Abstract
Glycerophosphodiesters are the products of phospholipase-mediated deacylation of phospholipids. In Saccharomyces cerevisiae, a single gene, GIT1, encodes a permease responsible for importing glycerophosphodiesters, such as glycerophosphoinositol and glycerophosphocholine, into the cell. In contrast, the Candida albicans genome contains four open reading frames (ORFs) with a high degree of similarity to S. cerevisiae GIT1 (ScGIT1) Here, we report that C. albicans utilizes glycerophosphoinositol (GroPIns) and glycerophosphocholine (GroPCho) as sources of phosphate at both mildly acidic and physiological pHs. Insertional mutagenesis of C. albicans GIT1 (CaGIT1) (orf19.34), the ORF most similar to ScGit1, abolished the ability of cells to use GroPIns as a phosphate source at acidic pH and to transport [3H]GroPIns at acidic and physiological pHs, while reintegration of a GIT1 allele into the genome restored those functions. Several lines of evidence, including the detection of internal [3H]GroPIns, indicated that GroPIns is transported intact through CaGit1. GroPIns transport was shown to conform to Michaelis-Menten kinetics, with an apparent Km of 28 ± 6 μM. Notably, uptake of label from [3H]GroPCho was found to be roughly 50-fold greater than uptake of label from [3H]GroPIns and roughly 500-fold greater than the equivalent activity in S. cerevisiae. Insertional mutagenesis of CaGIT1 had no effect on the utilization of GroPCho as a phosphate source or on the uptake of label from [3H]GroPCho. Growth under low-phosphate conditions was shown to increase label uptake from both [3H]GroPIns and [3H]GroPCho. Screening of a transcription factor deletion set identified CaPHO4 as required for the utilization of GroPIns, but not GroPCho, as a phosphate source.
INTRODUCTION
Glycerophosphodiesters result from the complete deacylation of glycerophospholipids via phospholipase-mediated hydrolysis. Most fungal cells, including those of Candida albicans and Saccharomyces cerevisiae, contain multiple phospholipase B (PLB)-encoding genes (25) that act on both fatty acyl ester groups to produce glycerophosphodiesters, such as glycerophosphocholine (GroPCho) and glycerophosphoinositol (GroPIns) (34). The potential role of C. albicans PLBs as virulence factors has been explored by others (21, 26, 29, 30, 40). For example, C. albicans strains exhibiting elevated PLB activity have been shown to be associated with increased virulence in mouse models of disseminated candidiasis (22). Disruption of PLB1 was subsequently shown to result in attenuated virulence in a mouse model (26), and reintroduction of a functional PLB1 into this mutant to restore virulence to levels observed for the parental strain (30). Also, inactivation of another PLB gene, PLB5, has been shown to result in attenuated virulence (41). Notably, the fate and potential function of the products of PLB turnover, the glycerophosphodiesters, have not been addressed.
Although glycerophosphodiesters are produced via C. albicans PLB activity, the organism, being an opportunistic commensal, is also likely to be exposed to sources of glycerophosphodiesters that are present in the host as a result of host phospholipase activity. Indeed, the literature indicates that glycerophosphodiesters, especially GroPIns and GroPCho, are present in serum, as well as other mammalian fluids and tissues. For example, GroPCho is an abundant organic osmolyte found in the renal medulla of the kidney (15, 16), and both GroPIns and GroPCho are found in other parts of the urinary tract, including renal proximal tubules (38). GroPCho has also been found in organs of the gastrointestinal tract, including the small and large intestines (2, 43, 44). Serum, cerebrospinal fluid, and brain tissue contain GroPCho, in addition to lysophosphatidylcholine and phosphatidylcholine that can be converted to GroPCho via phospholipases B (24, 31, 42). GroPIns has also been noted in other cells and tissues, including brain, kidney, and others (6).
In S. cerevisiae, extracellular GroPIns and, with less affinity, GroPCho, are transported into the cell via the ScGit1 transporter. Once inside the cell, they are metabolized and used as sources of nutrients such as phosphate, inositol, and choline (1, 13, 35). C. albicans contains four open reading frames (ORFs) (CaGIT1 to -4) predicted to encode transporters with a high degree of similarity to the S. cerevisiae GIT1 (ScGIT1) product (4). ScGit1 and CaGit1 to -4 are classified as members of the major facilitator superfamily (MFS) (17). The MFS is present in all kingdoms of life. Most MFS proteins are between 400 and 600 amino acids in length and contain either 12 or 14 membrane-spanning segments. MFS proteins facilitate symport, antiport, or uniport of various substrates. Those substrates include nutrients, drugs, nucleotides, nucleosides, and other metabolites (17, 32). Using the Transport Commission (TC) system (37), 95 potential MFS proteins clustering into 17 families have been predicted for C. albicans (17), but only a handful have been characterized. Like ScGit1, CaGit1 to -4 are predicted to belong to the phosphate:H+ symporter (PHS) family (TC no. 2.A.1.9) of the MSF (17). In total, the C. albicans genome is predicted to have 5 PHS family members: CaGit1 to -4 and the homolog of the S. cerevisiae high-affinity phosphate transporter, Pho84. No member of this family has been characterized in C. albicans.
Here, we investigate the ability of C. albicans to transport GroPIns and GroPCho into the cell and to utilize those compounds as sources of phosphate. In addition, we identify CaGIT1 (orf19.34) as a GroPIns permease.
MATERIALS AND METHODS
Strains and media.
Strains were grown aerobically at either 30°C or 37°C with shaking. Turbidity was monitored by measurement of optical density at 600 nm (OD600) on a Biomate 3 Thermo Spectronic spectrophotometer. Synthetic complete (yeast nitrogen base [YNB]) medium was prepared as described previously (33). High-Pi and low-Pi media were made by replacing the KH2PO4 (1 g/liter) in synthetic complete medium with KCl (1 g/liter) and adding KH2PO4 to 10 mM (high Pi) or 0.2 mM (low Pi). All media for C. albicans were supplemented with 80 μg/ml of uridine. For some experiments, media lacking KH2PO4 (no-Pi medium) contained GroPIns (Sigma no. G1891), GroPCho (Sigma no. G5291), or glycerol-3-phosphate (GroP) (Sigma no. G7886) at the indicated concentrations. Where indicated, YNB was buffered to pH 7.5 using 150 mM HEPES. Strains were maintained on yeast extract-peptone-dextrose (YEPD) medium consisting of 20 g glucose, 10 g yeast extract, and 20 g Bacto peptone per liter. The genotypes of C. albicans strains are indicated in Table 1. The S. cerevisiae strain, BY4741 is MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0.
Table 1.
C. albicans strains
| Strain | Genotype | Reference |
|---|---|---|
| BWP17 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG | 45 |
| DAY185 | ura3Δ::λimm434/ura3Δ::λimm434 pARG4::URA3::arg4::hisG/arg4::hisG pHIS1::his1::hisG/his1::hisG | 8 |
| JPV 512 | git1::UAU1/git1::URA3 + pDDB78GIT1 | This study |
| JPV 526 | git1::UAU1/git1::URA3 + pDDB78 | This study |
| WT-TF | arg4Δ/arg4Δ LEU2/leu2Δ HIS1/his1Δ URA3/ura3Δ::imm434 IRO1/iro1Δ::imm434 | 20 |
| pho4Δ/Δ-X1 mutant | arg4Δ/arg4Δ leu2Δ/leu2Δ his1Δ/his1Δ URA3/ura3Δ::imm434 IRO1/iro1Δ::imm434pho4Δ::LEU2/pho4Δ::HIS1 | 20 |
| pho4Δ/Δ-Y1 mutant | arg4Δ/arg4Δ leu2Δ/leu2Δ his1Δ/his1Δ URA3/ura3Δ::imm434 IRO1/iro1Δ::imm434pho4Δ::LEU2/pho4Δ::HIS1 | 20 |
Construction of a homozygous insertion mutant.
