Abstract
Nuclear proteins usually contain specific peptide sequences, referred to as nuclear localization signals (NLSs), for nuclear import. These signals remain unexplored in the protozoan pathogen, Trichomonas vaginalis. The nuclear import of a Myb2 transcription factor was studied here using immunodetection of a hemagglutinin-tagged Myb2 overexpressed in the parasite. The tagged Myb2 was localized to the nucleus as punctate signals. With mutations of its polybasic sequences, 48KKQK51 and 61KR62, Myb2 was localized to the nucleus, but the signal was diffusive. When fused to a C-terminal non-nuclear protein, the Myb2 sequence spanning amino acid (aa) residues 48 to 143, which is embedded within the R2R3 DNA-binding domain (aa 40 to 156), was essential and sufficient for efficient nuclear import of a bacterial tetracycline repressor (TetR), and yet the transport efficiency was reduced with an additional fusion of a firefly luciferase to TetR, while classical NLSs from the simian virus 40 T-antigen had no function in this assay system. Myb2 nuclear import and DNA-binding activity were substantially perturbed with mutation of a conserved isoleucine (I74) in helix 2 to proline that altered secondary structure and ternary folding of the R2R3 domain. Disruption of DNA-binding activity alone by point mutation of a lysine residue, K51, preceding the structural domain had little effect on Myb2 nuclear localization, suggesting that nuclear translocation of Myb2, which requires an ordered structural domain, is independent of its DNA binding activity. These findings provide useful information for testing whether myriad Mybs in the parasite use a common module to regulate nuclear import.
INTRODUCTION
Trichomonas vaginalis is a protozoan pathogen that causes the most widespread sexually transmitted disease of nonviral origin, especially affecting reproductive age females (9, 43). Although the symptoms are usually mild or asymptomatic and easy to cure, the infection can be an imminent threat to public health since it is a major risk factor in promoting cervical cancer and transmission of widespread human immunodeficiency virus and because of the emergence of drug-resistant clinical isolates (9, 39). Unlike most other parasites with multistage-life cycle, T. vaginalis trophozoites persistently colonize human urogenital tract without a dormant stage. Cytoadherence of the trophozoites to human vaginal epithelial cells is critical for successful infection and survival of the pathogen.
Iron is a major environmental factor that modulates virulence expression of the parasite. Two transcription activators, Myb2 and Myb3, and a Myb1 repressor were demonstrated to coregulate inducible transcription of the ap65-1 gene (15, 34, 35, 44), which encodes a 65-kDa malic enzyme that may also serve as an adhesion protein under iron-replete conditions (21, 31). The parasite genome encodes a super family of Myb proteins comprising >400 members (3), most of which share a conserved R2R3 DNA-binding domain (DBD), which comprises six α-helices spanning ∼100 amino acid (aa) residues (19, 28, 33), and highly variable N and C termini. It is conceivable that they are major regulators of gene transcription in T. vaginalis. To better understand transcription regulation of the parasite, it is desirable to know how the functions of myriad Myb proteins are regulated.
Nuclear import of transcription factors is a crucial checkpoint in regulating transcription in eukaryotic cells (2, 18, 45). Protein nuclear import usually requires specific entities, referred to as nuclear localization signals (NLSs), in cargo proteins or their interacting partners to be transported by the proteins in the β-karyopherin family (4). A small GTPase, Ran, which alternates between the GDP- and GTP-bound forms through various cofactors, is crucial in the regulation of protein nuclear translocation (40, 42). When fused to non-nuclear proteins, an NLS is both sufficient and essential for nuclear import of the fusion proteins. Most known NLSs are diverse in the peptide sequences and may contain structure elements, with the exceptions of those referred to as classical NLSs (cNLSs), which comprises a loose consensus polybasic sequence of K(K/R)X(K/R) (14, 22), and those referred to as PY-NLS, which comprises some ∼30 aa with a central motif rich in hydrophobic or basic amino acids and a C-terminal R/H/KX(2-5)PY in an overall basic context (25). Some cNLSs are bipartite with an additional dibasic sequence located 9 to 30 aa residues away (23, 46).
Our knowledge of protein nuclear import in T. vaginalis is limited. To date, only a few nuclear proteins have been studied in this parasite (15, 24, 27, 34, 35, 38). The molecular masses of these nuclear proteins are <40 kDa, the sizes of which may allow free diffusion across the nuclear pores (42). Since they are usually enriched in the nucleus, it is also likely that their nuclear import involves active transportation. In the present study, a highly organized structure with the sequence (aa 48 to 143) embedded in the DBD (aa 40 to 156), which comprises six α-helices and a short stretch of N- and C-terminal random coils to retain full DNA-binding activity (19), was shown to mediate nuclear localization of Myb2, while the polybasic cNLS-like sequence, 48KKQK51, along with its downstream dibasic sequence, 61KR62, were dispensable in this cellular process. Intriguingly, the cNLS from the simian virus 40 (SV40) T antigen (20) did not mediate protein nuclear import in the parasite when fused to non-nuclear proteins. Site-directed mutagenesis showed that point mutation of a lysine residue, K51, in the nonstructural domain of Myb2 that disrupted DNA-binding activity did not affect its nuclear localization, while point mutation of an isoleucine, I74, in the helix 2 of the DBD that slightly altered secondary helical contents and ternary folding perturbed both DNA-binding and nuclear localization of Myb2. These observations suggest that Myb2 relies on an ordered structure embedded in its DBD for nuclear import, and this cellular process is independent of the DNA-binding activity of the protein.
MATERIALS AND METHODS
Cultures.
T. vaginalis T1 cells and derived transfectants were maintained as previously described (44).
DNA transfection and selection of stable transfectants.
Plasmids were electroporated into T. vaginalis and paromomycin selection of stable transfectants were as previously described (44). Subcellular localization of overexpressed proteins in transfected cells were examined 2 weeks after drug selection of stable transfectants, unless otherwise specified in the text.
Construction of expression plasmids.
Sequences of the oligonucleotides used in the present study are listed in Table S1 in the supplemental material or were reported in previous studies (15, 35). The plasmid, pAP65-2.1-ha-myb2/TUBneo, which can overexpress Myb2 with a single copy of an N-terminal hemagglutinin (HA) tag was obtained from a previous study (35). To increase the detection sensitivity of HA-Myb2 in T. vaginalis, a modified vector, pAP65-2.1-4ha-myb2/TUBneo, which can overexpress Myb2 with four copies of the HA tag, was constructed by a two-step PCR as described below.
In the first-round PCR, the 5′-fragment was amplified from pAP65-2.1-ha-myb2/TUBneo using the primer pair ap65-2.2-sac2-5′ and ap65-2.2-3ha-bgl2-3′ (15), whereas the 3′-fragment was amplified using the primer pair bgl2-ha-myb2-5′ and SP6. The reaction products were gel purified, mixed, and denatured at 100°C for 5 min and slowly annealed at room temperature for the second-round PCR using the primer pair ap65-2.2-sac2-5′ and SP6. The product was cloned into a TA cloning vector, pGEM-T (Promega). The insert restricted by SacII/NsiI was cloned into the SacII/NsiI-restricted pAP65-2.1-ha-myb2/TUBneo backbone to generate pAP65-2.1-4ha-myb2/TUBneo. To delete the N terminus of Myb2 to a defined site, the PCR product amplified from pAP65-2.1-4ha-myb2/TUBneo using the forward primer bamh1-myb2(48)-5′ or bamh1-myb2(55)-5′ and the reverse primer, SP, were cloned into pGEM-T. The insert restricted by BamHI/NsiI was cloned into the BglII/NsiI-restricted pAP65-2.1-4ha-myb2/TUBneo backbone to generate ΔN48 or ΔN55, respectively.
A DNA fragment was amplified from pTv94Tet-Neo (36), which can overexpress TetR in T. vaginalis, using the primer pair not1-SV40-tetR-5′ and neo-3′ by PCR. The product was digested by NotI and SalI, and then cloned into NotI/SalI restricted pTv94TetNeo to generate the plasmid pcNLS-tetR, which can overexpress cNLS-TetR, in which the cNLS is derived from the cNLS sequence of SV40 T-antigen (20).
