Abstract
The green alga Chlamydomonas reinhardtii today is a premier model organism for the study of green algae and plants. Yet the efficient engineering of its nuclear genome requires development of new antibiotic resistance markers. We have recoded, based on codon usage in the nuclear genome, the AadA marker that has been used previously for chloroplast transformation. The recoded AadA gene, placed under the control of the HSP70A-RBCS2 hybrid promoter and preceded by the RbcS2 chloroplast-targeting peptide, can be integrated into the nuclear genome by electroporation, conferring resistance to spectinomycin and streptomycin. Transformation efficiency is markedly increased when vector sequences are completely eliminated from the transforming DNA. Antibiotic resistance is stable for several months in the absence of selection pressure. Shuttle markers allowing selection in both Chlamydomonas and Escherichia coli would also be a useful asset. By placing an artificial bacterial promoter and Shine-Dalgarno sequence in frame within the AadA coding sequence, we generated such a shuttle marker. To our surprise, we found that the classical AphVIII construct already functions as a shuttle marker. Finally, we developed a method to introduce the AadA and AphVIII markers into the vector part of the bacterial artificial chromosomes (BACs) of the Chlamydomonas genomic DNA library. Our aim was to facilitate complementation studies whenever the test gene cannot be selected for directly. After transformation of a petC mutant with a modified BAC carrying the AphVIII marker along with the PETC gene in the insert, almost half of the paromomycin-resistant transformants obtained showed restoration of phototrophy, indicating successful integration of the unselected test gene. With AadA, cotransformation was also observed, but with a lower efficiency.
INTRODUCTION
The unicellular green alga Chlamydomonas reinhardtii is a model organism widely used in the study of flagellar motility, chloroplast biology, photosynthesis, and lipid metabolism for biodiesel production (18). Among its major advantages in laboratory work, we cite its fast growth, metabolic plasticity, and fully tractable genetic system. All three genomes (chloroplast, mitochondrial, and nuclear) have been sequenced and carefully annotated. Transformation of the chloroplast and mitochondrial genomes involves homologous recombination (4, 5), while nuclear transformation usually occurs via illegitimate recombination (24). This has allowed the development of forward and reverse genetic approaches which make Chlamydomonas a premier system for functional genomics.
Yet the number of selection markers available for genetic engineering in Chlamydomonas is relatively small compared to those for other model systems. In practice, this limits the number of genes that can be manipulated in a single strain. The work presented here stemmed from our need to engineer genes in a strain that already contained the most usual resistance markers and lacked any usable auxotrophy. We have developed a new resistance marker which we hope can be useful to the Chlamydomonas community.
Two types of transformation markers have been used for nuclear transformation in Chlamydomonas: auxotrophy markers, which complement a mutation affecting the production of an essential metabolite, and antibiotic resistance markers. In the first category, the most popular marker is ARG7, encoding argininosuccinate lyase (9). This gene has been used for complementation studies (i.e., to form vegetative diploids), insertional mutagenesis, cointegration of transgenes or RNA interference (RNAi)/miRNA constructs, etc. One of its limitations is its large size (8 kb). Use of its cDNA (2.2 kb) has been proposed (1), but this shorter version unfortunately yields fewer transformants. Other genes that have been used in transformation experiments include NIT1, encoding nitrate reductase (24), the nitrate regulatory gene NIT2 (13), and THI10 and NIC7, involved in thiamine and nicotinamide biosynthesis (12).
A common limitation to the use of these auxotrophy markers is that recipients must carry the auxotrophy mutation, which can complicate experimental strategies. In this respect, antibiotic resistance markers are easier to use. The first marker available for nuclear transformation was the Chlamydomonas CRY1 gene, which encodes the ribosomal protein S14: a dominant mutant allele isolated from an emetine-resistant strain conferred resistance to the drug (27). Similarly, a mutated ALS gene encoding a sulfometuron-methyl-resistant variant of acetolactate synthase has also been used as a dominant selectable marker (25). A high frequency of transformation was achieved using the GC-rich ble gene from Streptoalloteichus hindustanus, conferring resistance to Zeocin (36). The original construct used the RBCS2 expression signals, including an enhancer in the first intron, and it was later improved by introduction of a fragment of the HSP70A promoter upstream of the RBCS2 promoter (32). The HSP70A fragment appears to protect the construct from silencing influences, resulting in a larger proportion of strains actually expressing the gene to a sufficient level for efficient selection. Unfortunately, Zeocin is light sensitive and can cause mutations, which prompted the development of new markers, such as aphVIII (34, 35) and aph7″ (2), conferring resistance to paromomycin (Pm) and hygromycin, respectively. AphVIII is currently the most widely used marker in Chlamydomonas.
The aadA gene, isolated from the Escherichia coli plasmid R538-1 (19), codes for an aminoglycoside 3″-adenylyltransferase and confers resistance to spectinomycin (Sp) and streptomycin (Sr). This gene is widely used in chloroplast transformation, particularly for site-directed mutagenesis. In the resistance cassette generally used in Chlamydomonas (15), the coding sequence (CDS) is placed under the control of the 5′ region of atpA (promoter, 5′-untranslated region [5′UTR], and start of the coding sequence) and is followed by the rbcL 3′UTR. The aadA gene has also been used for transformation of the Chlamydomonas nuclear genome (7), but the transformation efficiency was several orders of magnitude below that of classical nuclear markers, and half of the transformants rapidly lost resistance due to silencing (4). The low efficiency of the E. coli-derived aadA cassette can perhaps be explained by its exotic codon usage, inherited from its bacterial origin: the CDS is only 40% GC rich, compared to 65% for the average Chlamydomonas nuclear gene (26), and it uses many rare codons. Following the precedent of the green fluorescent protein (GFP) gene (14), we have recoded the aadA cassette and placed it under the control of the high-efficiency HSP70A-RBCS2 (AR) promoter construct (31, 32). We show that this CrAadA construct can be used as an Sp resistance cassette in nuclear transformation. To facilitate cloning of the cassette and to allow retrieval of flanking sequences by marker rescue, it is convenient to use so-called “shuttle markers” that can be used both in E. coli and in the organism under study. This strategy has been used with Chlamydomonas, using the ARG7 cDNA (1). We therefore engineered a version of the marker (CrEcAadA) that confers resistance in both E. coli and Chlamydomonas.