A clone of the GIT1 gene containing a Tn7-UAU transposon insertion produced via UAU1 methodology (8, 11) was obtained from Aaron Mitchell, Carnegie Mellon University. Plasmid CAGFN83 (clone 29331) bears the GIT1 gene containing the UAU1 insertion at bp 536. CAGFN83 was digested with NotI to release the Tn7-UAU1-mutagenized GIT1 gene and transformed into strain BWP17 (ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::his arg4::hisG/arg4::hisG) (45). Several Arg+ transformants were isolated and subjected to Arg+ Ura+ selection as described previously (8, 45). Genomic DNA was extracted from Arg+ Ura+ transformants, and PCR was performed using forward primers Arg4det (5′-GGAATTGATCAATTATCTTTTGAAC-3′) (11) and GITF2 (5′-TTCGGACAAGTGATTATTGGATTAACCGCT-3′) and reverse primer GITR3 (5′-TATAACTGACAAGCAGAAGAAAGGGGTTTA-3′). Heterozygous insertion mutants displayed two bands upon PCR: a 1.3-kb fragment corresponding to the GIT1 allele amplified with primers GITF2 and GITR3 and a 2.7-kb fragment corresponding to the presence of the git1::Tn7-UAU1 allele amplified with primers Arg4det and GITR3. Homozygous insertion mutants (git1::UAU1/git1::URA3; JPV484), displayed only the 2.7-kb band. The git1::UAU1/git1::URA3 mutant is also referred to here as the git1−/git1− mutant. Out of 25 Arg+ Ura+ transformants screened, 3 were found to be homozygous insertion mutants.
Construction of plasmid pDDB78GIT1.
GIT1 was amplified from genomic DNA using a forward primer incorporating a NotI restriction site (bold) located 900 bp upstream of the start site (5′-AATGTTAAATGCGGCCGCTGTACACGGCTTTATCGCACGGGATATGAA-3′) and a reverse primer incorporating an EcoRI restriction site (bold) located 540 bp downstream of the stop site (5′-AATGTTAAAGGGGAATTCGAAATTTGGTTATGTAGGGTTCAGTTAAAA-3′). The resulting PCR product and plasmid pDDB78 (39) were digested with NotI and EcoRI and ligated together to obtain plasmid pDDB78GIT1.
Insertional complementation of git1::UAU1/git1::URA3.
Plasmid pDDB78GIT1 was linearized by cutting within the HIS1 gene with NruI, and the resulting product was transformed into the git1::UAU1/git1::URA3 strain (JPV484) to produce JPV512 (git1::UAU1/git1::URA3 + pDDB78GIT1) (43). Several His+ transformants were selected and tested for complementation of the mutant phenotype. Empty plasmid pDDB78 was also linearized and transformed into the git1::UAU1/git1::URA3 strain to produce JPV526 (git1::UAU1/git1::URA3 + pDDB78).
Screening of the transcriptional regulator deletion set.
The deletion library (20) was purchased from the Fungal Genetics Stock Center (FGSC). The deletion strains were constructed in strain SN152 (arg4Δ/arg4Δ leu2Δ/leu2Δ his1Δ/his1Δ URA3Δ/ura3Δ IRO1Δ/iro1Δ). The auxotrophic markers HIS1 and LEU2 were used to delete the genes, as described previously (20). A wild-type control strain in which HIS1 and LEU2 were reintroduced into the parent strain (WT-TF) was included in the set. The deletion strains were screened by growth in liquid YNB medium containing either 200 μM GroPIns or 200 μM KH2PO4 as the source of phosphate. Growth after 48 h at 37°C was monitored.
[3H]inositol-GroPIns and [3H]choline-GroPCho uptake assays.
Label uptake assays (also referred to as transport assays) were performed essentially as described previously (1). For the standard assay, aliquots of the cultures were harvested and washed with sterile water. Each cell pellet was suspended in 100 mM sodium citrate buffer, pH 5.0, to an OD600 of 5. Following 10 min of incubation at 30°C with agitation, the reaction was started by the addition of 50 μl of 25 μM [3H]GroPIns or 50 μl of 1 mM [3H]GroPCho to 200 μl of the cell suspension to produce final concentrations of 5 μM [3H]GroPIns and 200 μM [3H]GroPCho. Following 10 min (for GroPIns transport) or 2 min (for GroPCho transport) of incubation at 30°C, the reaction was stopped by the addition of 10 ml ice-cold H2O. The samples were filtered through glass fiber (GF/C) filters, and the filters were washed with ice-cold H2O. Radioactivity on the filter was determined by liquid scintillation counting. Data are presented as pmol/min/optical density unit at 600 nm (ODU). For S. cerevisiae, transport assays were performed as described above for the standard C. albicans assays, with the exception that for both [3H]GroPIns and [3H]GroPCho a final concentration of 5 μM was used in 10-min assays. Tritium-labeled GroPIns ([3H]inositol-GroPIns) and tritium-labeled GroPCho ([3H]choline-GroPCho) were produced through the deacylation of phosphatidyl-myo-[2-3H]inositol (American Radiolabeled Chemicals) and phosphatidyl-methyl-[3H]choline (American Radiolabeled Chemicals) as described previously (18).
For the GroPIns transport competition assays, conditions were identical to those described above except that 25 mM HEPES buffer (pH 5) was used instead of citrate buffer and the transport assay mixtures included 1 mM GroPIns, 1 mM inositol, 1 mM KH2PO4, or 1 mM GroP, as indicated. The pH of the assays did not change during the course of the experiment. For the protonophore experiment, 100 mM carbonyl cyanide 3-chlorophenylhydrazone (CCCP) (Alfa Aesar no. L06932) stocks in ethanol were added to 25 mM HEPES buffer for a final concentration of 50 μM. For the experiments involving alterations in transport assay pH, 25 mM HEPES buffer was adjusted to pH 6.5, pH 7.5, or pH 8.5, as indicated.
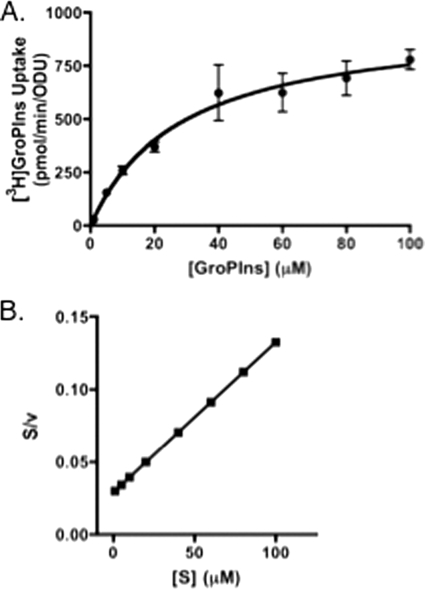
For the determination of GroPIns transport kinetics (see Fig. 5), assays were performed in 100 mM citrate buffer, pH 5. Transport was started with the addition of 50 μl of [3H]GroPIns ranging in concentration from 5 μM to 500 μM GroPIns, as indicated. Transport activity was linear with time at each substrate concentration used. Assays were stopped after 5 min. Saturation kinetics data for GroPIns were analyzed using the Levenberg-Marquardt algorithm for nonlinear regression in GraphPad Prism (version 4.0) to determine the apparent Km and Vmax. Values were determined by least-squares fitting of the data to the Michaelis-Menten equation V = Vmax[S]·(Km + [S])−1, where S represents GroPIns. The saturation kinetics data were manipulated to show linearization by a Hanes plot transformation (19).