The two-step PCR as described above was also used to construct the plasmid, p48∼143-tetR, which can overexpresses the 48∼143-TetR fusion protein in T. vaginalis. First, the 5′-fragment was amplified from pAP65-2.1-ha-myb2/TUBneo by using the primer pair not1-myb2(48)-5′ and myb2(143)-tetR-3′, whereas the 3′-fragment was amplified by using the primer pair myb2(143)-tetR-5′ and neo-3′. The reaction products were gel purified, mixed, denatured at 100°C for 5 min, and slowly annealed at room temperature for the second-round PCR using the primer pair, not1-myb2(48)-5′ and neo-3′. The product was cloned into a TA cloning vector, pGEM-T (Promega,). The insert restricted by NotI/SalI was cloned into the NotI/SalI-restricted pTv94Tet-Neo to generate p48∼143-tetR.
To fuse a firefly luciferase (Luc) C terminus to TetR, a luc DNA fragment was amplified from pAPluc+ (44) by using the primer pair xho1-luc-5′ and SP6. The DNA fragment was digested by XhoI/NsiI and cloned into the XhoI/NsiI-restricted p48∼143-tetR to generate p48∼143-tetR-luc, which produces 48∼143TetR-Luc fusion protein in the parasite. A two-step PCR was exploited to construct an expression plasmid, pcNLS-tetR-luc, which overexpresses HA-tagged cNLS-TetR-Luc. To do this, a 3′ DNA fragment was amplified from p48∼143-tetR-luc by using the primer pair bamh1-SV40-tetR-5′ and SP6, and a 5′ fragment was amplified by using the primer pair tub90f and bgl2-SV40-ha-3′. These two DNA fragments were denatured and annealed for PCR amplification using the primer pair tub90f and SP6. The PCR product was digested by SacII and NsiI and subcloned into SacII/NsiI-restricted p48∼143-tetR-luc to generate pcNLS-tetR-luc. pcNLS-tetR-luc was then digested by BglII and BamHI. The DNA backbone was ligated to generate ptetR-luc, which produces HA-tagged TetR-Luc fusion protein in the parasite.
Site-directed mutagenesis of selected amino acids in Myb2 was performed by using a two-step PCR strategy. In brief, a 5′-fragment was amplified from pAP65-2.1-4ha-myb2/TUBneo using the primer pair tub90f (35) and myb2(XnX′)-3′ (“X” is the amino acid to be mutated, “n” is the numeric location of the residue, and “X′” is the mutated counterpart, for example, I74A or I74P as shown in Table S1 in the supplemental material), whereas a 3′-fragment was amplified by using the primer pair myb2(XnX′)-5′ and SP6. The PCR products were gel purified, mixed, denatured, and annealed as described above for the second-round PCR using the primer pair tub-90f and SP6. The PCR product was cloned into pGEM-T. The insert restricted by SacII/NsiI was cloned into SacII/NsiI-restricted pAP65-2.1-4ha-myb2/TUBneo backbone. When the amino acid, I144 or T133, was mutated to a stop codon (ochre), the C-terminal deletion mutant, ΔC143 or ΔC132, was produced.
For clustered mutations in the polybasic sequences, the plasmid, pKK48∼49AA, which can overexpress the mutant protein KK48∼49AA was constructed by using the primer pair tub90f and myb2(KK48∼49AA)-3′ and the primer pair myb2(KK48∼49AA)-5′ and SP6, respectively, to clone the 5′- and 3′-fragments from the template pAP65-2.1-4ha-myb2/TUBneo. The plasmid, pKK48∼49AA/KR61∼62AA, which can overexpress KK48∼49AA/KR61∼62AA mutant protein, was constructed by using the primer pair tub90f and myb2(KR61∼62AA)-3′ and the primer pair myb2(KR61∼62AA)-5′ and SP6, respectively, to clone the 5′- and 3′-fragments from the template pKK48∼49AA. In each case, the second step PCR was performed as described above.
To produce the recombinant protein, the sequence spanning aa 48 to 148 of Myb2 was amplified from pAP65-2.1-ha-myb2/TUBneo by PCR using the primer pair sac1-myb2(48)-5′ and myb2(148)-bamh1-3′ and cloned into pGEM-T. The insert restricted by SacI/BamHI was cloned into SacI/BamHI-restricted pET6H (a gift from Tai-Huang Huang, Institute of Biomedical Sciences, Academia Sinica, Taiwan), which was modified from pET11d (Novagen) by the insertion of six histidines directly downstream of the first methionine in the coding sequence to produce pET6H/Myb2(48∼148). Site-directed mutagenesis using two-step PCR as described above and primer pairs listed in Table S1 in the supplemental material was performed to mutate the I74 isoleucine to a proline or alanine to produce pET6H/Myb2-I74P(48∼148) or pET6H/Myb2-I74A(48∼148). The plasmids that encode rMyb2(40∼156) and the related mutant protein, rK51A(40∼156), were obtained from our previous study (19).
IFA.
Subcellular localization of the target proteins was performed by an immunofluorescence assay (IFA) as previously described (34). For the detection of HA-tagged proteins, a mouse monoclonal anti-HA antibody (400×, HA-7; Sigma) was used. For the detection of TetR or its fusion proteins, a mouse monoclonal anti-TetR antibody (200×, TET03; Molecular Biologische Technologie) was used.
Western blotting.
The cytoplasmic and nuclear fractions of T. vaginalis lysate were prepared using the cellular fractionation kit, NE-PER, as described by the supplier (Pierce). In a Western blot assay, protein samples from total cell lysates or cytosolic/nuclear fractions equivalent to 5 × 105 or 106 cells, respectively, were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride membranes, Immobilon-P (Millipore), by a semidry electroblotter. Sequential immunoreactions were performed, and an enhanced chemiluminescence system was used for signal detection as instructed by the supplier (Pierce). Reaction conditions for the antibodies from commercial sources, including mouse monoclonal anti-TetR antibody (1,000×, TET03; Molecular Biologische Technologie), rabbit anti-acetyl-histone H3(Lys9) (3,000×; Upstate), and rat monoclonal anti-HA antibody (2,000×, 3F10; Roche), were as described by the supplier. The mouse anti-Cyp19 antibody (2,000×) was raised against recombinant protein derived from a 19-kDa cytosolic cyclophilin, TvCypA1 (gene accession number 123503967) from T. vaginalis (unpublished data).
Expression and purification of recombinant proteins.
pET6H/Myb2(48∼148), pET6H/Myb2-I74P, and pET6H/Myb2-I74A were each transformed into the Escherichia coli BL21-Codon Plus DE3-RIL strain (Stratagene). Transformed E. coli was incubated in shaking cultures at 37°C until the optical density at 600 nm reached 0.6. Protein induction was performed with the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 2 h. Recombinant proteins were each purified by using a His-binding nickel column as described by the supplier (Novagen).
EMSA.
Probe labeling and an electrophoretic mobility shift assay (EMSA) for testing DNA-binding activity of proteins were performed as previously described (44).
CD spectroscopy.
All circular dichroism (CD) spectra were measured using an Aviv CD 202 spectrometer (Aviv) calibrated with (+)-10-camphorsulfonic acid. The spectra were acquired at 25°C with ∼15 μM protein samples in phosphate-buffered saline (PBS; pH 7.4) with 1 mM dithiothreitol (DTT), and signals in the far-UV region (195 to 260 nm) were recorded three times with a wavelength step of 0.5 nm and a bandwidth of 1 mm. All average signals were converted from CD signals (millidegree) into a mean residue ellipticity after subtracting background signals. The fraction of the secondary structure in each CD spectrum was calculated using the reference set, SMP56, by the CONTINLL program packaged in CDPro software (http://lamar.colostate.edu/∼sreeram/CDPro).
Fluorescence spectroscopy.
Fluorescence emission spectra were recorded in a Fluorolog-3 spectrofluorimeter (Horiba; Jobin Yvon). Measurements were performed at 300 to 400 nm with 0.1-nm increments by using a 4-by-10-mm quartz cell at 20°C. The excitation wavelength was set to 280 nm with both the excitation and the emission slit widths set to 5 nm. The concentration of each target protein was 1 μM in PBS containing 1 mM DTT with or without addition of 6 M guanidine hydrochloride. Spectra were baseline corrected by subtracting the blank spectrum of the corresponding solution in the absence of protein.