We also present a new method for inserting resistance markers into bacterial artificial chromosomes (BACs). The BAC library prepared from Chlamydomonas DNA by the laboratory of P. Lefebvre (22) is extremely useful, for example, to show that a specific gene complements a mutation. Unfortunately, the BAC vector used does not contain a selection marker that is usable in Chlamydomonas transformation: the NAR2/NIT8 gene that was introduced into the vector when the library was created was later found to be inefficient as an auxotrophy marker (P. Lefebvre, personal communication). Using a suicide plasmid strategy, we introduced the AadA and AphVIII markers into the vector portion of the BAC. Upon transformation of the BAC into Chlamydomonas, a sizeable fraction of the antibiotic-resistant clones had integrated an unselected gene present in the BAC insert.
MATERIALS AND METHODS
Strains and growth conditions.
The C. reinhardtii strains used in this study were XS1 (cw15 arg7 mt+) (20) and petC-Δ1 (cw15 petC-Δ1 mt−) (10), both of which are available on request. Strains were grown at 23°C under continuous light on TAP or minimal (Min) medium (17, 18), supplemented with 100 μg/ml arginine for XS1.
Escherichia coli strain DH5α (supE44 lacU169 φ80dlacZΔM15 hsdR17 recA1 endA1 gyrA96 thi-1 relA1) was used as a host strain for plasmid construction, except for the R6K plasmids, which were built in strain S17-1 λpir (C600::RP-4 2-Tc::Mu-Km::Tn7 hsdR hsdM recA thi pro λpir). BL21 [B F− dcm ompT hsdS(rB− mB−) gal (malB+)K-12(λS)] was used as a RecA+ recipient strain for the BAC modification. All strains were grown on LB medium (30). Transformation was done by electroporation. After the electric shock, cells were shaken in 1 ml of LB medium at 37°C and 180 rpm for 2 or 3 h. For selection, 100 μg/ml of ampicillin (Ap), 50 μg/ml of spectinomycin (Sp), 2 μg/ml of paromomycin (Pm), or 25 μg/ml of chloramphenicol (Cm) was added. All plasmids and BACs were purified using a NucleoBond PC 100 kit (Macherey Nagel).
Plasmids.
pBC1 was derived from pSI103 (32) by cloning its entire insert into pBluescript KS(+) cut with XhoI and KpnI so that the AphVIII CDS was in opposite orientation to the lacZ promoter. The sequence of the recoded aadA gene was designed by Genecust (Dudelange, Luxembourg), based on Chlamydomonas nuclear codon usage, and further modified to eliminate or introduce restriction enzyme sites and to avoid unwanted polyadenylation or splicing signals. The gene was provided (by Genecust) cloned into pBluescript II SK(+) between the EcoRI and HindIII sites (plasmid pBSKS+CrAadA). To construct pALM32 (Fig. 1A), the insert was excised using AatII and cloned into AatII-restricted pMS188 (31), placing the CrAadA CDS between the AR promoter and the RBCS2 3′UTR. pALM34 was constructed by cutting pALM32 with SfiI and FspAI, recessing the 3′ overhang with T4 polymerase and religation. For pALM33 (Fig. 2A), we synthesized an artificial E. coli promoter by using the complementary oligonucleotides ProEcoli_sens and ProEcoli_antisens (see Table 1 for primer sequences) annealed to yield a double-stranded fragment that was cloned into FspAI-digested pALM32. To construct the pALM30 series of plasmids, we first modified plasmid pCVD442 (11) (a gift from J. M. Ghigo and C. Beloin, Institut Pasteur) to remove 500 bp of an IS1 repetitive element which generates unwanted chromosomal recombination (28). The plasmid was digested with NdeI and AsiSI and ligated with the bla gene, which was amplified by PCR using primers Bla_5NAA and Bla_3SS. Into this new plasmid (pCVD442modif), we cloned a 3-kbp fragment obtained by PCR from pBeloBAC11-Nit8, using primers pBeloBAC_AscI_ter and pBeloBAC_Age1_ter and the AscI and AgeI sites downstream of the bla gene. The resulting plasmid was further modified by cloning various versions of the AadA marker, amplified with Hsp_AadA_BAC5 and AadA_BAC3, into the FseI site. pAML30-Sp (see Fig. 4A) carries the shuttle marker from pALM33, while pALM30-CrSpA and pALM30-CrSpS carry the Chlamydomonas-only marker from pALM32, in antisense and sense orientation, respectively, with respect to NAR2. pALM30-Pm was generated in the same manner, except that we cloned (in sense orientation) a fragment amplified from pBC1 by use of the same primers.
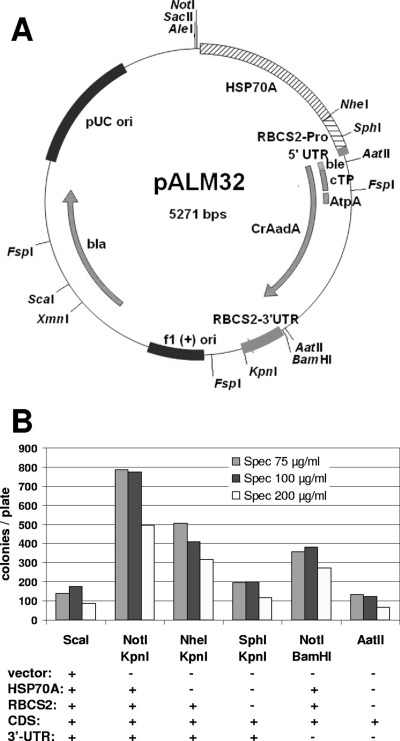
Fig. 1.
(A) Map of pALM32. (B) Transformation of strain XS1 by pALM32. The plasmid was digested by the indicated enzymes, releasing CrAadA flanked by the indicated functional elements.
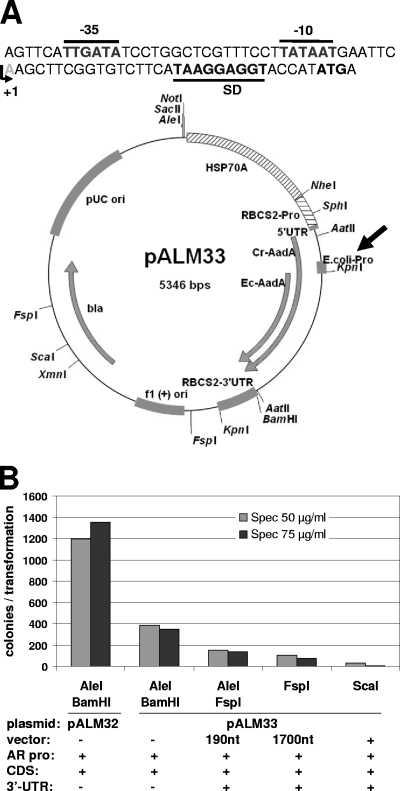
Fig. 2.