Fig. 5.
Kinetics of GroPIns transport. (A) Strains grown in 0.2 mM KH2PO4 (low Pi)-containing medium were harvested, washed, and assayed for GroPIns transport in the presence of concentrations of 1 μM to 100 μM [3H]GroPIns in 100 mM citrate buffer, pH 5.0, as described in the text. Substrate uptake was plotted against initial concentrations of GroPIns. (B) Data were linearized using a Hanes plot transformation. Values represent means ± SE of duplicate determinations.
LC-MS analysis of GroPIns uptake.
A 250-μl portion of each medium sample was diluted 5-fold in methanol-water (90:10) and centrifuged at 10,000 × g for 5 min. The supernatants were transferred into high-pressure liquid chromatography (HPLC) vials, dried down by nitrogen gas, resuspended in 250 μl of 75:25 acetonitrile-methanol, and placed in the autosampler of the liquid chromatography (LC) system. Samples were analyzed using an Agilent 6460 triple-quadrupole mass spectrometer (MS) coupled to an Agilent 1200 LC system. The scan mode was set to multiple-reaction monitoring (MRM) targeted for fragmentation of GroPIns (333→153) in negative-ionization mode. The capillary voltage was set to −3.5 kV, the fragmentor voltage to 50 V, and the collision energy to 20 eV, with a dwell time of 200 ms. The drying gas flow rate was set to 8 liters/min, and the nebulizer pressure was 50 lb/in2. Hydrophilic interaction liquid chromatography (HILIC) was performed with a 5-μm XBridge column (150 by 4.60 mm; Waters, Milford, MA). The mobile phase was 50:50 acetonitrile-water containing 10 mM NH4 acetate (NH4OAc), with a resulting pH of 7.19. The chromatography was performed using isocratic elution at a flow rate of 0.5 ml/min. An injection volume of 10 μl was used, and each sample was run in triplicate. Data were analyzed with MassHunter workstation software. The peak area of GroPIns was compared with the external calibration curve to calculate its relative concentration in the medium.
Analysis of internal [3H]GroPIns metabolites.
Following a standard [3H]GroPIns transport assay, internal counts were isolated via trichloroacetic acid (TCA) extraction (9) and separated by ion-exchange chromatography essentially as described previously (33). Briefly, at the conclusion of a 5-min [3H]GroPIns transport assay, 750 μl of sterile water was added to the 250-μl cell suspension and the cells were pelleted. The cells were washed with 500 μl of sterile water and repelleted. The pelleted cells were then suspended in 100 μl of a 5% TCA solution and incubated on ice for 10 min. After the incubation, the cells were pelleted, and the supernatant, containing the intracellular water-soluble metabolites, was removed to a fresh tube. An equal volume of 1 M Tris (pH 8) was added to the supernatant to neutralize it. Neutralized samples were diluted and applied to 1-ml Dowex 1X8-400 anion-exchange columns. Potential inositol-containing metabolites (inositol, GroPIns, and inositol phosphate) were eluted from the column as described previously (33). Appropriate tritium-labeled standards were used to validate this procedure. Radioactivity was determined using scintillation counting.
RNA Extraction and quantitative reverse transcriptase PCR (qRT-PCR) gene expression analysis.
Cultures were grown in either low-Pi or high-Pi medium supplemented with 80 μg/ml uridine to an OD of between 0.8 and 1.2. RNA was extracted using a hot phenol-chloroform extraction (10). RNA was DNase treated using the Turbo DNA-free kit (Applied Biosystems). A 5-μg sample of RNA was treated with 2 units of DNase and incubated at 37°C for 30 min. Samples were stored at −80°C until analysis. Primer 3 software (http://frodo.wi.mit.edu/primer3/) was used to design primers for CaGIT1 (forward, 5′-CGCATCTTTGTCAACTCAAG-3′; reverse, 5′-TAGCAGCTTCACTTGCTGTC-3′). Primer sequences for the endogenous control (CaTDH3) 5′-TGCTAAAGCCGTTGGTAAGG-3′ (forward) and 5′-AAATCGGTGGAGACAACAGC-3′ (reverse) (3). Real-time RT-PCR was performed using the Power SYBR green RNA-to-CT 1-Step kit (Applied Biosystems). Each reaction mixture consisted of 0.2 μl of a 125× RT enzyme mix, 12.5 μl of a 2× RT-PCR mix, 500 nM primers, and 1.5 μl DNase-treated RNA in a total volume of 25 μl. Experimental samples were analyzed in triplicate on an Applied Biosystems StepOnPlus instrument. Reverse transcription was carried out at 48°C for 30 min, followed by 95°C for 10 min for RT inactivation and polymerase activation, followed by 40 cycles of 95°C for 15 s, 50°C for 30 s, and 72°C for 40 s for amplification, followed by the melt curve to check for primer specificity. A no-template control reaction and a reaction without reverse transcriptase were performed to confirm lack of contamination in the RNA samples and/or the reagents. CaGIT1 expression was analyzed using the ΔΔCT method and normalized to the expression of the endogenous control, CaTDH3. The result for the wild-type strain grown in low-Pi medium was normalized to 1 and used as a comparison for fold change.
RESULTS
Potential ScGIT1 homologs found in the C. albicans genome.
Four ORFs with high similarity to ScGIT1, the gene that encodes the S. cerevisiae GroPIns and GroPCho transporter, have been identified in the C. albicans genome (4, 17). CaGIT1 corresponds to orf19.34. Currently, the Candida Genome Database (CGD) (http://www.candidagenome.org) has CaGIT1 as the primary designation for orf19.34 but has noted a nomenclature conflict in that both orf19.34 and orf19.1979 have previously been called CaGIT1. A WU-BLAST2 search reveals that CaGit1 displays 53% identity and 69% similarity to ScGit1 over 504 amino acids (97% of the protein). CaGIT1 resides on chromosome 2, while CaGIT2 to -4 (orf19.1978 to orf19.1980) lie in a tandem repeat on the left arm of chromosome 5 and are highly similar to each other. CaGit2 (orf19.1978) displays 75% identity and 86% similarity to CaGit3 (orf19.1979) and 69% identity and 81% similarity to CaGit4 (orf19.1980) in regions covering at least 95% of the predicted proteins.
Role of CaGit1 in utilization of GroPIns and GroPCho as phosphate sources.
