in children <36 months in age. In addition, severe malaria in previous studies was defined by mixed clinical phenotype (24), as opposed to our population in which SMA was the primary clinical outcome. It will be important to replicate the study in a different pediatric population experiencing SMA as the primary clinical outcome.
To determine if the relationship between IL-18 and anemia outcomes was conditioned by genotype, we first compared (cross-sectionally) the variant frequencies of the two loci independently. Although the SNPs at −137 were not different between the non-SMA and SMA groups, there was a marginal difference in frequency distribution for the −607 variants. In addition, the overall genotypic frequencies for the −137 and −607 SNPs in the non-SMA and SMA groups were consistent with HWE. Genotype frequencies for the −137 variants reported here are comparable to those documented in the HapMap database for the African ancestry population (GG, 0.696; GC, 0.304). Notably, the population investigated here possessed the homozygous CC (mutant) at 0.02, which was absent in the Yoruba population (sub-Saharan African descent) whose data are contained in HapMap (17). Single-locus minor allele frequencies (0.12 for −137C) closely resemble those for the Yoruban population (i.e., 0.15). Similarly, the −607 variant frequencies in the Kenyan children examined here are comparable to those for the Yoruban population (0.43 for CC, 0.44 for CA, and 0.12 for AA). The minor allele frequency (−607A) in the Kenyan children (0.40) showed a trend similar to that reported for the Yoruba population (0.34). Taken together, these results suggest that IL-18 promoter alleles and genotypes have been maintained in ethnic groups of African descent.
Functional assays have previously demonstrated that the −137C and −607A alleles are associated with decreased production of IL-18 (3, 13). Variant −137 is located within multiple nuclear binding sites, which include the human histone H4 gene-specific transcription factor 1 (H4TF1), hepatocyte nuclear factor-3β (forkhead box A2), and the Th2-specific transcription factor GATA binding protein 3 (6, 16, 52). In 2001, Giedraitis et al. (13) observed that transversion from G to C at position −137 changes the H4TF1 nuclear binding site into that for an unknown factor located within the granulocyte-macrophage colony-stimulating factor (GM-CSF) promoter, potentially reducing production of IL-18. Furthermore, the C-to-A change at the second loci we investigated (−607) mediates transcriptional activation in response to cAMP by disrupting the binding site, which may also result in lower IL-18 production.
Since some studies have shown an association between elevated IL-18 levels and severe P. falciparum malaria (20, 29), we hypothesized that single-locus substitutions resulting in decreased IL-18 production would potentially protect against SMA. However, multivariate modeling revealed that single-locus variants at position −137 were not significantly associated with susceptibility to SMA. Consistent with this finding, IL-18 mRNA expression did not appear to be conditioned by −137 variants. Previous studies showing that −137 variants may differentially regulate IL-18 production in liver hepatocyte cells (6, 16), presence of IL-18 in liver cells (40), and the role of IL-18 in T cell-mediated liver injury (45) suggest that −137 variants could potentially be important in the preerythrocytic (liver) stage of malaria. However, the study design employed here, examining the erythrocytic stage of disease, cannot address this hypothesis.
Cross-sectional investigation of variation at −607 demonstrated that the −607AA (mutant) genotype was significantly associated with protection against SMA and showed a tendency toward protection when SMA was defined as <6.0 g/dl Hb. In addition, although the differences in IL-18 mRNA levels between the genotypic groups did not significantly differ across the groups, carriers of the AA genotype had the lowest IL-18 levels. Post hoc analyses showed that IL-18 transcripts were significantly lower in the AA group than for both CC and CA carriers. Taken together, these findings support the notion that reduced levels of IL-18, conditioned, at least in part, by variation at −607, are associated with protection against SMA.