(A) Map of pALM33 and sequence of the introduced artificial promoter. The −35 and −10 boxes, Shine-Dalgarno sequence, and start codon are indicated, as well as the transcription start site. The AadA CDS expressed in Chlamydomonas and E. coli are indicated by Cr-AadA and Ec-AadA, respectively. (B) Transformation of Chlamydomonas petC-Δ1 by pALM33. The plasmid was digested by the indicated enzymes, releasing CrEcAadA flanked by the indicated functional elements.
Table 1.
Primers used in this study
| Primer | Sequence |
|---|---|
| ProEcoli_sens | AGTTCATTGATATCCTGGCTCGTTTCCTTATAATGAATTCAAGCTTCGGTGTCTTCATAAGGAGGTACCATATGA |
| ProEcoli_antisens | TCATATGGTACCTCCTTATGAAGACACCGAAGCTTGAATTCATTATAAGGAAACGAGCCAGGATATCAATGAACT |
| Bla_5NAA | GGAGCATATGGCGCGCCGGTACCGGTGCACTTTTCGGGGAAATGTGCGCGG |
| Bla_3SS | GGAGCCCGGGCGATCGCTGACGCTCAGTGGAACGAAAACTCACG |
| pBeloBAC_AscI_ter | GGAGGGCGCGCCGACGCCGAAGGAGTAGCCGTAGGTCAGGGCCAG |
| pBeloBAC_Age1_ter | CACGGTTACACACAAACCATCAGCATTGCGATTCAAC |
| Hsp_AadA_BAC5 | GGCGAATTCCGTACGGGAGCTCGCTGAGGCTTGACATG |
| AadA_BAC3 | GGCGAATTCCGTACGGGAGAAAGAGGCCAAAATCAACGGAGG |
| Hsp_CrAadA5 | CTAGAGTCGACAGCCATATCGCCGCCGCTTTGGCCACCTC |
| Hsp_CrAadA3 | AGCTGAGTGGTTATGTATAGCGGCAGAATAGTCGCGTATGTATAAGTGCT |
| CrAadA_5 | GCTTCGGTGTCTTCATAAGGAGGTACCATATGAGCACCCC |
| CrAadA_3 | ATTACTTGCCCACGACCTTCGTGATCTCGCCCTTGACGTA |
| RBCS3_5 | ACCCACTCTAGAGGATCCCCGCTCCGTGTA |
| RBCS3_3 | TGGGTACCCGCTTCAAATACGCCCAGCCCG |
Fig. 4.
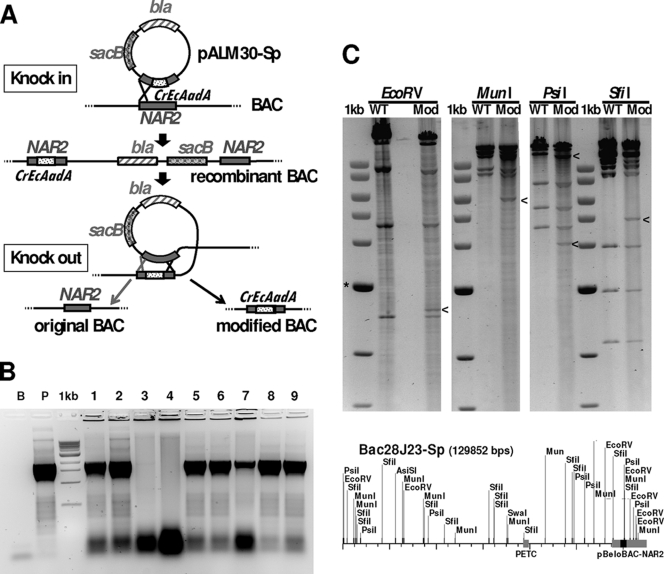
(A) Schematic representation of the two-step BAC modification system with a suicide plasmid. The Sp resistance marker (CrEcAadA) is shown as a dotted rectangle. (B) E. coli colony PCR using primers Hsp_AadA_BAC5 and AadA_BAC_3, indicating integration of CrEcAadA into the NAR2 gene of the resident BAC in 7 of 9 Cmr sucroser Aps colonies shown. Sp resistance correlated with the presence of CrEcAadA. Controls are the purified BAC 28J23 (B) and plasmid pALM30-Sp (P), shown next to the NEB 1-kb ladder. (C) Restriction digestion analysis of BAC 28J23 (WT) and the modified BAC 28J23-Sp (Mod), shown next to the NEB 1-kb ladder (*, 3-kb band). The differences, indicated by arrowheads, are as expected from the map of the modified plasmid, shown below. Note that the introduced cassette (black) contains new sites for EcoRV, MunI, PsiI, and SfiI.
Transformation of C. reinhardtii.
Nuclear transformation was performed by electroporation as described previously (33), with minor modifications. Cells were pelleted at 3,500 rpm for 4 min and resuspended in ToS (TAP plus 40 mM sucrose) at 108 cells/ml. Three hundred microliters of cells was used for each transformation. We added 4 μl (10 μg/μl) salmon sperm DNA (Sigma) and 2 μg transforming DNA. Digestion mixtures were used without further purification. Cells were incubated for 20 min at 16°C, and electroporation was performed at 10 μF and 720 V, using a Gene Pulser II and a Pulse Controller Plus apparatus (Bio-Rad). After electroporation, cells were incubated for 20 min at 16°C and plated on TAP plus arginine containing Ap to prevent bacterial infection and also containing the selection antibiotic. Plating was performed immediately for Sp selection, while for Pm selection, the cells were first diluted in 20 ml ToS in Falcon tubes and incubated for 3 h in a rotating wheel. For plating, cells were mixed with a 20% starch suspension. Plates were placed under illumination at 7 microeinsteins m−2 s−1, and transformants were counted after 15 days.
DNA analysis of C. reinhardtii transformants.