RESULTS
Nuclear localization of Myb2.
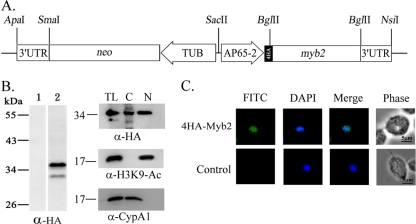
To study nuclear translocation of Myb2 in T. vaginalis by an IFA, an expression plasmid, pAP65-2-4ha-myb2/TUBneo (Fig. 1A), which encodes Myb2 fused to four copies of a HA tag at the N terminus, was constructed. The plasmid was transfected into T. vaginalis, and cells overexpressing 4HA-Myb2 were selected. 4HA-Myb2 was detected as a major 35-kDa band and a minor faster-migrating band in total cell lysate by Western blotting with a rat anti-HA antibody in samples only from transfected cells (Fig. 1B, left panel). This expression profile is similar to that of HA-Myb2 in our previous report (35), suggesting that the minor band is likely a degradation product of the major one. Lysates from transfected cells were then fractionated for Western blotting. 4HA-Myb2 was detected using the anti-HA antibody to a level substantially higher in the nuclear than cytosolic fractions (Fig. 1B, right panel), whereas an ∼18-kDa acetyl-histone H3(Lys9) was only detected in the nuclear fractions and an ∼19-kDa cyclophilin, CypA1, was only detected in the cytosolic fractions using respective antibodies. The purity of the cytosolic and nuclear fractions as described in subsequent experiments was examined in the same way and will not be further discussed in the text. 4HA-Myb2 was visualized as punctate signals in the nucleus of nearly all transfected cells by IFA with a mouse anti-HA antibody (Fig. 1C). The signal remained in the nucleus over ∼44 h of growth (data not shown). No signal was detected in nontransfected cells.
Fig. 1.
Overexpression of 4HA-Myb2 in Trichomonas vaginalis. (A) In pAP65-2-4ha-myb2, an ap65-2 promoter drives the expression of the myb2 gene fused to four copies of a HA tag, and the β-tubulin (TUB) promoter drives the expression of the neo gene. (B) Total lysates of nontransfected control (lane 1) and transfected cells (lane 2) were separated in 12% gel for Western blotting with a rat anti-HA antibody (left panel). Total cell lysates (TL) were then fractionated into the nuclear (N) and cytosolic (C) fractions. Protein samples were separated in 12% gel for Western blotting with a rat anti-HA antibody (right panel). Duplicate blots were examined by the anti-acetyl-histone H3(Lys9) (α-H3K9-Ac) and anti-CypA1 (α-CypA1) antibodies. (C) 4HA-Myb2 in transfected cells (top row) and nontransfected control (bottom row) was detected by IFA with a mouse anti-HA-antibody. The signal was shown as green fluorescence (FITC). The nucleus was stained with DAPI, and cell morphology was recorded by phase-contrast microscopy. Bar, 5 μm.
Mapping the NLS in Myb2.
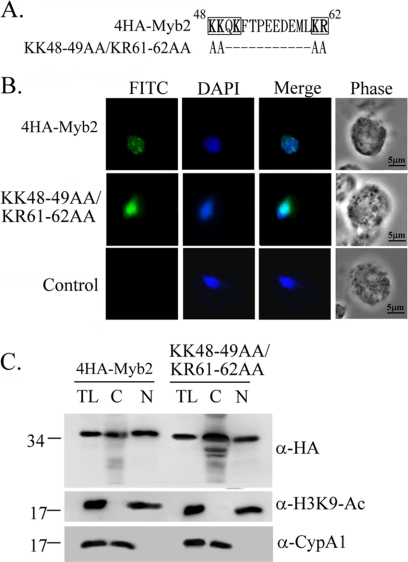
Myb2 sequence contains a cNLS-like polybasic sequence, 48KKQK51, and a downstream dibasic sequence 61KR62 (Fig. 2A). Clustered mutations were introduced into these sequences to test whether they may participate in nuclear translocation of Myb2. When overexpressed, the signal of KK48∼49AA/KR61∼62AA was largely diffusive in the nucleus of transfected cells in contrast to the punctate signal of 4HA-Myb2. The mutant protein, but not 4HA-Myb2, was also detected as punctate signal in the cytoplasm although the signal was weak. When examined by Western blotting (Fig. 2C), 4HA-Myb2 and the mutant protein were detected to similar levels in total lysates. 4HA-Myb2 was consistently detected to a higher level in the nuclear than cytosolic fractions, while a higher amount of the mutant protein was detected in the cytosolic than in the nuclear fractions. These results suggest that the polybasic sequences, 48KKQK51 and 61KR62, may play certain roles in Myb2 nuclear translocation and subnuclear localization, and yet they are not essential for nuclear import of Myb2.
Fig. 2.
Contribution of the polybasic sequences in nuclear localization of Myb2. (A) The sequence spanning aa 48 to 62 of Myb2 contains two polybasic clusters, as shown in boldface. The polybasic residues 48KK49 and 61KR62 in 4HA-Myb2 were mutated to polyalanines as shown. Hyphens (−) indicate unchanged amino acid sequences. (B) Subcellular localization of 4HA-Myb2 and KK48-49AA/KR61-62AA was detected by IFA using a mouse anti-HA-antibody. The signal was shown as green fluorescence (FITC). The nucleus was stained with DAPI, and cell morphology was recorded by phase-contrast microscopy. Bar, 5 μm. (C) Total lysates (TL) from cells overexpressing 4HA-Myb2 and KK48-49AA/KR61-62AA were fractionated into nuclear (N) and cytosolic (C) fractions. Protein samples were separated in 12% gel for Western blotting with a rat anti-HA antibody. Duplicate blots were examined by the anti-acetyl-histone H3(Lys9) (α-H3K9-Ac) and anti-CypA1 (α-CypA1) antibodies.
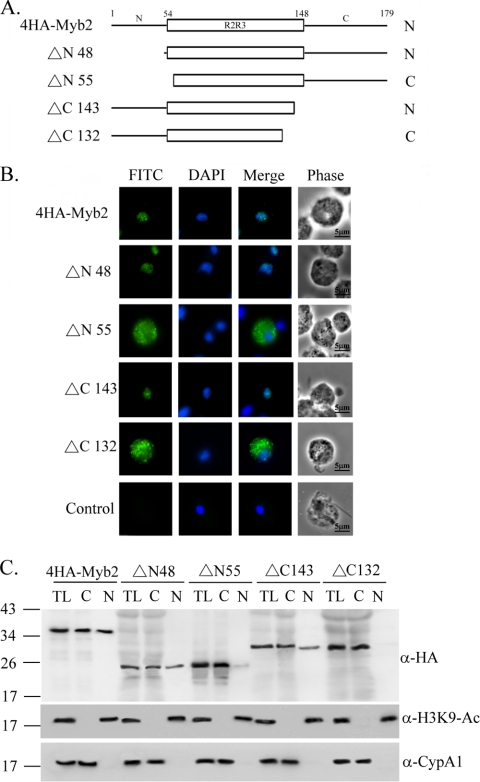
Serial deletions were performed from the N or C terminus of 4HA-Myb2 to delineate the region required for its nuclear localization (Fig. 3). When overexpressed in T. vaginalis, the mutant protein ΔN48, with aa 1 to 47 in Myb2 deleted, and ΔC143, with aa 144 to 179 deleted, were each detected as punctate signals and localized to the nucleus like 4HA-Myb2. In contrast, ΔN55, with aa 1 to 54 deleted, and ΔC132, with aa 133 to 179 deleted, were each localized mostly to the cytoplasm as punctate signals. When examined by Western blotting (Fig. 3C), 4HA-Myb2 was detected in both cytosolic and nuclear fractions to a similar level, whereas ΔN48 and ΔC143 were detected to a level slightly higher in the cytosolic than nuclear fractions. ΔN55 was detected primarily in the cytosolic fractions, with only a minor amount in the nuclear fractions, while ΔC132 was only detected in the cytosolic fractions. These observations suggest that the flanking sequences in the regions from aa 48 to 54 and aa 133 to 143 are each essential for protein nuclear import.