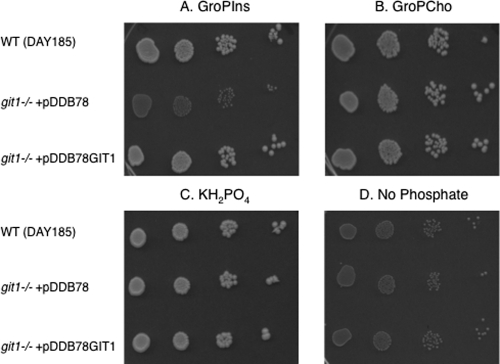
As shown in Fig. 1, a wild-type C. albicans strain (DAY185) is able to utilize both GroPIns (Fig. 1A) and GroPCho (Fig. 1B) as sources of phosphate. As expected, the wild-type strain grew when KH2PO4 was supplied (Fig. 1C) but did not grow when no phosphate source was supplied (Fig. 1D). To investigate the role of CaGIT1 in the utilization of GroPIns and GroPCho, a homozygous insertion mutant of CaGIT1 was constructed using UAU methodology (8, 11) to produce the git1:UAU1/git1::URA3 strain, as described in Materials and Methods. For insertional complementation, plasmid pDDB78 (39) containing CaGIT1 pDDB78GIT1 was linearized and transformed into the git1:UAU1/git1::URA3 strain to produce the git1:UAU1/git1::URA3 + pDDB78GIT1 strain. As evident in Fig. 1A, insertional mutagenesis of CaGIT1 abolished the utilization of GroPIns as a phosphate source on YNB plates, while reintegration of CaGIT1 restored that ability. The plates shown were grown at 37°C, but identical results were obtained at 30°C. Also evident is that CaGIT1 is not required for the utilization of GroPCho as a phosphate source.
Fig. 1.
C. albicans utilizes GroPIns and GroPCho as sources of phosphate. Strains pregrown in 200 μM KH2PO4 (low Pi)-containing medium were harvested, washed, and spotted in a series of 10× dilutions onto plates containing 200 μM GroPIns (A), 200 μM GroPCho (B), 200 μM KH2PO4 (C), or medium containing no phosphate (D). Data are representative of multiple experiments.
Uptake of label from [3H]inositol-GroPIns requires CaGit1 and is regulated by phosphate availability.
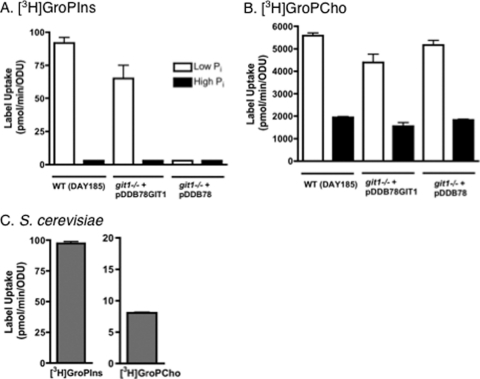
Since GroPIns and GroPCho can act as sources of phosphate, we next monitored the ability of cells to take up label from [3H]inositol-GroPIns and [3H]choline-GroPCho. Label uptake was monitored in cells grown in YNB medium containing either a high (10 mM KH2PO4) or low (0.2 mM KH2PO4) phosphate level (Fig. 2), as phosphate availability has been shown to regulate the expression of ScGIT1 (1). The transport assays (2 min for GroPCho and 10 min for GroPIns) were optimized to ensure that uptake was linear with time at the given substrate concentration. As shown in Fig. 2A, [3H]GroPIns uptake is completely absent in cells grown under high-Pi conditions, and the label uptake that occurs in cells grown in low-Pi medium requires CaGIT1. In contrast, growth under high-phosphate conditions decreased, but did not abolish, the uptake of label from [3H]GroPCho (Fig. 2B). As expected, CaGIT1 played no role in [3H]GroPCho uptake under high- or low-phosphate conditions.
Fig. 2.
[3H]GroPIns uptake requires CaGit1 and is regulated by phosphate availability. (A and B) Strains grown in either 10 mM KH2PO4 (high Pi) or 0.2 mM KH2PO4 (low Pi)-containing medium were harvested, washed, and assayed for either GroPIns transport in the presence of 5 μM [3H]GroPIns (A) or GroPCho transport in the presence of 200 μM [3H]GroPCho (B), as described for the standard transport assays. (C) S. cerevisiae transport was determined in the presence of 5 μM [3H]GroPIns or [3H]GroPCho, as described in Materials and Methods. Values represent means ± standard errors (SE) of triplicate determinations. The experiment was repeated with similar results.
Notably, uptake of label from [3H]GroPCho is quite robust, being roughly 50-fold greater than that from [3H]GroPIns under low-phosphate conditions. Furthermore, it is roughly 500-fold greater than that observed in S. cerevisiae cells grown under low-phosphate conditions (compare Fig. 2B and C). In contrast, [3H]GroPIns transport is roughly the same in C. albicans as it is in S. cerevisiae cells grown under low-phosphate conditions (compare Fig. 2A and C).
Glycerophosphodiester utilization at physiological pH.
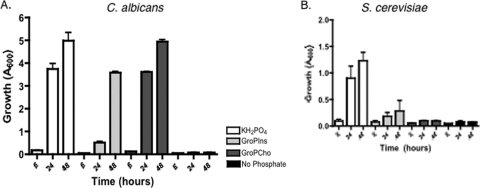
In order to determine if the observed growth on GroPIns and GroPCho might also occur under nonacidic conditions that may be encountered in a human host, we monitored growth of the wild-type strain at pH 7.5. We also compared the growth to that obtained for the nonpathogenic S. cerevisiae (Fig. 3). As seen in Fig. 3A, C. albicans grew when 200 μM KH2PO4, GroPIns, or GroPCho was provided as the phosphate sources. Growth on GroPCho was just as robust as when KH2PO4 was supplied as the phosphate source after 24 and 48 h. Growth on GroPIns was somewhat slower, requiring 48 h for the cells to reach an OD600 of between 3 and 4. In contrast, S. cerevisiae grew at pH 7.5 when KH2PO4 was supplied but displayed little or no growth when GroPIns or GroPCho was supplied (Fig. 3B). Thus, C. albicans is clearly more adept than S. cerevisiae at using glycerophosphodiesters as phosphate sources at pH 7.5. Serum pH, typically between 7.3 and 7.5, it is often referred to as physiological pH.
Fig. 3.
C. albicans and S. cerevisiae vary in their ability to grow on glycerophosphodiesters at physiological pH. Wild-type C. albicans (DAY185) (A) and S. cerevisiae (BY4741) (B) strains were grown in YNB medium, buffered to pH 7.5, containing 200 μM GroPIns, 200 μM GroPCho, or 200 μM KH2PO4 as phosphate sources or containing no phosphate source. Values represent means ± SE of duplicate determinations. The experiment was repeated with similar results.
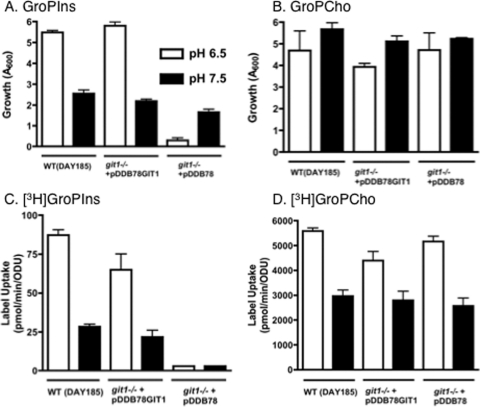
A direct comparison of the utilization of glycerophosphodiesters by C. albicans at pH 6.5 versus pH 7.5 is shown in Fig. 4A and B. These mildly acidic and mildly alkaline conditions could both be encountered by C. albicans in a mammalian host. For these experiments, cells were grown under low-phosphate conditions. First, it is clear that C. albicans reaches a lower optical density at pH 7.5 than at pH 6.5 when grown on GroPIns (Fig. 4A). At the same time, [3H]GroPIns uptake at pH 7.5 is lessened by approximately two-thirds compared to that by cells grown at pH 6.5 (Fig. 4C) and is dependent upon CaGIT1. Loss of CaGIT1 also results in the inability of cells to utilize GroPIns as a phosphate source at pH 6.5 (Fig. 4A), as expected from the plate results seen in Fig. 1. At pH 7.5, however, the growth that occurs on GroPIns after 48 h is primarily independent of CaGIT1. Thus, a CaGit1-independent mechanism exists for scavenging phosphate from GroPIns at pH 7.5.