Southern blotting was performed following migration of 2 μg PstI-digested genomic DNA in 0.7% agarose-Tris-acetate-EDTA (TAE) gels and transfer to Hybond N+ nylon membranes (GE Healthcare) (30). DNA probes were amplified from pALM33 by PCRs using Hsp_CrAadA5 and Hsp_CrAadA3 (for HSP70), CrAadA_5 and CrAadA_3 (for CrAadA CDS), and RBCS3_5 and RBCS3_3 (for RBCS2 3′UTR). Probes were labeled with [33P]dATP (Perkin Elmer) by use of a Rediprime II kit (GE Healthcare), purified on MicroSpin S200 HR columns (GE Healthcare), and denatured for 2 min at 100°C prior to hybridization. Membranes were prehybridized for 1 h in deionized formamide (50%), 20× SSPE (to 1×; 1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 20% SDS (to 0.2%), and 100× Denhardt's solution (to 5×), followed by the addition of denatured probe and hybridization for 16 h at 42°C. Following hybridization, membranes were washed twice for 15 min in 2× SSPE-0.5% SDS (wt/vol) at 42°C.
Nucleotide sequence accession numbers.
The plasmids described in this study, and their sequences, can be obtained from the Chlamydomonas Resource Center (http://chlamycollection.org/).
RESULTS
Recoded CrAadA marker.
In initial experiments (not shown), we observed that plasmids containing the E. coli aadA CDS flanked by the AR promoter and the RBCS2 3′UTR, with or without the RbcS2 chloroplast transit peptide, failed to yield spectinomycin-resistant transformants even at 50 μg/ml Sp, the lowest concentration usable for selection. We therefore decided to recode the AadA marker (renamed “CrAadA”) by using the codon bias specific for Chlamydomonas nuclear genes (26). At the N terminus of the recoded CDS, we placed the RbcS2 chloroplast transit peptide (ending at position 62) to direct the protein to the chloroplast stroma. We also kept a 29-residue fragment of AtpA which now forms the N terminus of the mature CrAadA protein because it was present in the highly efficient chloroplast cassette (15). This CDS was placed under the control of the HSP70A-RBCS2 (AR) promoter (32), with the 3′UTR being provided by the RBCS2 downstream region. The resulting plasmid, pALM32, is depicted in Fig. 1A.
Transformation of the wall-less strain XS1 with pALM32 yielded a large number of transformants on plates containing 75, 100, or 200 μg/ml spectinomycin (Fig. 1B). At these concentrations, no colonies appeared on control plates transformed in the absence of plasmid DNA. Lower concentrations could not be used with XS1 because of an unacceptable number of clones in control transformations. However, a lower Sp concentration of 50 μg/ml could be used with another strain, petC-Δ1, without the appearance of background colonies. In our transformation experiments, colonies usually appeared 5 days after transformation, and their number increased for up to 12 days. Regardless of the antibiotic concentration used for selection or the time of their appearance, almost all clones were found to be resistant to 500 μg/ml Sp (198 of 207 clones) or 10 μg/ml Sr (183 of 207 clones), and 206 of 207 clones were resistant to 100 μg/ml Sp. A fraction was also resistant to 100 μg/ml Sr (40 of 207 clones), with a slight bias in favor of clones selected on higher concentrations of Sp. Overall, resistance was found to be stable over time: 200 transformants were challenged with 200 μg/ml Sp after 2 months of propagation in the absence of antibiotics, and only 7% were found to have lost resistance to Sp.
In the experiment described in the legend to Fig. 1B, we compared the transformation efficiencies of fragments of various lengths derived from pALM32 by restriction enzyme digestion. The highest efficiency was obtained with NotI/KpnI double digestion, which extracts the entire AR-CrAadA-RBCS2 3′UTR cassette from the plasmid. As expected (31), lower transformation rates were obtained with NheI and KpnI digestion, which eliminates the HSP70A fragment. Interestingly, we also obtained transformants when no promoter was included (SphI/KpnI digestion). In these transformants, the CrAadA construct must have integrated downstream of an endogenous promoter that allows its expression. We also noticed that the 3′UTR was not essential for expression of the marker: in both the presence and absence of the AR promoter (NotI/BamHI or AatII digestion), omission of the 3′UTR resulted in only about 50% reductions of the transformation yield. Surprisingly, the lowest efficiency was observed with ScaI digestion, which simply linearizes the plasmid by cutting in the vector part (Fig. 1A) and thus preserves all the expression signals of the marker. Similar results were obtained with other enzymes that linearize pALM33 (see below) or with other plasmids containing the AphVIII marker (data not shown): a simple linearization consistently yielded lower transformation rates than double digestions that completely extracted the cassette.
In order to test the influence of the intracellular localization of the AadA protein, we created pALM34, where the RbcS2 cTP has been excised. As a result, the protein is predicted to remain in the cytosol instead of being targeted to the chloroplast. The resistance cassette (AR-CDS-3′UTR) excised from pALM34 produced a satisfactory yield of transformants. Compared to an identical fragment from pALM32, the efficiency was almost as high at 75 μg/ml Sp (94% ± 4% of control level; n = 2) but lower at 200 μg/ml Sp (52% ± 0%; n = 2).
Shuttle markers functioning in Chlamydomonas and E. coli.
One of the goals of this study was to develop a marker that confers resistance in both Chlamydomonas and E. coli. Plasmid pBSKS+CrAadA, in which the recoded CrAadA gene is cloned in frame within the sequence encoding the LacZ alpha peptide, conferred high-level resistance in E. coli, even in the absence of IPTG (isopropyl-β-d-thiogalactopyranoside) (not shown). Unfortunately, this property was lost in pALM32, where the CDS is placed downstream of a Chlamydomonas, not E. coli, promoter. We therefore decided to introduce E. coli expression signals immediately upstream of the AadA protein gene. We designed an artificial DNA fragment of 75 bp, comprising the canonical −35 and −10 promoter elements, a short 5′UTR, and a Shine-Dalgarno sequence upstream of an ATG start codon. This sequence was cloned into pALM32, just downstream of the RbcS2 cTP and AtpA fragment (Fig. 2A). We took care to avoid in-frame stop codons or polyadenylation or splicing signals that would disrupt expression in Chlamydomonas. As we had hoped, the new plasmid (pALM33) could be selected directly in E. coli by plating the transformation mixture on LB agar plates containing 50 μg/ml Sp in addition to Ap. We also found that it could be used in Chlamydomonas transformation. Figure 2B shows the results of a typical experiment, performed on strain petC-Δ1. When we used AleI and BamHI, which extract the promoter and CDS only (without the 3′UTR), the transformation efficiency with pALM33 was about one-third of that obtained with pALM32. We found a similar ratio when strain XS1 was used (not shown). Note that, again, simple linearization or even inclusion of 200 nucleotides (nt) of vector sequence downstream of the RBCS 3′UTR (using FspI) decreased the transformation rate significantly.