Since haplotypes often reveal how combinations of different functional polymorphic alleles interact to amplify, or moderate, their individual effects (38, 51), we constructed haplotypes for the two loci. Consistent with previous reports (39, 42), there was linkage disequilibrium between the −137 and −607 loci in our study population. Stratification of children according to haplotypes showed that the group with the −137G/−607C (GC) haplotype had a significantly higher proportion of SMA patients and significantly lower Hb concentrations. Additional analyses using multivariate logistic regression, controlling for confounders, revealed that GC carriers had twice the risk of developing SMA compared to children without the haplotype. Further investigation using the modified definition of SMA (Hb < 6.0 g/dl) showed a similar trend toward increased risk.
Investigation of IL-18 mRNA expression in the haplotypic groups revealed that children with the GC haplotype also had significantly higher levels of IL-18 transcripts. These results are consistent with previous studies demonstrating that the GC and GA haplotypes had higher transcriptional activity than the CA and CC haplotypes (13, 53). Given the significantly elevated transcript levels of IL-18 in the SMA group, coupled with results showing that the GC haplotype is associated with increased risk of SMA and significantly higher IL-18 levels, we propose that elevated IL-18 augments SMA pathogenesis. This hypothesis is supported by previous investigations showing that elevated IL-18 levels are associated with severe malaria outcomes (20, 29).
Investigation of the relationship between haplotypes and susceptibility to SMA over the 3-year follow-up did not show any significant association between haplotypes and repeated episodes of SMA. These results suggest that although IL-18 promoter haplotypes are important in conditioning susceptibility to SMA in children once they acquire falciparum malaria, they do not appear to influence either the acquisition of repeated malaria infections (data not presented) or future SMA episodes. However, there was a 4-fold-higher all-cause mortality rate among carriers of the −137C/−607C (CC) haplotype according to hierarchical logistic regression modeling. Additional investigation of the relationship between carriage of the CC haplotype and mortality by using Cox regression modeling, controlling for the same confounding variables, demonstrated that the probability of mortality over time was 5.72-fold higher in carriers of the CC haplotype. The high rate of mortality in this group is consistent with the exceedingly low CC haplotypic frequency, which represented 1% of the total haplotypic constructs. Although it remains to be determined, the low frequency of the CC haplotype in the children examined here, and the complete absence of the CC haplotype in the adult Yoruban population, may indicate a selection bias due to its association with mortality.
In conclusion, we report for the first time that IL-18 promoter haplotypes (IL-18 −137G→C/−607C→A) are important in conditioning the development of SMA in children residing in an area where P. falciparum transmission is holoendemic. Based on data presented here, carriers of the GC haplotype appear to have a genetic predisposition to develop SMA once they acquire a P. falciparum infection, at least in part, through overproduction of IL-18. In addition, although the haplotypes examined here do not appear to influence susceptibility to either malaria or SMA longitudinally, carriage of the CC haplotype appears to be an important genetic factor that impacts on childhood mortality. Future longitudinal investigations aimed at determining the exact cause of death in carriers of the CC haplotype will be required to determine how inheritance of this haplotype impacts on childhood mortality.
ACKNOWLEDGMENTS
We offer our sincere gratitude and appreciation to all parents, guardians, and children from the Siaya District community for their participation in this study. We also thank the staff at University of New Mexico/KEMRI laboratories, including Anne A. Ong'ondo, Chrispine W. Ochieng, Joan L. A. Ochieng, Nicholas K. Otieno, Rodney B. Mongare, Salome A. Yala, Moses Epungure, Fabian Oduor, and Joseph Oduor, as well as the Siaya District Hospital management team, for their support during the study. These data presented are published with the permission and approval of the Director, Kenya Medical Research Institute.
The study was funded from National Institutes of Health (NIH) grant 1 R01A151305 (D.J.P.) and Fogarty International Center (FIC) training grant 1 D43TW05884 (D.J.P.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We declare that no competing interests exist.
Footnotes
Published ahead of print on 3 October 2011.