Fig. 4.
GroPIns and GroPCho utilization by C. albicans at pH 6.5 versus pH 7.5. (A and B) Strains were grown in either YNB medium at pH 6.5 or YNB medium buffered to pH 7.5 containing 200 μM GroPIns (A) or 200 μM GroPCho (B) as sole phosphate sources. (C and D) Strains grown in low-Pi YNB medium and low-Pi YNB medium buffered to pH 7.5 were harvested, washed, and assayed for either GroPIns transport in the presence of 5 μM [3H]GroPIns (C) or GroPCho transport in the presence of 200 μM [3H]GroPCho (D), as described for standard transport assays. Values represent means ± SE of duplicate determinations. The experiment was repeated with similar results.
Interestingly, Fig. 4B shows that C. albicans grows equally well at pH 6.5 as at pH 7.5 when GroPCho is supplied as the phosphate source. At both pH 6.5 and pH 7.5, growth on GroPCho is independent of CaGIT1 (Fig. 4B). Although uptake of label from [3H]GroPCho into cells grown at pH 7.5 is decreased by about one-third compared to those grown at pH 6.5 (Fig. 4D), it is still quite high. These experiments were performed at 30°C, because the filamentation that occurs at pH 7.5 and 37°C causes the cells to flocculate and makes it difficult to get accurate optical density readings and repeatable transport assays. However, the following major findings were repeated at 37°C (data not shown): (i) CaGIT1 is required for liquid growth on GroPIns at pH 6.5, and (ii) CaGIT1 is not required for growth on GroPCho at either pH 6.5 or pH 7.5.
GroPIns kinetics, specificity, and proton dependence.
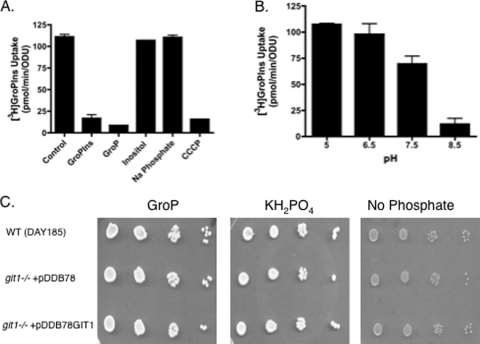
Since our data indicated that CaGit1 is a transporter for GroPIns, but not GroPCho, we focused the remainder of our studies on characterizing GroPIns transport through CaGit1. For analyzing the kinetic parameters and specificity of [3H]inositol- GroPIns uptake, cells were grown under low-Pi conditions on unbuffered YNB medium, the conditions under which GroPIns transport was shown to be greatest. Uptake of label from [3H]inositol-GroPIns conformed to Michaelis-Menton kinetics (Fig. 5), as expected for saturable carrier-mediated transport. The apparent Vmax for GroPIns transport was 960 ± 70 pmol/min/ODU, and the apparent Km was 28 ± 6 μM. This compares well with the apparent Km determined for GroPIns transport by ScGit1 (ca. 20 μM) (33). Transformation of the data (Fig. 5B) into a Hanes plot (19) results in a straight line, suggesting a single transport system under the conditions tested. These results provided added confidence that the observed label uptake from [3H]GroPIns was indeed the result of “transport.”
We next tested the transport specificity of CaGit1 by performing competition experiments. For these experiments, a 40-fold excess of unlabeled compounds was added to the transport assay and the effect upon GroPIns transport activity monitored (Fig. 6A). Compounds able to compete with GroPIns for transport through CaGit1 or to bind to the permease with some specificity should decrease transport activity. As expected, an excess of unlabeled GroPIns decreased the transport activity (Fig. 6A). Interestingly, glycerol-3-phosphate (GroP) also decreased transport activity. This finding suggested that GroP was transported by CaGit1 or that it bound to CaGit1 with enough affinity to inhibit GroPIns transport but that it was not transported by CaGit1. To address these possibilities, we first performed [3H]glycerol-GroP transport assays using our standard GroPIns transport conditions (data not shown), but we were unable to detect any activity. Next, we monitored the ability of C. albicans to utilize GroP as a phosphate source and found that it does but that the growth is not dependent on CaGIT1 (Fig. 6C). Thus, the most likely interpretation of these experiments is that GroP can compete with GroPIns for binding to CaGit1 but that GroP is not transported to an appreciable extent by CaGit1. Since GroP can clearly act as a phosphate source for C. albicans, there must be another mechanism for its utilization. Two possibilities are that GroP has a transporter distinct from CaGit1 or that GroP is hydrolyzed extracellularly to free phosphate and glycerol before being used as a phosphate source.
Fig. 6.
Specificity and proton dependence of GroPIns transport. (A and B) Wild-type strain (DAY185) was grown in low-Pi YNB medium to logarithmic phase, harvested, washed and assayed for transport activity in the presence of the indicated compounds at 1 mM for GroPIns, GroP, inositol, and sodium phosphate and 50 μM for CCCP (A) or as a function of increasing pH (B), as described in the text. (C) Strains grown in low-Pi medium were harvested, washed, and spotted in a series of 10× dilutions onto plates containing 200 μM GroP, KH2PO4, or no Pi. Values for panels A and B represent means ± SE of duplicate determinations. The experiments were repeated with similar results.
GroPIns transport was not affected by an excess of either unlabeled inositol or unlabeled phosphate, indicating that those compounds have little or no affinity for the permease. In particular, the fact that excess unlabeled inositol did not decrease label incorporation from [3H]inositol-GroPIns is evidence that our transport assays measure intact [3H]inositol-GroPIns transport and not the uptake of free [3H]inositol following hydrolysis of [3H]inositol-GroPIns outside the cell.
If CaGit1 is a proton symporter, as predicted by in silico analysis (17), the protonophore carbonyl cyanide 3-chlorophenylhydrazone (CCCP) should drastically reduce GroPIns transport activity, and it did (Fig. 6A). In addition, if proton motive force is important to transport, altering the pH of the assay buffer should affect transport activity. As shown in Fig. 6B, GroPIns transport activity decreased with increasing pH, consistent with CaGit1 being a proton symporter.
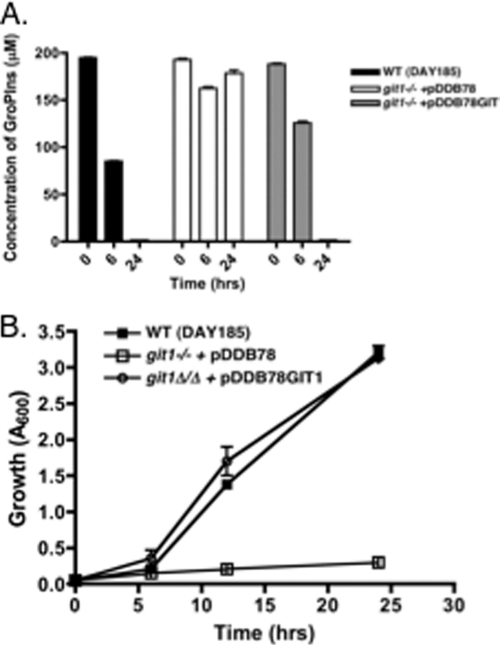
Depletion of GroPIns from the medium corresponds to the presence of CaGit1 and intact GroPIns detected intracellularly.