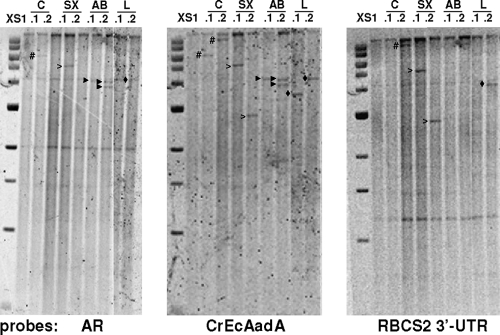
To gain an idea of the number of marker copies integrated into the genomes of the transformants, we chose eight pALM33 transformants obtained from strain XS1 by use of various enzyme combinations and subjected them to Southern blotting (Fig. 3). Using the CrEcAadA CDS as a probe, we found seven transformants with a single insertion and one with two insertions. Some of the bands were also detected with probes from the HSP70A promoter and from the RBCS2 3′UTR, but this was not always true. For example, strains C.2, SX.2, and L.1 lacked the HSP70A signal, while C.1 and L.1 lacked the RBCS2 3′UTR signal. This indicates that the transforming DNA did not always integrate entirely.
Fig. 3.
Southern blot analysis of selected transformants obtained with pALM33 in recipient strain XS1. DNA was digested by PstI, which does not cut the plasmid. The transformants studied were obtained with the uncut circular plasmid (C) or the plasmid cut with SacII/XmnI (SX) or AleI/BamHI (AB) or linearized with AleI (L). Relevant bands are marked by #, unfilled arrowheads, filled black arrowheads, and black diamonds. The 1-kb ladder from NEB is shown on the left (with bands at 0.5, 1, 1.5, 2, 3, 4, 5, 6, 8, and 10 kbp). Note the common bands corresponding to the endogenous HSP70A (2 kb) and RBCS2 (4.2 kb) promoters and to the RBCS2 3′UTR (1.2 kb).
We also examined the possibility of using AphVIII as a shuttle marker. To our surprise, we found that plasmid pBC1, where the AphVIII CDS is placed under the same control elements as the AadA CDS in pALM32, conferred resistance of E. coli to Pm (2 μg/ml). The only bacterial promoters present in the plasmid (for lacZ and bla) are both on the opposite strand. Yet it is clear that the existing AphVIII construct can be used as such for selection in E. coli.
Two-step modification of BACs.
A Chlamydomonas BAC library has been prepared by the laboratory of P. Lefebvre, using genomic DNA from the cw92 mutant (strain CC-503) (22). The DNA was cloned into the HindIII sites of a modified vector (pBeloBAC11) carrying chloramphenicol resistance (23). We decided to introduce the CrEcAadA and AphVIII genes into the BACs to allow direct selection of the transformants in Chlamydomonas. Because BACs are not easily manipulated in vitro, we used homologous recombination in vivo. We created the suicide plasmids pALM30-Sp and pALM30-Pm, where the CrEcAadA and AphVIII genes (including the AR promoter and 3′UTR) are flanked by the 5′ and 3′ halves of the NAR2 gene (1.5 kb on each side). In addition, these plasmids contain the bla gene conferring resistance to Ap, the sacB gene necessary for knockout (see below), and a λpir-dependent origin of replication, from plasmid R6K. Because this origin is nonfunctional in E. coli K-12, the plasmids can replicate only in a λpir strain such as S17-1 λpir. When this kind of plasmid is transformed into a strain devoid of λpir, Ap resistance is observed only when the bla gene (in fact the whole plasmid) integrates into the genome or into a resident BAC. In the case of pALM30-Sp and pALM30-Pm, integration is directed to the BAC because the NAR2 gene is present in the vector part of the BAC (Fig. 4A).
We transformed pALM30-Sp and pALM30-Pm into an E. coli strain carrying BAC 28J23, which contains the PETC gene in the middle of a 110-kb insert. Note that because homologous recombination is inefficient in the absence of the rec system, we first had to transfer the BAC from the library host (HB10B, a recA mutant) into a rec+ strain (BL21). We obtained Cmr Apr Spr and Cmr Apr Pmr transformants where the targeted vector region of the BAC had indeed been modified. Using PCR and restriction digestion, we verified that the entire suicide plasmid had been integrated by a single-crossover event (data not shown). After this “knock-in” step, we went on to eliminate the unnecessary part of the integrated plasmid, aiming to retain only the insertion of the CrEcAadA or AphVIII marker. The “knockout” also used homologous recombination, this time between the NAR2 fragments that were now present as two copies in the recombinant BAC (Fig. 4). To select for recombination events, we took advantage of the presence in the integrated plasmid of the Bacillus subtilis sacB gene, which confers sensitivity to sucrose in Gram-negative bacteria such as E. coli. By plating serial dilutions of an overnight culture on LB plates containing Cm (to select for the BAC), 5% sucrose (to select for recombinants), and either Sp or Pm (to select for the desired recombination geometry), we were able to retrieve clones carrying the 28J23 BAC with the CrEcAadA or AphVIII marker integrated into the NAR2 gene. For CrEcAadA, about half (19 of 40 clones) of the sucrose-resistant clones tested were sensitive to Ap and resistant to Cm and Sp, and they carried the CrEcAadA CDS based on colony PCR (Fig. 4B). One of these BACs (28J23-Sp) was checked by restriction analysis and found to display the expected changes in digestion pattern (Fig. 4C). Using the same procedure, we modified BAC 28J23 to introduce the more efficient CrAadA marker (in both orientations with respect to NAR2 [BACs 28J23-CrSpA and 28J23-CrSpS]) and the AphVIII marker (BAC 28J23-Pm).
Finally, we transformed the modified BACs into the Chlamydomonas petC-Δ1 strain. This strain is nonphotosynthetic because of a short deletion in the PETC gene for the Rieske Fe-S protein of cytochrome b6f (10). This ac− phenotype is stable, as we have never seen it reverse in dozens of control transformation experiments. Using 28J23-Pm, we obtained a large number of transformants on TAP-Pm plates (Table 2). When 24 randomly picked clones were tested on Min plates, 10 (42%) were found to have recovered phototrophy. This shows that a BAC modified to carry the AphVIII gene can be used to screen Pmr transformants for cointegration of a gene present in the insert, even when this gene is not selected for. We could also obtain cotransformants on Min-Pm plates, albeit fewer than on TAP-Pm plates (Table 2). When we asked whether clones selected on Min medium had acquired Pm resistance, we found, to our surprise, that none of the 24 clones tested were Pmr.