Fig. 3.
Mapping the nuclear localization domain of Myb2. The sequence of Myb2 is depicted as the N terminus, R2R3 DNA-binding domain, and C terminus. Series deletions from the N and C termini of Myb2 were performed, and the mutant proteins are referred to as ΔN or ΔC, respectively, with a number to show the deletion boundary. (B) Subcellular localization of 4HA-Myb2, Δ48, Δ55, Δ143, and Δ132 was detected by IFA with a mouse anti-HA antibody. The signal was shown as green fluorescence (FITC). The nucleus was stained with DAPI, and cell morphology was recorded by phase-contrast microscopy. Bar, 5 μm. (C) Total lysates (TL) from cells overexpressing 4HA-Myb2 and Δ48, Δ55, Δ143, and Δ132 were fractionated into the nuclear (N) and cytosolic (C) fractions. Protein samples were separated in 12% gel for Western blotting using a rat anti-HA antibody. Duplicate blots were examined by the anti-acetyl-histone H3(Lys9) (α-H3K9-Ac) and anti-CypA1 (α-CypA1) antibodies.
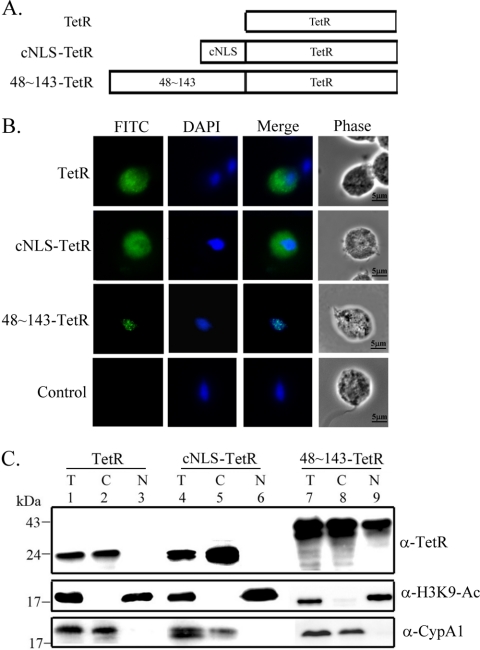
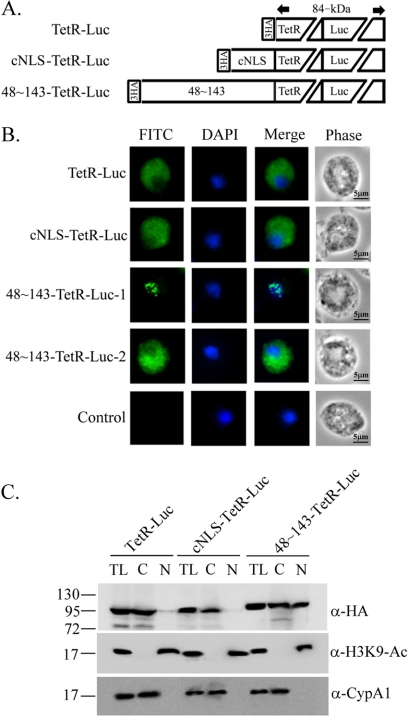
To test whether aa 48 to 143, which overlaps with a highly ordered structure in the R2R3 DNA-binding domain (aa 54 to 156) of Myb2 (19), is sufficient for the nuclear import of a non-nuclear protein, a bacterial tetracycline repressor (TetR) was fused to the C terminus of aa 48 to 143 (Fig. 4A). When overexpressed in T. vaginalis, the fusion protein, 48∼143-TetR, was detected in the nucleus as punctate signal by IFA using the anti-TetR antibody, but TetR alone was detected as diffusive signal only in the cytoplasm (Fig. 4B). When TetR was fused to an N-terminal cNLS from the SV40 T-antigen, the fusion protein, cNLS-TetR, was detected only in the cytoplasm. When examined by the Western blotting with the mouse anti-TetR antibody, 48∼143-TetR was detected as an ∼45-kDa protein with a higher amount in the nuclear than cytosolic fractions, while TetR and cNLS-TetR were detected only in the cytosolic fractions (Fig. 4C). These results suggest that aa 48 to 143, but not the cNLS from SV40, is sufficient for the nuclear import of a non-nuclear protein in T. vaginalis.
Fig. 4.
Region in Myb2 sufficient for nuclear import of a TetR fusion protein. (A) The region spanning aa 48 to 143 of Myb2 or the cNLS from the SV40-T antigen was fused with a C-terminal bacterial tetracycline repressor, TetR. (B) Subcellular localization of TetR, cNLS-TetR, and 48∼143-TeR was examined by IFA with the anti-TetR antibody. The signal was shown as green fluorescence (FITC). The nucleus was stained with DAPI, and cell morphology was recorded by phase-contrast microscopy. Bar, 5 μm. (C) Total lysates (T) from cells overexpressing TetR, cNLS-TetR, and 48∼143-TeR were fractionated into the nuclear (N) and cytosolic (C) fractions. Protein samples were separated in 12% gel for Western blotting using the anti-TetR antibody. Duplicate blots were examined by the anti-acetyl-histone H3(Lys9) (α-H3K9-Ac) and anti-CypA1 (α-CypA1) antibodies.
The molecular mass of 48∼143-TetR is ∼40.5 kDa, which is at the margin for diffusion of macromolecules through nuclear pores, raising a concern that it may be passively diffused into the nucleus and retained there through binding to chromosome DNA. To address the first issue, an ∼60-kDa firefly luciferase (Luc) was fused to the N-terminal 48∼143-TetR, cNLS-TetR, or TetR alone to create a larger fusion protein, 48∼143-TetR-Luc (∼100 kDa), cNLS-TetR-Luc (∼90 kDa), or TetR-Luc (∼89 kDa), respectively (Fig. 5A). In the stable transfectants, 48∼143-TetR-Luc was localized to either nucleus or cytoplasm to similar amount of transfected cells by IFA. Intriguingly, the signal in the nucleus was much weaker than that in the cytoplasm (data not shown). Subcellular localization of these proteins was again examined by the IFA ∼16 h posttransfection (Fig. 5B). Among 264 cells examined in two separate experiments, 48∼143-TetR-Luc was localized to the nucleus of 144 cells (55%) and to the cytoplasm of 120 cells (45%). The ratio of the nuclear and cytoplasmic signals was consistent in these two experiments, with transfection efficiency varied between 18 and 33%. In contrast, TetR-Luc and cNLS-TetR-Luc were localized to the cytoplasm of all transfected cells examined. By 2 days posttransfection, the signal intensity of 48∼143-TetR-Luc in the nucleus was substantially weaker than that in the cytoplasm, but the proportion of these two cell populations changed little. When examined by Western blotting (Fig. 5C), the ∼100-kDa 48∼143-TetR-Luc was detected in both the cytosolic and the nuclear fractions to a similar level, while the ∼90-kDa cNLS-TetR-Luc was detected only in the cytosolic fractions. The ∼89-kDa TetR-Luc was mostly detected in the cytosolic fractions, with a very faint band seen in the nuclear fractions, likely through a minor contamination from the cytosolic fractions. These results suggest that aa 48 to 143 is sufficient for the nuclear transport of 48∼143-TetR-Luc, but with a reduced efficiency compared to that of 48∼143-TetR.
Fig. 5.