Further evidence for the transport of GroPIns across the plasma membrane was obtained by monitoring the levels of the metabolite in the medium by liquid chromatography-mass spectroscopy (LC-MS) as a function of growth. At time zero, cultures were spiked with 200 μM GroPIns, and after 6 and 24 h of growth, the GroPIns remaining in the medium was monitored by LC-MS (Fig. 7). The levels of GroPIns in the media of the wild-type strain and the homozygous insertion mutant bearing a reintegrated copy of CaGIT1 decreased with time (Fig. 7A) and cell growth (Fig. 7B). In contrast, the homozygous git1−/git1− mutant grew very little, and there was no decrease in the GroPIns peak. The fact that the levels of GroPIns in the medium remained the same in the git1−/git1− mutant is another indication that extracellular hydrolysis of the compound does not occur under these conditions.
Fig. 7.
Depletion of GroPIns from the medium as a function of growth. (A) The indicated strains were inoculated in YNB medium lacking KH2PO4 and containing 200 μM GroPIns. The GroPIns concentration in the medium was determined by LC-MS after 0, 6, and 24 h. Values represent means ± SE of triplicate determinations. (B) Growth curves of the strains.
As final confirmation that GroPIns is transported intact into the cell, we analyzed the labeled compounds in the intracellular fraction of the cell following a short-term transport assay with [3H]inositol-GroPIns. The internal water-soluble counts were extracted, separated, and identified, and the amount of label present in the particulate membrane fraction of the cell was also determined. Importantly, we detected intact [3H]GroPIns as 21% ± 6% of the internalized label. In addition, we detected free [3H]inositol (69% ± 6%), as would be expected if GroPIns is rapidly hydrolyzed once it enters the cell. Finally, we also detected 11% ± 2% of the label in the membrane fraction, presumably in phosphatidylinositol or the inositol-containing sphingolipids derived from phosphatidylinositol.
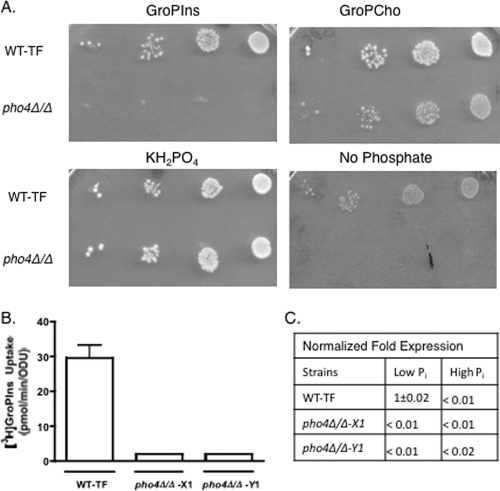
CaPHO4 is required for GroPIns transport, utilization, and expression.
In order to gain insight into the transcriptional regulation of CaGIT1, a library of transcription factor mutants (20) was screened. A total of 143 strains, each bearing homozygous deletions in a single transcriptional regulator, were assayed for growth when GroPIns was supplied as the phosphate source. Only one strain, that bearing a deletion in CaPHO4, displayed a clear inability to grow on GroPIns (Fig. 8 A). The deletion set contained two isolates of each strain, and the isolates behaved identically. The pho4Δ/Δ mutants were able to grow on GroPCho, although slightly less well than the wild type (Fig. 8A). Note that little or no background growth was seen in the pho4Δ/Δ mutant compared to the wild-type strain when no phosphate source was provided. This result is likely due to the fact that CaPHO4 is required to induce genes involved in mobilization of internal stores of phosphate and phosphate scavenging when phosphate is limiting, as is the case with S. cerevisiae (28, 36). As expected, transport assays confirmed that little or no transport activity exists in either pho4Δ/Δ mutant isolate (Fig. 8B). RT-PCR gene expression analysis was also performed on both pho4Δ/Δ isolates and the reference wild type (Fig. 8C). These results confirm that CaPho4 is required for CaGIT1 expression.
Fig. 8.
Effect of PHO4 deletion on growth and transport activity. (A) The wild-type strain and a pho4Δ/Δ deletion mutant were spotted in a series of 10× dilutions onto plates containing the indicated sources of phosphate at a concentration of 200 μM. A second pho4Δ/Δ isolate gave identical results. (B) Uptake activities of the wild type and the two pho4Δ/Δ deletion mutant isolates. Values represent means ± SE of triplicate determinations. The experiment was repeated with similar results. (C) Expression of CaGIT1 in the pho4Δ/Δ mutant and reference wild-type strain. Strains were grown on synthetic YNB supplemented with 200 μM (low Pi) or 10 mM (high Pi) KH2PO4. RNA was extracted and expression measured by quantitative real-time PCR analysis. Values were normalized to the CaTDH3 control and to 1. Values represent the means of triplicate determinations ± SE, where applicable.
DISCUSSION
Fungal cells scavenge nutrients from the environment to support their cellular activities. Potential nutrients can arise through the organism's own cellular activities or may be provided by the host environment. The glycerophosphodiesters produced through phospholipase-mediated hydrolysis of phospholipids are utilized by the nonpathogenic S. cerevisiae as sources of phosphate, inositol, and choline (1, 13, 35). Here we have shown that the pathogenic organism C. albicans is also capable of transporting and utilizing glycerophosphodiesters as nutrients, but a number of differences between the organisms are evident.
Whereas a single transporter is responsible for both GroPIns and GroPCho transport in S. cerevisiae (35), our data indicate that at least two glycerophosphodiester transporters exist in C. albicans, with CaGit1 being a GroPIns permease. CaGit1 is required for the utilization of GroPIns as a phosphate source on standard YNB solid and liquid media at pH 6.5. In addition, CaGit1 is required for GroPIns transport when cells are grown at both acidic (pH 6.5) and physiological (pH 7.5) pHs. The increased transport activity seen at lower pH is consistent with a recent study in which high-throughput sequencing of cDNA revealed much greater expression of CaGIT1 at pH 4 than at pH 8 (5). Although no GroPIns transport activity occurs in the absence of CaGit1, C. albicans is still able to use GroPIns as a phosphate source at pH 7.5, albeit slowly. Thus, a second, CaGit1-independent, mechanism must exist for utilizing GroPIns as a phosphate source at physiological pH. That mechanism may involve an unidentified transport activity with slow enough kinetics that it was not detected under the short-term transport assay employed here or, perhaps, external hydrolysis of GroPIns to liberate free phosphate. Indeed, starvation for phosphate is known to induce the production of extracellular phosphatases and phosphoesterases for hydrolyzing and scavenging phosphate in S. cerevisiae (28, 36), although none that hydrolyze GroPIns have been identified. Note that the extracellular liberation and subsequent transport of free phosphate that could theoretically occur at pH 7.5 would not be detected in our uptake assays, as the phosphate group of [3H]inositol-GroPIns is not labeled.