Table 2.
Transformation of Chlamydomonas with modified BACs
| Variablea | Value for: |
||||
|---|---|---|---|---|---|
| Expt A |
Expt B |
||||
| 28J23-Pm | 28J23-Sp | 28J23-Sp | 28J23-CrSpA | 28J23-CrSpS | |
| No. of colonies per transformation with TAP plus antibiotic | 238 | 10 | 48 | 192 | 178 |
| No. of colonies showing phototrophy (ac+)/no. of colonies tested | 10/24 | 0/9 | 2/24 | 6/48 | 5/48 |
| No. of colonies per transformation with Min | 231 | 235 | >300 | >300 | ∼200 |
| No. of colonies showing Pmr or Spr/no. of colonies tested | 0/24 | 0/24 | 0/48 | 0/48 | 0/89 |
| No. of colonies per transformation with Min plus antibiotic | 74 | 0 | 0 | 0 | 0 |
Selection and tests for antibiotic resistance used 5 μg/ml Pm or 75 μg/ml Sp.
Using the AadA marker, we also obtained cotransformation, i.e., Spr clones that had recovered phototrophy, but more screening was needed. Our first experiment (Table 2, experiment A) even failed to show cotransformation, probably because we had too few transformants to screen. By taking cells as early as possible in the exponential phase, we finally managed to observe cotransformation (Table 2, experiment B). Comparing the three modified BACs that we had constructed, we found, as expected, that the yield of Spr transformants was higher with the CrAadA marker (28J23-CrSpA and -B) than with CrEcAadA (28J23-Sp). The cotransformation rates (proportion of Spr transformants that had recovered phototrophy) were about the same, with a 10% rate overall. We were unable to obtain transformants on Min-Sp, and all of the ac+ transformants tested were found to be sensitive to Sp, indicating that cotransformation worked in one direction but not the other.
DISCUSSION
In this study, we present CrAadA, a recoded version of the classical AadA marker that can be used to transform the Chlamydomonas nuclear genome to Sp and Sr resistance. Working concentrations for Sr are lower than those for Sp, as observed for the chloroplast AadA cassette. Compared to chloroplast transformants, which are perfectly healthy with up to 5 mg/ml Sp or 500 μg/ml Sr, nuclear transformants with CrAadA showed a lesser degree of resistance, in spite of the fact that the presence of the RbcS2 cTP should direct the protein to the chloroplast stroma. Because the predicted mature sequence of the protein is the same as that for the chloroplast cassette, the level of accumulation of AadA protein must be limiting in many of our nuclear transformants. Indeed, when we analyze transformation plates by fluorescence induction kinetics, which probes the state of the electron transfer chain (21), we often observe that colonies have a reduced variable fluorescence and an altered induction profile, similar to photoinhibited cells. This effect disappears when the colonies are transferred to antibiotic-free plates. This suggests that chloroplast protein translation is affected by the drug, even when the presence of CrAadA allows growth of colonies on Sp-containing medium. This phenomenon is also observed with chloroplast transformants immediately after transformation but disappears when the cassette becomes homoplasmic, we assume because the cells now express higher levels of the enzyme. In accordance with this hypothesis, the transformation rate of pALM34, where the chloroplast transit peptide of CrAadA has been removed, is almost identical to that of pALM32 at 75 μg/ml Sp but reduced by a factor of 2 at 200 μg/ml Sp, suggesting that detoxification is less efficient when the enzyme is localized in the cytosol than when it is in the chloroplast. Cerutti et al. found that chloroplast targeting was not necessary for AadA function, but they tested a single concentration of 90 μg/ml (7).
Usage of the codon bias specific for Chlamydomonas nuclear genes was essential for obtaining an efficient marker, because a similar construct using the original bacterial aadA sequence flanked by the same promoter and 3′UTR failed to give transformants. This could mean that the recoded marker is translated more efficiently or is less susceptible to silencing, or even that it is integrated more efficiently into the genome. Our experiments do not allow us to distinguish between these hypotheses. We note that silencing is a major issue when the bacterial AadA marker is used as a nuclear marker, since about half of the resistant clones rapidly lose (and regain) resistance over time (6). It is not clear how codon usage and compositional bias influence silencing pathways in Chlamydomonas. We serendipitously observed a loss of resistance in XS1 transformants obtained with AphVIII, in spite of its GC-richness (not shown). At any rate, it was necessary to check that resistance conferred by CrAadA was stable over time. When pALM32 transformants were propagated for 2 months in the absence of antibiotics, resistance was found to be maintained in 93% of the transformants, sometimes with a slight reduction in the maximum concentration tolerated. Based on this finding, we think that the CrAadA marker is at least as good as AphVIII in terms of long-term stability.
One of the surprising aspects of our results was the strong dependence of transformation efficiency upon the nature of the DNA transformed. Whenever we were able to compare fragments that contained only the marker cassette (promoter-CDS-3′UTR) with fragments that also contained vector DNA, we found that the latter gave fewer transformants (for example, see Fig. 2B). This was observed for CrAadA, CrEcAadA, and AphVIII by use of restriction digestion and PCR fragments. The linearized plasmid consistently gave the lowest yields, sometimes even below those obtained with the CDS alone. This could be due to a lower efficiency of DNA integration or, more likely, to increased transgene silencing. The pBluescript DNA is only about 50% GC-rich, which could promote silencing in the GC-rich genome of Chlamydomonas. The G+C content of the BamHI-FspI segment, which reduced the transformation efficiency >2-fold (Fig. 2B), is 54.5%, compared to 67.2% for the sequence of the same length upstream of the BamHI site. Similarly, the FspI-AleI fragment, which further decreased the transformation efficiency, is only 49.5% G+C. In addition, we cannot rule out that functional elements in the vector, such as the T7 promoter downstream of the insert, also adversely affect expression of the cassette. Whatever the reasons, it is clear that a clean excision of the cassette is preferable when large numbers of transformants are desired.