Region in Myb2 sufficient for nuclear import of a TetR-Luc fusion protein. (A) Firefly luciferase (Luc) was fused to the C terminus of the HA-TetR, cNLS-TetR, or 48∼143-TetR, as depicted. (B) Subcellular localization of HA-TetR-Luc, cNLS-TetR-Luc, and 48∼143-TetR-Luc was examined by IFA with a mouse anti-HA antibody. The signal was shown as green fluorescence (FITC). The nucleus was stained with DAPI, and cell morphology was recorded by phase-contrast microscopy. Bar, 5 μm. (C) Total lysates (T) from cells overexpressing TetR-Luc, cNLS-TetR-Luc, and 48∼143TeR-Luc were fractionated into the nuclear (N) and cytosolic (C) fractions. Protein samples were separated by SDS-PAGE in 10% gel for Western blotting with a rat anti-HA antibody. Duplicate blots were examined by the anti-acetyl-histone H3(Lys9) (α-H3K9-Ac) and anti-CypA1 (α-CypA1) antibodies.
Structure integrity of Myb2 nuclear localization domain.
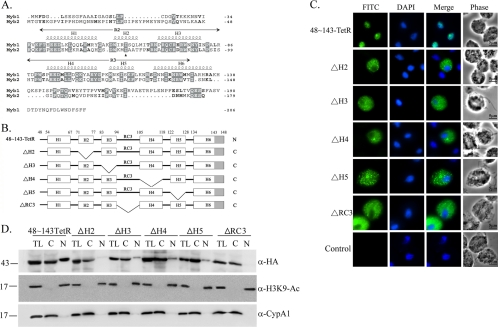
Secondary structure of Myb2 R2R3 domain comprises six α-helices (19), referred to as H1 (aa 54 to 67), H2 (aa 71 to 77), H3 (aa 83 to 94), H4 (aa 105 to 118), H5 (aa 122 to 128), and H6 (aa 134 to 148) (19, 28) (Fig. 6A). To study minimal sequence requirement for Myb2 nuclear import, individual internal α-helices (referred to as ΔH2∼ΔH5) or the random coil, RC3, connecting H3 and H4 (referred to as ΔRC3) from 48∼143-TetR were deleted (Fig. 6B). When overexpressed in T. vaginalis, ΔH2, ΔH3, ΔH4, ΔH5, and ΔRC3 were all localized to the cytoplasm (Fig. 6C). When examined by Western blotting (Fig. 6D), 48∼143-TetR was detected in the cytosolic and nuclear fractions to similar extents. ΔH2 and ΔRC3 were each detected only in the cytosolic fractions, whereas ΔH3, ΔH4, and ΔH5 were each detected primarily in the cytosolic fractions, with a minor amount in the nuclear fractions. These observations suggest that overall structure of Myb2 may play a critical role in its nuclear import.
Fig. 6.
Internal deletion to define the essential regions for Myb2 nuclear import. (A) Sequence alignments of Myb1 (AY948338) and Myb2 (AY948337). Secondary structures of aa 40 to 156 of Myb2 and the R2R3 domain of Myb2 are shown on the top of the amino acid sequence. Respective helices with numbers defining their boundaries are indicated as H1 to H6. A random coil connecting H3 and H4 is referred to as RC3. (B) The internal regions in 48∼143TetR were individually deleted as depicted. The gray-shaded area indicates the deleted region of helix 6. C and N indicate the cytoplasmic and nuclear signals, respectively, as examined below. (C) Subcellular localization of 48∼143-TetR, ΔH2, ΔH3, ΔH4, ΔH5, and ΔRC3 was detected by IFA with a mouse anti-HA antibody. The signal was shown as green fluorescence (FITC). The nucleus was stained with DAPI, and cell morphology was recorded by phase-contrast microscopy. Bar, 5 μm. (D) Total lysates (TL) from cells overexpressing 48∼143-TetR, ΔH2, ΔH3, ΔH4, ΔH5, and ΔRC3 were fractionated into the nuclear (N) and cytosolic (C) fractions. Protein samples were separated in 12% gel for Western blotting with the rat anti-HA antibody (right panel). Duplicate blots were examined by the anti-acetyl-histone H3(Lys9) (α-H3K9-Ac) and anti-CypA1 (α-CypA1) antibodies.
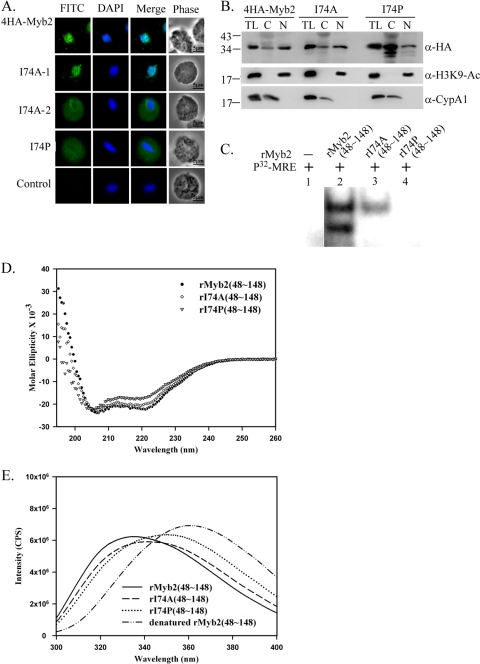
To examine this possibility, a conserved isoleucine (I74) residue in H2 was mutated to either proline (I74P) or alanine (I74A). In contrast to the nuclear localization of 4HA-Myb2, I74P was apparently localized to the cytoplasm by IFA (Fig. 7A). Among 337 cells examined in two separate experiments, I74A was localized either to the nucleus (230/337) or to the cytoplasm (107/337), the latter of which often exhibited a stronger signal in the region surrounding nuclear membrane. When examined by Western blotting (Fig. 7B), the expression profile of I74A was similar to that of 4HA-Myb2, which was detected more in the nuclear than cytosolic fractions. In contrast, overall expression of I74P as detected in total lysates was substantially higher than that of 4HA-Myb2. I74P was detected to a much higher level in the cytosolic than nuclear fractions, implying that the nuclear import of I74P is substantially perturbed.
Fig. 7.
Effects of I74 on the nuclear localization, DNA-binding activity, and structure integrity of Myb2. (A) A conserved isoleucine, I74, in 4HA-Myb2 was mutated to alanine (I74A) or proline (I74P). Subcellular localization of 4HA-Myb2, I74A, and I74P was examined by IFA with a mouse anti-HA antibody. The signal was shown as green fluorescence (FITC). The nucleus was stained with DAPI, and cell morphology was recorded by phase-contrast microscopy. Bar, 5 μm. Two distinct forms of I74A are indicated as I74A-1 and I74A-2. (B) Total lysates (TL) from cells overexpressing 4HA-Myb2, I74A, and I74P were fractionated into the nuclear (N) and cytosolic (C) fractions. Protein samples were separated in 12% gel for Western blotting with a rat anti-HA antibody (right panel). Duplicate blots were examined by the anti-acetyl-histone H3(Lys9) (α-H3K9-Ac) and anti-CypA1 (α-CypA1) antibodies. (C) I74, in the recombinant protein rMyb2(48∼148) (lane 2) was mutated to alanine or proline to produce rI74A(48∼148) (lane 3) or rI74P(48∼148) (lane 4), respectively. EMSA was performed using coincubation of the recombinant proteins with a γ-32P-labeled MRE-1 probe (lane 1). The reaction mixtures were separated in 12% gels. The signal was detected by autoradiogram. (D) The secondary structures of the recombinant proteins rMyb2(48∼148), rI74A(48∼148), and rI74P(48∼148) were monitored by far-UV CD spectra. E. The ternary folding of rMyb2(48∼148), rI74A(48∼148), and rI74P(48∼148) was examined by fluorescence spectroscopy. All fluorescence emission spectra were measured at 300 to 400 nm and 25°C. Protein samples were dissolved in PBS buffer (pH 7.4) with 1 mM DTT. rMyb2(48∼148) was also denatured with 6 M guanidine hydrochloride.