REFERENCES
- 1. Abramoff M. D., Magelhaes P. J., Ram S. J. 2004. Image processing with ImageJ. Biophotonic Int. 11:36–42 [Google Scholar]
- 2. An P., et al. 2008. Regulatory polymorphisms in the interleukin-18 promoter are associated with hepatitis C virus clearance. J. Infect. Dis. 198:1159–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arimitsu J., et al. 2006. IL-18 gene polymorphisms affect IL-18 production capability by monocytes. Biochem. Biophys. Res. Commun. 342:1413–1416 [DOI] [PubMed] [Google Scholar]
- 4. Awandare G. A., et al. 2009. MIF (macrophage migration inhibitory factor) promoter polymorphisms and susceptibility to severe malarial anemia. J. Infect. Dis. 200:629–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beier J. C., et al. 1994. Plasmodium falciparum incidence relative to entomologic inoculation rates at a site proposed for testing malaria vaccines in western Kenya. Am. J. Trop. Med. Hyg. 50:529–536 [DOI] [PubMed] [Google Scholar]
- 6. Bingle C. D., Hackett B. P., Moxley M., Longmore W., Gitlin J. D. 1995. Role of hepatocyte nuclear factor-3 alpha and hepatocyte nuclear factor-3 beta in Clara cell secretory protein gene expression in the bronchiolar epithelium. Biochem. J. 308(Part 1):197–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bloland P. B., et al. 1999. Longitudinal cohort study of the epidemiology of malaria infections in an area of intense malaria transmission II. Descriptive epidemiology of malaria infection and disease among children. Am. J. Trop. Med. Hyg. 60:641–648 [DOI] [PubMed] [Google Scholar]
- 8. Chaisavaneeyakorn S., et al. 2003. Relationship between plasma interleukin-12 (IL-12) and IL-18 levels and severe malarial anemia in an area of holoendemicity in western Kenya. Clin. Diagn. Lab. Immunol. 10:362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chomczynski P., Sacchi N. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156–159 [DOI] [PubMed] [Google Scholar]
- 10. Cox-Singh J., et al. 2008. Plasmodium knowlesi malaria in humans is widely distributed and potentially life threatening. Clin. Infect. Dis. 46:165–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dzeing-Ella A., et al. 2005. Severe falciparum malaria in Gabonese children: clinical and laboratory features. Malar. J. 4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gaunt T. R., Rodriguez S., Zapata C., Day I. N. 2006. MIDAS: software for analysis and visualisation of interallelic disequilibrium between multiallelic markers. BMC Bioinformatics 7:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Giedraitis V., He B., Huang W. X., Hillert J. 2001. Cloning and mutation analysis of the human IL-18 promoter: a possible role of polymorphisms in expression regulation. J. Neuroimmunol. 112:146–152 [DOI] [PubMed] [Google Scholar]
- 14. Hedrick P. W. 2011. Population genetics of malaria resistance in humans. Heredity 107:283–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hollegaard M. V., Bidwell J. L. 2006. Cytokine gene polymorphism in human disease: on-line databases, supplement 3. Genes Immun. 7:269–276 [DOI] [PubMed] [Google Scholar]
- 16. Huang M. C., Li K. K., Spear B. T. 2002. The mouse alpha-fetoprotein promoter is repressed in HepG2 hepatoma cells by hepatocyte nuclear factor-3 (FOXA). DNA Cell Biol. 21:561–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. International HapMap Consortium 2003. The International HapMap Project. Nature 426:789–796 [DOI] [PubMed] [Google Scholar]
- 18. Kantele A., Jokiranta T. S. 2011. Review of cases with the emerging fifth human malaria parasite, Plasmodium knowlesi. Clin. Infect. Dis. 52:1356–1362 [DOI] [PubMed] [Google Scholar]