Several lines of evidence support the notion that GroPIns is transported intact across the plasma membrane via CaGit1, even though a second, CaGit1-independent, mechanism also exists for acquiring phosphate from GroPIns at pH 7.5. First, competition assays indicate that neither free inositol nor free phosphate, potential hydrolysis products of GroPIns, has significant affinity for the permease. Although GroP, another potential hydrolysis product of GroPIns, did compete for transport activity, we were unable to detect GroP transport (data not shown), and the ability of GroP to support growth was not dependent on CaGIt1. Thus, GroP likely has affinity for the transporter but is not a substrate for it. Second, the ability of cells to utilize GroPIns as a phosphate source correlates with the depletion of the compound from the medium as measured by LC-MS. Importantly, the LC-MS data also show that GroPIns is stable in the medium at pH 6.5, since GroPIns levels did not change, even after 24 h of incubation in medium inoculated with a git1−/git1− mutant. Finally, we detected intact GroPIns in the intracellular fraction of the cell following a short-term transport assay.
Several aspects of uptake of label from [3H]GroPCho by C. albicans are worth noting. First, uptake of label from [3H]GroPCho is much greater (approximately 50-fold) than that from [3H]GroPIns. Also, when comparing uptake between S. cerevisiae and C. albicans, an enormous difference exists, with the activity of C. albicans being approximately 500-fold greater when the organisms are grown under identical conditions. The fact that C. albicans has three remaining ORFs with high similarity to CaGIT1 leads to the possibility that one or more of those may be involved in GroPCho transport. It is also possible that other, unidentified, transporters are involved.
Not unexpectedly, phosphate levels regulate GroPIns transport activity, as they do in S. cerevisiae (1). In addition, we identified CaPHO4, a homolog of ScPHO4, as being required for the transport and utilization of GroPIns as a phosphate source. The pho2Δ/Δ mutant was not picked up in our screen, and CaPHO2 was not required for growth on GroPIns when tested individually (data not shown). In S. cerevisiae, ScPHO2 and ScPHO4 are both involved in the transcriptional regulation of a number of phosphate-responsive genes (28, 36), including ScGIT1. However, others have shown that CgPHO2 is not important for the transcriptional response to low phosphate in Candida glabrata and, specifically, for the induction of the C. glabrata homolog of GIT1 (23). Interestingly, a study in which the property differences of the major clades of C. albicans were investigated found an association between phosphate-related metabolism and virulence (27). Specifically, CaGIT1 (numbered orf19.34 but not named in the paper) was one of 18 genes, 5 involved in phosphate metabolism, whose expression profile differed significantly in isolates of high, medium, and low virulence (27), being the highest in the most virulent strains.
Our results show that C. albicans has an expanded ability to transport and utilize glycerophosphodiesters compared to S. cerevisiae. Notably, C. albicans is able to use GroPIns and GroPCho as phosphate sources at both acidic pH and physiological pH (the approximate pH of serum, pH 7.5), whereas S. cerevisiae does so only marginally at pH 7.5. This fact gains importance when considering that C. albicans may be exposed to a range of pHs, from acidic to alkaline, in its mammalian host (7, 12). In fact, when considering the gastrointestinal tract alone, pH can vary from pH 2 in the stomach to pH 6 to 7.4 in the intestine and terminal ileum to pH 6.7 in the rectum (12). The vagina is also considered to be an acidic environment. Finally, human saliva can range from pH 6.5 to pH 7.5 (46). Thus, our focus on mildly acidic (pH 6.5) and mildly basic (pH 7.5) conditions is relevant to conditions that could be encountered during infection. Another expansion of abilities when comparing glycerophosphodiester utilization between the two organisms is not only that GroPIns and GroPCho utilization occurs via separate mechanisms in C. albicans but that those mechanisms are regulated differentially by both pH and phosphate availability. Like pH, phosphate levels are likely to vary in the human host. For example, while phosphate levels in a healthy human serum are normally between 0.8 and 1.45 mM, those levels can drop to 0.3 mM and lower (the low-phosphate range for the experiments performed here), depending upon diet and various disease states that induce hypophosphatemia (14). Taken together, our findings lead us to speculate that the expanded ability of C. albicans to utilize GroPIns and GroPCho results from the organism's pathogenic nature and its need to occupy a variety of environments within its host organism. This possibility is buttressed by the fact that GroPIns and GroPCho are present and abundant in human fluids, as mentioned in the introduction (2, 6, 15, 16, 24, 31, 38, 42–44).
Future studies will focus on identifying the transporter(s) responsible for GroPCho utilization and on determining the importance of glycerophosphodiester metabolism in C. albicans virulence and survival in a mammalian host.
ACKNOWLEDGMENTS
We thank Qi Zhao and William C. Nierman (TIGR) and Frank J. Smith and Aaron P. Mitchell (Carnegie Mellon University) for the gift of plasmid CAGFN83. We thank Claudia Almaguer and Beth Surlow for technical assistance. The pho4Δ/Δ-X1 and pho4Δ/Δ-Y1 strains (obtained from the FGSC) were prepared by Oliver Homann.
We thank the National Science Foundation for providing support (MRIDBI-0821401) toward purchase of mass spectrometers. NIH grant 1R01AI057804 supported our use of plasmid CAGFN83.
Footnotes
Published ahead of print on 7 October 2011.
REFERENCES
- 1. Almaguer C., Cheng W., Nolder C., Patton-Vogt J. 2004. Glycerophosphoinositol, a novel phosphate source whose transport is regulated by multiple factors in Saccharomyces cerevisiae. J. Biol. Chem. 279:31937–31942 [DOI] [PubMed] [Google Scholar]
- 2. Backshall A., et al. 2009. Detection of metabolic alterations in non-tumor gastrointestinal tissue of the Apc(Min/+) mouse by (1)H MAS NMR spectroscopy. J. Proteome Res. 8:1423–1430 [DOI] [PubMed] [Google Scholar]
- 3. Blankenship J. R., Fanning S., Hamaker J. J., Mitchell A. P. 2010. An extensive circuitry for cell wall regulation in Candida albicans. PLoS Pathog. 6:e1000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Braun B. R., et al. 2005. A human-curated annotation of the Candida albicans genome. PLoS Genet. 1:36–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bruno V. M., et al. 2010. Comprehensive annotation of the transcriptome of the human fungal pathogen Candida albicans using RNA-seq. Genome Res. 20:1451–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Corda D., Iurisci C., Berrie C. P. 2002. Biological activities and metabolism of the lysophosphoinositides and glycerophosphoinositols. Biochim. Biophys. Acta 1582:52–69 [DOI] [PubMed] [Google Scholar]
- 7. Davis D. 2003. Adaptation to environmental pH in Candida albicans and its relation to pathogenesis. Curr. Genet. 44:1–7 [DOI] [PubMed] [Google Scholar]
- 8. Davis D. A., Bruno V. M., Loza L., Filler S. G., Mitchell A. P. 2002. Candida albicans Mds3p, a conserved regulator of pH responses and virulence identified through insertional mutagenesis. Genetics 162:1573–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dowd S. R., Bier M. E., Patton-Vogt J. L. 2001. Turnover of phosphatidylcholine in Saccharomyces cerevisiae. The role of the CDP-choline pathway. J. Biol. Chem. 276:3756–3763 [DOI] [PubMed] [Google Scholar]