To further study physical and biochemical properties of the domain mediating nuclear localization of Myb2, the recombinant protein, rMyb2(48∼148), and the related proteins rI74P(48∼148) and rI74A(48∼148) were produced and purified to near homogeneity as examined by SDS-PAGE (data not shown). The DNA-binding activities of these recombinant proteins were examined by EMSA. When protein samples were reacted with the 32P-MRE-1 DNA probe (34), two retarded bands were detected in the reaction with rMyb2(48∼148) (Fig. 7C), as previously reported for the binding activity of full-length rMyb2 and nuclear extract (35). The binding profile was not changed with the addition of DTT in the binding reactions (data not shown), indicating that the DNA-binding activity is independent of a possible intra- or intermolecular disulfide bond formation through the conserved cysteine that may serve as a redox sensor to regulate DNA-binding activity in mammalian or plant Myb proteins (10, 11, 13). Since the DNA sequence of MRE-1 contains only a single site for Myb2 binding (35), it is likely that rMyb2(48∼148) exists as two different conformations, but one of them is lost in I74A(48∼148), to bind DNA. A retarded band with a lower signal intensity was detected in the reaction with rI74A(48∼148), indicating that this mutation resulted in partial reduction of the DNA-binding activity. The DNA-binding activity was lost in rI74P(48∼148). Since the conserved I74 is unlikely to be involved in protein-DNA interactions (19, 28, 33), these observations suggest that the abolished DNA-binding activity caused by the mutation of I74 to proline may have been due to a perturbation of Myb2's structure.
The secondary structure of the recombinant proteins was examined by circular dichroism spectroscopy (Fig. 7D). Although the far-UV spectra of rMyb2(48∼148) and rI74A(48∼148) were similar and both of them showed absorbance minima at ∼222 and 208 nm, the values of minima are still different. The absorbance minimum of rI74P(48∼148) was slightly shifted. The estimated α-helical contents for rMyb2(48∼148), rI74A(48∼148), and rI74P(48∼148) were 0.548, 0.485, and 0.408, respectively, based on the CONTINLL program by analyzing the wavelength range of 205 to 240 nm. These results suggest that the α-helical content, presumably at H2, was significantly reduced in I74P(48∼148).
When examined by fluorescence spectroscopy (Fig. 7E), the emission maximum of the denatured rMyb2(48∼148) was at 360.7 nm, a value almost identical to that of free tryptophan (360 nm), indicating that all five of the tryptophan residues in denatured rMyb2(48∼148) were exposed to the solvent. In contrast, the emission maximum for rMyb2(48∼148) was at 335.3 nm with blue shifts of about ∼25 nm relative to that of the denatured form, indicating that all tryptophans were located in the hydrophobic cores. The emission maximum of rI74A(48∼148) was 340.5 nm, which was slightly greater than that of rMyb2(48∼148), suggesting that the ternary structure of Myb2 may become looser with the mutation of I74 to alanine. For rI74P(48∼148), the emission maximum at 350.3 nm was blue-shifted 10 nm relative to that of denatured rMyb2(48∼148), implying that some of the tryptophan residues in the mutant protein were exposed to the solvent and some were still located in the hydrophobic cores, and those exposed to the solvent in the mutant protein may be important to maintain structural integrity for Myb2 nuclear localization. Together, our observations suggest that a moderate change in the structure of R2R3 drastically alters both the DNA-binding activity and the nuclear translocation of Myb2.
DNA-binding activity-independent nuclear localization.
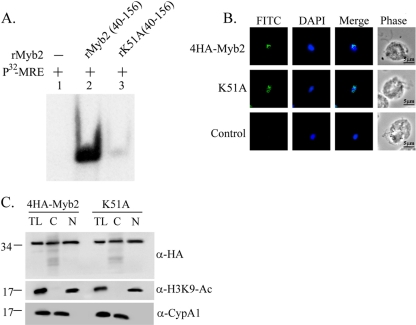
An amino acid residue, K51, which is located in the flexible random coil preceding helix 1 of the R2R3 domain away from the hydrophobic core of R2, was previously shown to be critical for the DNA-binding activity of a recombinant Myb2 protein, rMyb2(40∼156), by a isothermal titration calorimetry assay (19). To confirm this observation, rK51A(40∼156) with K51 in rMyb2(40∼156) mutated to alanine was used for EMSA. rK51A(40∼156) formed a very faint retarded product in reaction with the 32P-MRE-1 DNA in contrast to the retarded band of rMyb2(40∼156) (Fig. 8A). K51 in 4HA-Myb2 was mutated to alanine, and the mutant protein K51A was overexpressed in T. vaginalis. Similar to 4HA-Myb2, K51A was detected in the nucleus when examined by IFA (Fig. 8B). When examined by Western blotting (Fig. 8C), the expression profile of K51A was similar to that of 4HA-Myb2, with similar distributions in the cytosolic and nuclear fractions. These observations suggest that the nuclear localization of Myb2 does not rely on its DNA-binding activity.
Fig. 8.
Effect of K51 on the DNA-binding activity and nuclear localization of Myb2. (A) A lysine residue, K51, in rMyb2(40∼156) (lane 2) was mutated to alanine to produce rK51A(40∼156) (lane 3). EMSA was performed with coincubation of the recombinant proteins with a γ-32P-labeled MRE-1 probe (lane 1). The reaction mixtures were separated in 12% gel. The signal was detected by autoradiogram. (B) The lysine residue, K51, in 4HA-Myb2 was mutated to alanine. Subcellular localization of 4HA-Myb2 and K51A was examined by IFA with a mouse anti-HA antibody. The signal was shown as green fluorescence (FITC). The nucleus was stained with DAPI, and cell morphology was recorded by phase-contrast microscopy. Bar, 5 μm. (C) Total lysates (TL) from cells overexpressing 4HA-Myb2 and K51A were fractionated into the nuclear (N) and cytosolic (C) fractions. Protein samples were separated in 12% gel for Western blotting with a rat anti-HA antibody (right panel). Duplicate blots were examined by using anti-acetyl-histone H3(Lys9) (α-H3K9-Ac) and anti-CypA1 (α-CypA1) antibodies.
DISCUSSION
Protein nuclear import in T. vaginalis is relatively unexplored, without the identification of an NLS on cargo proteins. In the present study, the nuclear import of Myb2 was examined in the hope of elucidating fundamental module mediating nuclear import of myriad Myb proteins in the parasite. Toward this goal, a transgenic gene overexpression system was established to study subcellular localization and distribution of Myb2. Ideally, a microscopic assay to observe dynamic flow of a target protein fused to the green fluorescent protein (GFP) in living cells is preferable to using the IFA to detect signals in fixed cells. Unfortunately, conditions allowing this microaerophilic parasite to emit detectable and lasting signals from overexpressed GFP, which requires auto-oxidation to emit fluorescence (12), have not been established. Moreover, fusion of aa 48 to 143 with GFP resulted in the retention of the 48∼143-GFP fusion protein in the cytoplasm of ∼80% cells as examined by the IFA (unpublished observations), rendering GFP fusion unsuitable for the study of Myb2 nuclear import. We thus relied on IFA to detect Myb2 fused to a HA tag in transfected cells. The overexpressed protein, with four copies of the HA tag fused to the N terminus of Myb2, allowed unambiguous detections of Myb2 in the nucleus of nearly all transfected cells (Fig. 1C). Subcellular distribution of 4HA-Myb2 in transfected cells was similar to that of endogenous Myb2 (35), indicating that the transgenic system reflects nuclear translocation of endogenous Myb2.
Surprisingly, cNLS from SV40-T antigen when fused with a C-terminal TetR or TetR-Luc fusion protein did not serve as an NLS in T. vaginalis (Fig. 4 and 5). cNLS-mediated protein nuclear import is evolutionally conserved in eukaryotes. It was also demonstrated to exist in several parasitic protozoa, such as Trypanosome brucei, T. cruzi, and Toxoplasma gondii (1, 6, 29). In Leishmania major, an atypical NLS was identified, which has a central cNLS-like sequence and several additional basic amino acids dispersed in a context of 42 aa (6). Like many other eukaryotic systems, the cNLS from SV40-T antigen fused to a non-nuclear protein is sufficient for the nuclear import of the fusion protein in Giardia lamblia (7). In vertebrates, B-Myb and c-Myb also use a cNLS for nuclear import (5, 16, 41). However, the cNLS-like polybasic sequence, 48KKQK51, and downstream dibasic sequence, 61KR62, are dispensable for Myb2 nuclear import (Fig. 2), and the cNLS from the SV40 T-antigen did not serve as an NLS in T. vaginalis (Fig. 4 and 5), suggesting that T. vaginalis either uses a cNLS deviates from that of SV40 T-antigen or it might not possess canonical cNLS-mediated nuclear import.