- 19. Koch O., et al. 2005. Investigation of malaria susceptibility determinants in the IFNG/IL26/IL22 genomic region. Genes Immun. 6:312–318 [DOI] [PubMed] [Google Scholar]
- 20. Kojima S., Nagamine Y., Hayano M., Looareesuwan S., Nakanishi K. 2004. A potential role of interleukin 18 in severe falciparum malaria. Acta Trop. 89:279–284 [DOI] [PubMed] [Google Scholar]
- 21. Kwiatkowski D. P., Luoni G. 2006. Host genetic factors in resistance and susceptibility to malaria. Parassitologia 48:450–467 [PubMed] [Google Scholar]
- 22. Lee H. M., et al. 2006. Interleukin-18/-607 gene polymorphism in allergic rhinitis. Int. J. Pediatr. Otorhinolaryngol. 70:1085–1088 [DOI] [PubMed] [Google Scholar]
- 23. Liang X. H., Cheung W., Heng C. K., Wang D. Y. 2005. Reduced transcriptional activity in individuals with IL-18 gene variants detected from functional but not association study. Biochem. Biophys. Res. Commun. 338:736–741 [DOI] [PubMed] [Google Scholar]
- 24. Malaguarnera L., Pignatelli S., Musumeci M., Simpore J., Musumeci S. 2002. Plasma levels of interleukin-18 and interleukin-12 in Plasmodium falciparum malaria. Parasite Immunol. 24:489–492 [DOI] [PubMed] [Google Scholar]
- 25. Marsh K., et al. 1995. Indicators of life-threatening malaria in African children. N. Engl. J. Med. 332:1399–1404 [DOI] [PubMed] [Google Scholar]
- 26. McElroy P. D., et al. 1999. Analysis of repeated hemoglobin measures in full-term, normal birth weight Kenyan children between birth and four years of age. III. The Asemobo Bay Cohort Project. Am. J. Trop. Med. Hyg. 61:932–940 [DOI] [PubMed] [Google Scholar]
- 27. McInnes I. B., Gracie J. A., Leung B. P., Wei X. Q., Liew F. Y. 2000. Interleukin 18: a pleiotropic participant in chronic inflammation. Immunol. Today 21:312–315 [DOI] [PubMed] [Google Scholar]
- 28. Mockenhaupt F. P., et al. 2004. Manifestation and outcome of severe malaria in children in northern Ghana. Am. J. Trop. Med. Hyg. 71:167–172 [PubMed] [Google Scholar]
- 29. Nagamine Y., et al. 2003. Involvement of interleukin-18 in severe Plasmodium falciparum malaria. Trans. R. Soc. Trop. Med. Hyg. 97:236–241 [DOI] [PubMed] [Google Scholar]
- 30. Nakanishi K., Yoshimoto T., Tsutsui H., Okamura H. 2001. Interleukin-18 is a unique cytokine that stimulates both Th1 and Th2 responses depending on its cytokine milieu. Cytokine Growth Factor Rev. 12:53–72 [DOI] [PubMed] [Google Scholar]
- 31. Nakanishi K., Yoshimoto T., Tsutsui H., Okamura H. 2001. Interleukin-18 regulates both Th1 and Th2 responses. Annu. Rev. Immunol. 19:423–474 [DOI] [PubMed] [Google Scholar]
- 32. Obonyo C. O., Vulule J., Akhwale W. S., Grobbee D. E. 2007. In-hospital morbidity and mortality due to severe malarial anemia in western Kenya. Am. J. Trop. Med. Hyg. 77:23–28 [PubMed] [Google Scholar]
- 33. Okamura H., Tsutsui H., Kashiwamura S., Yoshimoto T., Nakanishi K. 1998. Interleukin-18: a novel cytokine that augments both innate and acquired immunity. Adv. Immunol. 70:281–312 [DOI] [PubMed] [Google Scholar]
- 34. Okiro E. A., Alegana V. A., Noor A. M., Snow R. W. 2010. Changing malaria intervention coverage, transmission and hospitalization in Kenya. Malar. J. 9:285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ong'echa J. M., et al. 2006. Parasitemia, anemia, and malarial anemia in infants and young children in a rural holoendemic Plasmodium falciparum transmission area. Am. J. Trop. Med. Hyg. 74:376–385 [PubMed] [Google Scholar]