- 10. Elion E. A., Warner J. R. 1984. The major promoter element of rRNA transcription in yeast lies 2 kb upstream. Cell 39:663–673 [DOI] [PubMed] [Google Scholar]
- 11. Enloe B., Diamond A., Mitchell A. P. 2000. A single-transformation gene function test in diploid Candida albicans. J. Bacteriol. 182:5730–5736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fallingborg J. 1999. Intraluminal pH of the human gastrointestinal tract. Dan. Med. Bull. 46:183–196 [PubMed] [Google Scholar]
- 13. Fisher E., Almaguer C., Holic R., Griac P., Patton-Vogt J. 2005. Glycerophosphocholine-dependent growth requires Gde1p (YPL110c) and Git1p in Saccharomyces cerevisiae. J. Biol. Chem. 280:36110–36117 [DOI] [PubMed] [Google Scholar]
- 14. Gaasbeek A., Meinders A. E. 2005. Hypophosphatemia: an update on its etiology and treatment. Am. J. Med. 118:1094–1101 [DOI] [PubMed] [Google Scholar]
- 15. Gallazzini M., Burg M. B. 2009. What's new about osmotic regulation of glycerophosphocholine. Physiology 24:245–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gallazzini M., Ferraris J. D., Burg M. B. 2008. GDPD5 is a glycerophosphocholine phosphodiesterase that osmotically regulates the osmoprotective organic osmolyte GPC. Proc. Natl. Acad. Sci. U. S. A. 105:11026–11031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gaur M., et al. 2008. MFS transportome of the human pathogenic yeast Candida albicans. BMC Genomics 9:579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hama H., Takemoto J. Y., DeWald D. B. 2000. Analysis of phosphoinositides in protein trafficking. Methods 20:465–473 [DOI] [PubMed] [Google Scholar]
- 19. Hanes C. S. 1932. Studies on plant amylases: The effect of starch concentration upon the velocity of hydrolysis by the amylase of germinated barley. Biochem. J. 26:1406–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Homann O. R., Dea J., Noble S. M., Johnson A. D. 2009. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 5:e1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hoover C. I., Jantapour M. J., Newport G., Agabian N., Fisher S. J. 1998. Cloning and regulated expression of the Candida albicans phospholipase B (PLB1) gene. FEMS Microbiol. Lett. 167:163–169 [DOI] [PubMed] [Google Scholar]
- 22. Ibrahim A. S., et al. 1995. Evidence implicating phospholipase as a virulence factor of Candida albicans. Infect. Immun. 63:1993–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kerwin C. L., Wykoff D. D. 2009. Candida glabrata PHO4 is necessary and sufficient for Pho2-independent transcription of phosphate starvation genes. Genetics 182:471–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Klein J., Gonzalez R., Koppen A., Loffelholz K. 1993. Free choline and choline metabolites in rat brain and body fluids: sensitive determination and implications for choline supply to the brain. Neurochem. Int. 22:293–300 [DOI] [PubMed] [Google Scholar]
- 25. Kohler G. A., et al. 2006. Phospholipase A2 and phospholipase B activities in fungi. Biochim. Biophys. Acta 1761:1391–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leidich S. D., et al. 1998. Cloning and disruption of caPLB1, a phospholipase B gene involved in the pathogenicity of Candida albicans. J. Biol. Chem. 273:26078–26086 [DOI] [PubMed] [Google Scholar]
- 27. MacCallum D. M., et al. 2009. Property differences among the four major Candida albicans strain clades. Eukaryot. Cell 8:373–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mouillon J. M., Persson B. L. 2006. New aspects on phosphate sensing and signalling in Saccharomyces cerevisiae. FEMS Yeast Res. 6:171–176 [DOI] [PubMed] [Google Scholar]
- 29. Mukherjee P. K., Chandra J., Kuhn D. M., Ghannoum M. A. 2003. Differential expression of Candida albicans phospholipase B (PLB1) under various environmental and physiological conditions. Microbiology 149:261–267 [DOI] [PubMed] [Google Scholar]
- 30. Mukherjee P. K., et al. 2001. Reintroduction of the PLB1 gene into Candida albicans restores virulence in vivo. Microbiology 147:2585–2597 [DOI] [PubMed] [Google Scholar]
- 31. Paban V., Fauvelle F., Alescio-Lautier B. 2010. Age-related changes in metabolic profiles of rat hippocampus and cortices. Eur. J. Neurosci. 31:1063–1073 [DOI] [PubMed] [Google Scholar]
- 32. Pao S. S., Paulsen I. T., Saier M. H., Jr 1998. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62:1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Patton J. L., Pessoa-Brandao L., Henry S. A. 1995. Production and reutilization of an extracellular phosphatidylinositol catabolite, glycerophosphoinositol, by Saccharomyces cerevisiae. J. Bacteriol. 177:3379–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Patton-Vogt J. 2007. Transport and metabolism of glycerophosphodiesters produced through phospholipid deacylation. Biochim. Biophys. Acta 1771:337–342 [DOI] [PubMed] [Google Scholar]
- 35. Patton-Vogt J. L., Henry S. A. 1998. GIT1, a gene encoding a novel transporter for glycerophosphoinositol in Saccharomyces cerevisiae. Genetics 149:1707–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Persson B. L., et al. 2003. Regulation of phosphate acquisition in Saccharomyces cerevisiae. Curr. Genet. 43:225–244 [DOI] [PubMed] [Google Scholar]
- 37. Saier M. H., Jr., Tran C. V., Barabote R. D. 2006. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 34:D181–D186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Senar S., Recio M. N., Perez-Albarsanz M. A. 1994. Lindane affects phosphoinositide turnover through a different mechanism of the phosphatidylinositol synthesis inhibition in rat renal proximal tubule cell culture. Cell Signal. 6:433–438 [DOI] [PubMed] [Google Scholar]
- 39. Spreghini E., Davis D. A., Subaran R., Kim M., Mitchell A. P. 2003. Roles of Candida albicans Dfg5p and Dcw1p cell surface proteins in growth and hypha formation. Eukaryot. Cell 2:746–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sugiyama Y., et al. 1999. Molecular cloning of a second phospholipase B gene, caPLB2 from Candida albicans. Med. Mycol. 37:61–67 [PubMed] [Google Scholar]
- 41. Theiss S., et al. 2006. Inactivation of the phospholipase B gene PLB5 in wild-type Candida albicans reduces cell-associated phospholipase A2 activity and attenuates virulence. Int. J. Med. Microbiol. 296:405–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Walter A., et al. 2004. Glycerophosphocholine is elevated in cerebrospinal fluid of Alzheimer patients. Neurobiol. Aging. 25:1299–1303 [DOI] [PubMed] [Google Scholar]
- 43. Wang Y., et al. 2007. Topographical variation in metabolic signatures of human gastrointestinal biopsies revealed by high-resolution magic-angle spinning 1H NMR spectroscopy. J. Proteome Res. 6:3944–3951 [DOI] [PubMed] [Google Scholar]
- 44. Wang Y., et al. 2005. Biochemical characterization of rat intestine development using high-resolution magic-angle-spinning 1H NMR spectroscopy and multivariate data analysis. J. Proteome Res. 4:1324–1329 [DOI] [PubMed] [Google Scholar]
- 45. Wilson R. B., Davis D., Mitchell A. P. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yosipovitch G., et al. 2001. Distribution of mucosal pH on the bucca, tongue, lips and palate. A study in healthy volunteers and patients with lichen planus, Behcet's disease and burning mouth syndrome. Acta Derm. Venereol. 81:178–180 [DOI] [PubMed] [Google Scholar]