Intriguingly, the protein sequence mediating nuclear localization of Myb2 was mapped to a contiguous region spanning aa 48 to 143, referred to as the nuclear localization domain (NLD), which encompasses nearly the entire DBD excluding the C-terminal half of its helix 6 and the flexible C-terminal tail (Fig. 3), suggesting that the NLD may comprise a highly structured entity distinct from that of the DBD. This NLD is sufficient for the nuclear import of a large fusion protein 48∼143-TetR-Luc (∼100 kDa) (Fig. 5). Expression of 48∼143-TetR-Luc in the nucleus, but not cytoplasm, was reduced to a relatively low level a few days after transfection, suggesting that this large fusion protein may be toxic to the parasite when overexpressed in the nucleus. The function of the NLD was largely lost with deletion of either the N-terminal random coil preceding H1 or the entire H6 of the C terminus (Fig. 3), suggesting that structural integrity of the NLD is indispensable for Myb2 nuclear import. This was supported by the observations that individual deletions of internal helices or a loop region connecting H3 in the R2 domain and H4 in the R3 domain in Myb2 that may have drastically disrupted the overall structure also resulted in the loss of function (Fig. 6). The importance of the structure elements of some NLSs has been reported in mammalian transcription factors, such as PDX-1, SREBP-2, and Sp1, which, respectively, require the structure elements H3, the dimer of helix-loop-helix, and zinc fingers (17, 30, 32). Like the NLD of Myb2, these NLSs are also embedded in their DNA-binding domains.
The structural integrity of the NLD is critical for the efficiency of nuclear translocation as revealed by the effect of point mutation of I74 in H2 on subcellular localization and distribution of overexpressed Myb2 (Fig. 7). The mutation to proline, which has the highest propensity to disrupt a helix (37), resulted in the accumulation of Myb2 in the cytoplasm as revealed by IFA (Fig. 7A) and the cytosolic fractions as revealed by Western blotting (Fig. 7B) and a more-altered helical content and ternary folding of Myb2 than the mutation to alanine (Fig. 7D and E), which has the lowest propensity to disrupt a helix. It is notable that with even a change in the structure as slight as that of I74A (Fig. 7D and E), nuclear translocation of Myb2 was substantially perturbed with an apparent accumulation of Myb2 near the nuclear membrane in a significant fraction of transfected cells (Fig. 7A), underscoring the importance of structural integrity of the NLD of Myb2. A major concern for the nuclear import of a small protein like Myb2 is its ability to freely diffuse into the nucleus and remain there through its binding to chromosomal DNA. Although structural changes due to mutation of I74 correlated with the changes in the DNA-binding activity, nuclear localization of Myb2 did not rely on its DNA-binding activity, as demonstrated by the critical role of K51 in the DNA binding, but not nuclear localization, of Myb2 (Fig. 8). Indeed, F52, K138, and N139, which are crucial for DNA-binding activity (19), had only a slight effect on the nuclear localization of Myb2 (unpublished observations). It is conceivable that the structural aspect of Myb2 NLD may only provide itself a suitable conformation to contact its interacting partner, be it a cargo protein or a karyopherin-like protein. The binding affinity must be strengthened through interactions of specific amino acid residues on the exposed surface of the NLD and its interacting import partner (4).
It is intriguing to observe different localizations of 48∼143-TetR-Luc (Fig. 5B) or I74A in two cell subpopulations (Fig. 7A) by IFA. In mammals, the tumor suppressor VHL protein was differentially localized to the cytoplasm and nucleus to different extents in various proportions of cells depending on the cell density, and the proportion of individual phenotypes may vary in the expression of different mutant proteins (26). Since 4HA-Myb2 was localized to the nucleus of individual transfected cells to similar levels for a period of 44 h, Myb2 nuclear import is unlikely regulated by its expression level or the progression of cell cycle in different cells. The phenomenon could not be explained by potential mutations of expression plasmids since no mutation was detected when the plasmids recovered from transfected cells were sequenced. However, T. vaginalis T1 isolate used in the present study is not a cloned cell line, the possibility that differential localization of the mutant proteins in two subpopulations of cells is originated from clonal variation cannot be excluded. It is also important to note that the NLD as identified herein cannot be defined as an NLS without the identification of an importin/karyopherin-like transporter that binds the NLD. It is also not clear whether Myb2 interacts with an NLS-containing nuclear protein, which binds to an importin/karyopherin, for nuclear translocation, or whether Myb2 nuclear translocation is as NLS independent and importin independent as that of the β-catenin in vertebrates (8). Our results do not exclude the possibility that a small fraction of Myb2 could enter the nucleus through diffusion since small amounts of some of the mutant proteins, such as ΔN55 (Fig. 3B versus 3C), ΔH3, ΔH4, ΔH5 (Fig. 6C versus 6D), and I74P (Fig. 7A versus 7B) that were localized to the cytoplasm by IFA were still detectable in the nuclear fractions by Western blotting.
The genome of T. vaginalis encodes ∼400-Myb proteins (3), most of which contain a conserved R2R3 DNA-binding domain that probably preserves a structural context resembling to that of the human c-Myb (19, 28, 33). The identification of NLD in Myb2 to span nearly the entire R2R3 domain infers that different Myb proteins in the parasite may share a structural module for their basal nuclear import. Although their functions are largely unknown, it is likely that members of the Myb super family are the major regulators of gene-specific transcription in T. vaginalis. It will be intriguing to test whether a similar NLD is present in Myb1 and Myb3 and see how their nuclear imports are differentially regulated to better understand gene-specific transcription regulation of the parasite.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Science Council (NSC97-2320-B-001-005-My3) and IBMS, Academia Sinica.
We thank Dan Chamberlin for correcting the manuscript and Patricia Johnson (UCLA) for providing the TetR expression plasmid, pTv94TetNeo (36).
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 21 October 2011.