Expression signals upstream and downstream of the CrAadA and CrEcAadA CDS were also found to boost transformation rates, but less than we anticipated. Removal of the AR promoter reduced the transformation efficiency by a factor of only 4. Expression from promoterless constructs has also been observed for several genes involved in chloroplast genome expression, for example, NAC2, TCA1, and MCA1 (3, 29), which are also believed to not need a high level of expression to fulfill their functions. In such cases, the DNA must have inserted downstream of an endogenous promoter or revealed the cryptic promoter activity of a hitherto silent region. This constitutes a promoter-trap experiment similar to those that have been performed before for many organisms, including Chlamydomonas (16). Note that our limited Southern blot analysis of pALM33 transformants indicates that the transforming DNA can be shortened substantially during its integration, in particular on the promoter side (Fig. 3C), so that even some transformants obtained with a full cassette must in fact use an endogenous promoter for CrEcAadA expression.
Similarly, we found that the 3′UTR was also dispensable for transformation (for example, compare the NotI/KpnI and NotI/BamHI fragments in Fig. 1B). Here, again, the reduction in efficiency was only a modest factor of 2. In this case, because we know that mRNA stability and cytosolic translation require the presence of a poly(A) tail and its interaction with the 5′ cap, we must assume that the Chlamydomonas genomic DNA downstream of the CrAadA CDS provides an artificial 3′UTR, yielding a chimeric mRNA. In a separate study, we will report how we can use this property to identify flanking sequence tags in insertional mutants.
Selection markers are useful not only to generate mutants but also to integrate into the genome constructs that cannot be selected for by themselves. This is the case, for example, when a BAC must be transformed into a mutant to complement a phenotype that is not a growth defect. Modified BACs carrying a resistance marker would be an asset here, especially if the marker could be used both in E. coli, to select for the modified BAC, and in Chlamydomonas, to select for transformants harboring the BAC DNA. We were able to engineer this property into CrAadA by adding a synthetic promoter and Shine-Dalgarno sequence. The new marker, CrEcAadA, was slightly less efficient in Chlamydomonas, while remaining in a comfortable working range for use in insertional mutagenesis. For AphVIII, no modification was necessary, since the Chlamydomonas sequence upstream of the marker apparently allowed a sufficient level of expression in E. coli to confer Pm resistance. As analyzed using the program BPROM (SoftBerry), the sequence upstream of the CDS, which by and large corresponds to the first intron of RBCS2, appears to contain a reasonably strong bacterial promoter (score of 2.93 versus 2.85 for the bla promoter and 5.91 for the synthetic promoter in pALM33), with −35 and −10 boxes placed 225 nt and 202 nt before the ATG, respectively.
To modify the BACs, we used a two-step suicide plasmid strategy requiring two homologous recombination events to first introduce the plasmid into the BAC and then remove the unnecessary plasmid sequence so as to leave only the marker in the BAC. Our attempts to simplify the protocol by directly plating the transformation mixture on sucrose-Sp-Cm plates, requiring a double-crossover event, failed, but we believe that the two-step method is sufficiently simple to be applied in routine work. Alternatively, recombineering (8) can be proposed as a simple and versatile way to introduce markers into BACs, including within the genes in the insert.
Upon transformation of Chlamydomonas with the modified 28J23-Pm BAC, we obtained high-frequency cotransformation of the AphVIII cassette and a target gene within the BAC insert: 40% of the Pmr transformants were phototrophic, indicating that they contained and expressed both AphVIII and PETC. Using the AadA marker, fewer transformants were obtained and a smaller proportion (about 10%) was found to have restored PETC function. Thus, in spite of the need for more screening, AadA can also be useful for complementation work, for example, in mutants that already carry the AphVIII cassette. Because the yield of transformants could be limiting in using a BAC modified with the CrEcAadA marker (see experiment A in Table 2), we recommend using CrAadA instead, even if it does not allow selection of the modified BACs in E. coli.
At present, we cannot explain why cotransformation was observed in a larger fraction of the Pmr than Spr transformants: because the PETC gene is not selected for, its cointegration should occur at similar frequencies regardless of the resistance marker used. Also, we note that no transformants were ever obtained on Min-Sp, while the yield was reasonable on Min-Pm. Our screening for cotransformants relied on full restoration of phototrophy, so we suspect that the level of expression of the PETC gene could be limiting in some cells that have integrated it along with the marker. For some reason, this type of nonproductive integration occurred more frequently during AadA transformations, maybe because this marker lacks the RBCS2 first intron present in the AphVIII construct. Because the BACs were transformed without prior linearization, the DNA must be cleaved in Chlamydomonas before integration can occur. If multiple cleavages occur, this could lead to the resistance marker and the gene of interest inserting at different loci in the genome, resulting in uncorrelated expression levels. Indeed, this may be the explanation for another troubling observation, namely, that none of the transformants selected on Min ever showed high-level resistance to Pm or Sp. This could indicate that the most robust PETC transformants are those that have not integrated the vector part of the BAC, leading to an underrepresentation of antibiotic-resistant clones in the transformants we screened. At any rate, it is clear that integration of the transforming DNA is not sufficient for its expression. We therefore advise the user who may have difficulties observing phenotypic complementation to screen transformants for the expression of the gene of interest rather than its sole integration.
ACKNOWLEDGMENTS
L.M.-C. was supported by the Agence Nationale de la Recherche (ALGOMICS grant).
We thank R. Kuras and all members of the Wollman laboratory for support and advice.
Footnotes
Published ahead of print on 14 October 2011.