- 36. Otieno R. O., et al. 2006. Increased severe anemia in HIV-1-exposed and HIV-1-positive infants and children during acute malaria. AIDS 20:275–280 [DOI] [PubMed] [Google Scholar]
- 37. Ouma C., et al. 2008. Polymorphic variability in the interleukin (IL)-1beta promoter conditions susceptibility to severe malarial anemia and functional changes in IL-1beta production. J. Infect. Dis. 198:1219–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ouma C., et al. 2008. Haplotypes of IL-10 promoter variants are associated with susceptibility to severe malarial anemia and functional changes in IL-10 production. Hum. Genet. 124:515–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shin H. D., et al. 2005. Association of interleukin 18 (IL18) polymorphisms with specific IgE levels to mite allergens among asthmatic patients. Allergy 60:900–906 [DOI] [PubMed] [Google Scholar]
- 40. Singh R. P., et al. 2002. The role of IL-18 in blood-stage immunity against murine malaria Plasmodium yoelii 265 and Plasmodium berghei ANKA. J. Immunol. 168:4674–4681 [DOI] [PubMed] [Google Scholar]
- 41. Snow R. W., et al. 1997. Relation between severe malaria morbidity in children and level of Plasmodium falciparum transmission in Africa. Lancet 349:1650–1654 [DOI] [PubMed] [Google Scholar]
- 42. Sugiura T., et al. 2006. A promoter haplotype of the interleukin-18 gene is associated with juvenile idiopathic arthritis in the Japanese population. Arthritis Res. Ther. 8:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Thompson S. R., Humphries S. E. 2007. Interleukin-18 genetics and inflammatory disease susceptibility. Genes Immun. 8:91–99 [DOI] [PubMed] [Google Scholar]
- 44. Torre D., et al. 2001. Serum levels of interleukin-18 in patients with uncomplicated Plasmodium falciparum malaria. Eur. Cytokine Netw. 12:361–364 [PubMed] [Google Scholar]
- 45. Tsutsui H., et al. 1999. Caspase-1-independent, Fas/Fas ligand-mediated IL-18 secretion from macrophages causes acute liver injury in mice. Immunity 11:359–367 [DOI] [PubMed] [Google Scholar]
- 46. Walker W., Aste-Amezaga M., Kastelein R. A., Trinchieri G., Hunter C. A. 1999. IL-18 and CD28 use distinct molecular mechanisms to enhance NK cell production of IL-12-induced IFN-gamma. J. Immunol. 162:5894–5901 [PubMed] [Google Scholar]
- 47. Were T., et al. 2011. Bacteremia in Kenyan children presenting with malaria. J. Clin. Microbiol. 49:671–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. WHO 2000. Severe falciparum malaria. World Health Organization, Communicable Diseases Cluster. Trans. R. Soc. Trop. Med. Hyg. 94(Suppl. 1):S1–S90 [PubMed] [Google Scholar]
- 49. WHO 2005. World malaria report 2005. World Health Organization/United Nations Children's Fund, Geneva, Switzerland: http://www.rollbackmalaria.org/wmr2005/pdf/WMReport_lr.pdf [Google Scholar]
- 50. WHO 2008. World malaria report 2008. World Health Organ. Tech. Rep. Ser. 1:1–80 [Google Scholar]
- 51. Wilson J. N., et al. 2005. Analysis of IL10 haplotypic associations with severe malaria. Genes Immun. 6:462–466 [DOI] [PubMed] [Google Scholar]
- 52. Zheng W., Flavell R. A. 1997. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 89:587–596 [DOI] [PubMed] [Google Scholar]
- 53. Zhou Y., Yamaguchi E., Hizawa N., Nishimura M. 2005. Roles of functional polymorphisms in the interleukin-18 gene promoter in sarcoidosis. Sarcoidosis Vasc. Diffuse Lung Dis. 22:105–113 [PubMed] [Google Scholar]
- 54. Zucker J. R., et al. 1997. Clinical signs for the recognition of children with moderate or severe anaemia in western Kenya. Bull. World Health Organ. 75(Suppl. 1):97–102 [PMC free article] [PubMed] [Google Scholar]