REFERENCES
- 1. Bhatti M. M., Sullivan W. J., Jr 2005. Histone acetylase GCN5 enters the nucleus via importin-alpha in protozoan parasite Toxoplasma gondii. J. Biol. Chem. 280:5902–5908 [DOI] [PubMed] [Google Scholar]
- 2. Boulikas T. 1994. Putative nuclear localization signals (NLS) in protein transcription factors. J. Cell. Biochem. 55:32–58 [DOI] [PubMed] [Google Scholar]
- 3. Carlton J. M., et al. 2007. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 315:207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chook Y. M., Süel K. E. 2010. Nuclear import by karyopherin-βs: recognition and inhibition. Biochim. Biophys. Acta doi: 10.1016/j.bbamcr.2010.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dang C. V., Lee W. M. F. 1989. Nuclear and nucleolar targeting sequences of c-erb-A, c-myb, N-myc, p53, HSP70, and HIV-tat proteins. J. Biol. Chem. 264:18019–18023 [PubMed] [Google Scholar]
- 6. Dubessay P., et al. 2006. Cell cycle-dependent expression regulation by the proteasome pathway and characterization of the nuclear targeting signal of a Leishmania major Kin-13 kinesin. Mol. Microbiol. 59:1162–1174 [DOI] [PubMed] [Google Scholar]
- 7. Elemendorf H. G., Singer S. M., Nash T. E. 2000. Targeting of proteins to the nuclei of Giardia lamblia. Mol. Biochem. Parasitol. 106:315–319 [DOI] [PubMed] [Google Scholar]
- 8. Fagotto F., Glück U., Gumbiner B. M. 1998. Nuclear localization signal-independent and importin/karyopherin-independent nuclear import of β-catenin. Curr. Biol. 8:181–190 [DOI] [PubMed] [Google Scholar]
- 9. Fichorova R. N. 2009. Impact of T. vaginalis infection on innate immune responses and reproductive outcome. J. Reprod. Immunol. 83:185–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grässer F. A., La Montagne K., Whittaker L., Stohr S., Lipsick J. S. 1992. A highly conserved cysteine in the v-Myb DNA-binding domain is essential for transformation and transcriptional transactivation. Oncogene 7:1005–1009 [PubMed] [Google Scholar]
- 11. Guehmann S., Vorbrueggen G., Kalkbrenner F., Moelling K. 1992. Reduction of a conserved Cys is essential for Myb DNA-binding. Nucleic Acids Res. 20:2279–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Heim R., Prasher D. C., Tsien R. Y. 1994. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc. Natl. Acad. Sci. U. S. A. 91:12501–12504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Heine G. F., Hernandez J. M., Grotewold E. 2004. Two cysteines in plant R2R3 MYB domains participate in redox-dependent DNA binding. J. Biol. Chem. 279:37878–37885 [DOI] [PubMed] [Google Scholar]
- 14. Hodel M. R., Corbett A. H., Hodel A. E. 2001. Dissection of a nuclear localization signal. J. Biol. Chem. 276:1317–1325 [DOI] [PubMed] [Google Scholar]
- 15. Hsu H. M., Ong S. J., Lee M. C., Liu H. W., Tai J. H. 2009. Transcriptional regulation of an iron-inducible gene by differential and alternate promoter entries of multiple Myb proteins in the protozoan parasite Trichomonas vaginalis. Eukaryot. Cell 8:362–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Humbert-Lan G., Pieler T. 1999. Regulation of DNA-binding activity and nuclear transport of B-Myb in Xenopus oocyte. J. Biol. Chem. 274:10293–10300 [DOI] [PubMed] [Google Scholar]
- 17. Ito T., Azumato M., Uwatoko C., Itoh K., Kuwahara J. 2009. Role of zinc finger structure in nuclear localization of transcription factor Sp1. Biochem. Biophys. Res. Commun. 380:28–32 [DOI] [PubMed] [Google Scholar]
- 18. Jans D. A. 1995. The regulation of protein transport to the nucleus by phosphorylation. Biochem. J. 311:705–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jiang I., et al. 2011. Molecular basis of the recognition of the ap65-1 gene transcription promoter elements by a Myb protein from the protozoan parasite Trichomonas vaginalis. Nucleic Acids Res. doi: 10.1093/nar/gkr558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kalderon D., Roberts B. L., Richardson W. D., Smith A. E. 1984. A short amino acid sequence able to specify nuclear location. Cell 39:499–509 [DOI] [PubMed] [Google Scholar]
- 21. Kucknoor A. S., Mundodi V., Alderete J. F. 2005. Heterologous expression in Tritrichomonas vaginalis of functional Trichomonas vaginalis AP65 adhesin. BMC Mol. Biol. 6:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lange A., et al. 2007. Classical nuclear localization signals: definition, function, and interaction with importin α. J. Biol. Chem. 282:5101–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lange A., McLane L. M., Mills R. E., Devine S. E., Corbett A. H. 2010. Expanding the definition of the classical bipartite nuclear localization signal. Traffic 11:311–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lau A. O., Liston D. R., Vanacova S., Johnson P. J. 2003. Trichomonas vaginalis initiator binding protein, IBP39, contains a novel DNA-binding motif. Mol. Biochem. Parasitol. 130:167–171 [DOI] [PubMed] [Google Scholar]
- 25. Lee B. J., et al. 2006. Rules for nuclear localization sequence recognition by karyopherin β2. Cell 126:543–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee S., et al. 1996. Nuclear/cytoplasmic localization of the von Hippel-Lindau tumor suppressor gene product is determined by cell density. Proc. Natl. Acad. Sci. U. S. A. 93:1770–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liston D. R., Lau A. O. T., Ortiz D., Smale S. T., Johnson P. J. 2001. Initiator recognition in a primitive eukaryote: IBP39, an initiator-binding protein from Trichomonas vaginalis. Mol. Cell. Biol. 21:7872–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lou Y. C., et al. 2009. NMR structural analysis of DNA recognition by a novel Myb1 DNA-binding domain in the protozoan parasite Trichomonas vaginalis. Nucleic Acids Res. 37:2381–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marchetti M. A., Tschudi C., Kwon H., Wolin S. L., Ullu E. 2000. Import of proteins into the trypanosome nucleus and their distribution at karyokinesis. J. Cell Sci. 113:899–906 [DOI] [PubMed] [Google Scholar]
- 30. Moede T., Leibiger B., Pour G. H., Berggren P. O., Leibiger I. B. 1999. Identification of a nuclear localization signal, RRMKWKK, in the homeodomain transcription factor PDX-1. FEBS Lett. 461:229–234 [DOI] [PubMed] [Google Scholar]
- 31. Mundodi V., Kucknoor A. S., Klumpp D. J., Chang T. H., Alderete J. F. 2004. Silencing the ap65 gene reduces adherence to vaginal epithelial cells by Trichomonas vaginalis. Mol. Microbiol. 53:1099–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nagoshi E., Imamoto N., Sato R., Yoneda Y. 1999. Nuclear import of sterol regulatory element-binding protein-2, a basic helix-loop-helix-leucine zipper (bHLH-Zip)-containing transcription factor, occurs through the direct interaction of importin beta with HLH-Zip. Mol. Biol. Cell 10:2221–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ogata K., et al. 1994. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell 79:639–648 [DOI] [PubMed] [Google Scholar]
- 34. Ong S. J., Hsu H. M., Liu H. W., Chu C. H., Tai J. H. 2006. Multifarious transcriptional regulation of adhesion protein gene ap65-1 by a novel Myb1 protein in the protozoan parasite Trichomonas vaginalis. Eukaryot. Cell 5:391–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ong S. J., Hsu H. M., Liu H. W., Chu C. H., Tai J. H. 2007. Activation of multifarious transcription of an adhesion protein ap65-1 gene by a novel Myb2 protein in the protozoan parasite Trichomonas vaginalis. J. Biol. Chem. 282:6716–6725 [DOI] [PubMed] [Google Scholar]
- 36. Ortiz D., Johnson P. J. 2003. Tetracycline-inducible gene expression in Trichomonas vaginalis. Mol. Biochem. Parasitol. 128:43–49 [DOI] [PubMed] [Google Scholar]
- 37. Pace C. N., Scholtz J. M. 1998. A helix propensity scale based on experimental studies of peptides and proteins. Biophys. J. 75:422–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schumacher M. A., Lau A. O. T., Johnson P. J. 2003. Structural basis of core promoter recognition in a primitive eukaryote. Cell 115:413–424 [DOI] [PubMed] [Google Scholar]
- 39. Shafir S. C., Sorvillo F. J., Smith L. 2009. Current issues and considerations regarding trichomoniasis and human immunodeficiency virus in African-Americans, Clin. Microbiol. Rev. 22:37–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sorokin A. V., Kim E. R., Ovchinnikov L. P. 2007. Nuclear cytoplasmic transport of proteins. Biochemistry (Moscow) 72:1439–1457 [DOI] [PubMed] [Google Scholar]
- 41. Takemoto Y., Tashiro S., Handa H., Ishii S. 1994. Multiple nuclear localization signals of the B-Myb gene products. FEBS Lett. 350:55–60 [DOI] [PubMed] [Google Scholar]
- 42. Terry L. J., Shows E. B., Wente R. S. 2007. Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science 318:1412–1416 [DOI] [PubMed] [Google Scholar]
- 43. Thurman A. R., Doncel G. F. 2010. Innate immunity and inflammatory response to Trichomonas vaginalis and bacterial vaginosis: relationship to HIV acquisition. Am. J. Reprod. Immunol. 65:89–98 [DOI] [PubMed] [Google Scholar]
- 44. Tsai C. D., Liu H. W., Tai J. H. 2002. Characterization of an iron-responsive promoter in the protozoan pathogen Trichomonas vaginalis. J. Biol. Chem. 277:5153–5162 [DOI] [PubMed] [Google Scholar]
- 45. Vandromme M., Gauthier-Rouviere C., Lamb N., Fernandez A. 1996. Regulation of transcription factor localization: fine-tuning of gene expression. Trends Biochem. Sci. 21:59–64 [PubMed] [Google Scholar]
- 46. Xiao Z., Latek R., Lodish H. F. 2003. An extended bipartite nuclear localization signal in Smad4 is required for its nuclear import and transcriptional activity. Oncogene 22:1057–1069 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.