REFERENCES
- 1. Auchincloss A. H., Loroch A. I., Rochaix J. D. 1999. The argininosuccinate lyase gene of Chlamydomonas reinhardtii: cloning of the cDNA and its characterization as a selectable shuttle marker. Mol. Gen. Genet. 261: 21–30 [DOI] [PubMed] [Google Scholar]
- 2. Berthold P., Schmitt R., Mages W. 2002. An engineered Streptomyces hygroscopicus aph 7″ gene mediates dominant resistance against hygromycin B in Chlamydomonas reinhardtii. Protist 153: 401–412 [DOI] [PubMed] [Google Scholar]
- 3. Boudreau E., Nickelsen J., Lemaire S. D., Ossenbuhl F., Rochaix J. D. 2000. The Nac2 gene of Chlamydomonas encodes a chloroplast TPR-like protein involved in psbD mRNA stability. EMBO J. 19: 3366–3376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boynton J. E., Gillham N. W. 1996. Genetics and transformation of mitochondria in the green alga Chlamydomonas. Methods Enzymol. 264: 279–296 [DOI] [PubMed] [Google Scholar]
- 5. Boynton J. E., et al. 1988. Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science 240: 1534–1538 [DOI] [PubMed] [Google Scholar]
- 6. Cerutti H., Johnson A. M., Gillham N. W., Boynton J. E. 1997. Epigenetic silencing of a foreign gene in nuclear transformants of Chlamydomonas. Plant Cell 9: 925–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cerutti H., Johnson A. M., Gillham N. W., Boynton J. E. 1997. A eubacterial gene conferring spectinomycin resistance on Chlamydomonas reinhardtii: integration into the nuclear genome and gene expression. Genetics 145: 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Copeland N. G., Jenkins N. A., Court D. L. 2001. Recombineering: a powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2: 769–779 [DOI] [PubMed] [Google Scholar]
- 9. Debuchy R., Purton S., Rochaix J. D. 1989. The argininosuccinate lyase gene of Chlamydomonas reinhardtii: an important tool for nuclear transformation and for correlating the genetic and molecular maps of the ARG7 locus. EMBO J. 8: 2803–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de Vitry C., Finazzi G., Baymann F., Kallas T. 1999. Analysis of the nucleus-encoded and chloroplast-targeted Rieske protein by classic and site-directed mutagenesis of Chlamydomonas. Plant Cell 11: 2031–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Donnenberg M. S., Kaper J. B. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59: 4310–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ferris P. J. 1995. Localization of the nic-7, ac-29 and thi-10 genes within the mating-type locus of Chlamydomonas reinhardtii. Genetics 141: 543–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ferris P. J., Woessner J. P., Goodenough U. W. 1996. A sex recognition glycoprotein is encoded by the plus mating-type gene fus1 of Chlamydomonas reinhardtii. Mol. Biol. Cell 7: 1235–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fuhrmann M., Oertel W., Hegemann P. 1999. A synthetic gene coding for the green fluorescent protein (GFP) is a versatile reporter in Chlamydomonas reinhardtii. Plant J. 19: 353–361 [DOI] [PubMed] [Google Scholar]
- 15. Goldschmidt-Clermont M. 1991. Transgenic expression of aminoglycoside adenine transferase in the chloroplast: a selectable marker of site-directed transformation of Chlamydomonas. Nucleic Acids Res. 19: 4083–4089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Haring M. A., Beck C. F. 1997. A promoter trap for Chlamydomonas reinhardtii: development of a gene cloning method using 5′ RACE-based probes. Plant J. 11: 1341–1348 [DOI] [PubMed] [Google Scholar]
- 17. Harris E. H. 1989. The Chlamydomonas source book: a comprehensive guide to biology and laboratory use. Academic Press, San Diego, CA: [DOI] [PubMed] [Google Scholar]
- 18. Harris E. H. 2009. The Chlamydomonas sourcebook. Academic Press, San Diego, CA [Google Scholar]
- 19. Hollingshead S., Vapnek D. 1985. Nucleotide sequence analysis of a gene encoding a streptomycin/spectinomycin adenylyltransferase. Plasmid 13: 17–30 [DOI] [PubMed] [Google Scholar]
- 20. Johnson X., et al. 2010. MRL1, a conserved pentatricopeptide repeat protein, is required for stabilization of rbcL mRNA in Chlamydomonas and Arabidopsis. Plant Cell 22: 234–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Joliot P., Beal D., Delosme R. 1998. In vivo measurements of photosynthetic activity: methods, p. 433–449 In Rochaix J.-D., Goldschmidt-Clermont M., Merchant S. (ed.), The molecular biology of chloroplast and mitochondria in Chlamydomonas, vol. 7 Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 22. Kathir P., et al. 2003. Molecular map of the Chlamydomonas reinhardtii nuclear genome. Eukaryot. Cell 2: 362–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim U. J., et al. 1996. Construction and characterization of a human bacterial artificial chromosome library. Genomics 34: 213–218 [DOI] [PubMed] [Google Scholar]
- 24. Kindle K. L., Schnell R. A., Fernandez E., Lefebvre P. A. 1989. Stable nuclear transformation of Chlamydomonas using the Chlamydomonas gene for nitrate reductase. J. Cell Biol. 109: 2589–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kovar J. L., Zhang J., Funke R. P., Weeks D. P. 2002. Molecular analysis of the acetolactate synthase gene of Chlamydomonas reinhardtii and development of a genetically engineered gene as a dominant selectable marker for genetic transformation. Plant J. 29: 109–117 [DOI] [PubMed] [Google Scholar]
- 26. Merchant S. S., et al. 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nelson J. A., Savereide P. B., Lefebvre P. A. 1994. The CRY1 gene in Chlamydomonas reinhardtii: structure and use as a dominant selectable marker for nuclear transformation. Mol. Cell. Biol. 14: 4011–4019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Philippe N., Alcaraz J. P., Coursange E., Geiselmann J., Schneider D. 2004. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51: 246–255 [DOI] [PubMed] [Google Scholar]
- 29. Raynaud C., et al. 2007. Evidence for regulatory function of nucleus-encoded factors on mRNA stabilization and translation in the chloroplast. Proc. Natl. Acad. Sci. U. S. A. 104: 9093–9098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 31. Schroda M., Beck C. F., Vallon O. 2002. Sequence elements within an HSP70 promoter counteract transcriptional transgene silencing in Chlamydomonas. Plant J. 31: 445–455 [DOI] [PubMed] [Google Scholar]
- 32. Schroda M., Blocker D., Beck C. F. 2000. The HSP70A promoter as a tool for the improved expression of transgenes in Chlamydomonas. Plant J. 21: 121–131 [DOI] [PubMed] [Google Scholar]
- 33. Shimogawara K., Fujiwara S., Grossman A., Usuda H. 1998. High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics 148: 1821–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sizova I., Fuhrmann M., Hegemann P. 2001. A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene 277: 221–229 [DOI] [PubMed] [Google Scholar]
- 35. Sizova I. A., et al. 1996. Stable nuclear transformation of Chlamydomonas reinhardtii with a Streptomyces rimosus gene as the selective marker. Gene 181: 13–18 [DOI] [PubMed] [Google Scholar]
- 36. Stevens D. R., Rochaix J. D., Purton S. 1996. The bacterial phleomycin resistance gene ble as a dominant selectable marker in Chlamydomonas. Mol. Gen. Genet. 251: 23–30 [DOI] [PubMed] [Google Scholar]