Abstract
The “secretory” Na+-K+-2Cl− cotransporter, NKCC1, belongs to the SLC12 gene family of electroneutral cation-chloride cotransporters. A number of these proteins, including NKCC1 itself, exist as homodimers in the membrane suggesting that this may be a common feature of the SLC12 family. We have previously demonstrated that replacing the C-terminus of NKCC1 with that of its close homologue NKCC2 produced a fully functional chimeric protein that formed homodimers but did not dimerize with NKCC1. Here we employ a novel co-immunoprecipitation assay to study the dimerization interaction of NKCC1 using additional NKCC1/NKCC2 C-terminal chimeras and point mutants. Our results indicate that the substitution of a number of regions of the C-terminus of NKCC1 with the corresponding sequence from NKCC2 results in weakened dimerization with wild-type NKCC1, demonstrating that various residues play a role in this interaction. Most interestingly, however, we find that the replacement of a single NKCC1 residue, G812, with cysteine, the corresponding amino acid in NKCC2, results in a point mutant that displays no significant dimerization with the wild-type protein. In addition to this effect on heterodimer formation we also find that G812 mutants can nevertheless form homodimers but that this interaction can be weaker than that observed for wild-type NKCC1. We demonstrate that our results are consistent with at least one established mechanism of protein dimer formation, that of “domain swapping”, as well as with a recently reported crystal structure of the C-terminus of a bacterial SLC12 homologue.
Keywords: membrane protein structure, cation-chloride cotransporter, protein dimers, SLC12A2
Members of the SLC12 gene family of electroneutral cation-chloride-coupled cotransporters were first identified at the molecular level in fish and then in mammals (1-3). Homologues have since been found in crustaceans, insects, worms, plants, fungi and some bacteria. In vertebrates there are nine family members (4), two Na+-K+-2Cl− cotransporters (NKCC1 and NKCC2), a Na+-Cl− cotransporter (NCC), four K+-Cl− cotransporters (KCC1, KCC2, KCC3 and KCC4), and two additional homologues of unknown function. SLC12 family members are known to play important roles in numerous physiological processes including exocrine fluid secretion, renal salt and water absorption, hearing, olfaction, spermatogenesis, regulation of blood pressure, pain perception, visual processing and other neuronal functions (4-12).
Experiments from our laboratory published in 2000 (13) established that the secretory Na+-K+-2Cl− cotransporter NKCC1 exists as a homodimer in the plasma membrane and that this dimer is sufficiently stable that it remains intact after membrane solubilization in mild detergents. Subsequent studies from other groups provided evidence that NKCC2 (14), NCC (15) and the KCCs (16-18) also occur as dimers suggesting that this may be a common feature of the SLC12 family. More recently we have demonstrated that the cytosolic 50 kDa C-terminus of NKCC1 is essential for dimer formation (19). In these experiments we showed that NKCC1 molecules lacking their C-termini failed to dimerize and that replacing the C-terminus of NKCC1 with that of its close homologue NKCC2 produced a fully functional chimeric protein that formed homodimers but did not dimerize with NKCC1. Using additional chimeras we showed that the residues required for dimer formation lie between amino acids 751 and 998 of rat NKCC1. In the present paper we have continued our studies of the role of the NKCC1 C-terminus in dimerization by systematically substituting NKCC1 C-terminal amino acids with the corresponding residues of NKCC2. Our results show that dimerization is a complex interaction apparently involving multiple regions of the NKCC1 C-terminus. But interestingly we also find that the relatively conservative substitution of a single NKCC1 amino acid (G812 in rat NKCC1) results in a fully functional point mutant that homodimerizes but has no detectable interaction with wild-type NKCC1. As discussed later in the paper, in addition to establishing a central role for this residue in the dimerization interaction, our observations also impose significant constraints on the configuration of the dimerization interface.
Methods and Materials
Materials
The homobifunctional, water-soluble, amino cross-linking reagent DTSSP (3,3′-Dithiobis(sulfosuccinimidylpropionate)) was purchased from Pierce. The rat NKCC2 clone (2) was kindly provided by Dr. Gerardo Gamba, Universidad Nacional Autonoma de Mexico. The antibody α-wCT(r) was raised in rabbits against the recombinant C-terminus of rat NKCC1 (20); the antibody α-wCT(g) was raised in goat against the same antigen. The antibody α-wNT(r) was raised in rabbits against the recombinant N-terminus (amino acids 3-202) of rat NKCC1 (21).
DNA Constructs
The mammalian expression vector pBK-CMVlac− (pBK−; (13)) was used for all expression studies. Full length rat NKCC1 in Bluescript SK (22) was cloned between the EcoRI and XhoI sites of pBK− in which the BamHI site of the multiple cloning site had been destroyed by blunt end ligation (19). The N-terminally truncated rat NKCC1 (nttNKCC1) in pBK− employed here, whose coding sequence begins at M209, has been described previously (13;19).
Chimeras in which regions of the C-terminus of rat NKCC1 (amino acids 750-1203) were replaced by the corresponding amino acids of rat NKCC2 are named according to the replaced NKCC1 amino acids. Thus the chimera in which the entire NKCC1 C-terminus is replaced by that of NKCC2 is called ‘aa750-1203’ (this chimera was previously described in ref. (19) where it was referred to as “1/2”). To construct additional chimeras the wild-type NKCC1 sequence was modified using the Quikchange site-directed mutagenesis kit (Stratagene) to incorporate a unique BspEI site at R912. This had the effect of introducing the mutation L913I into the NKCC1 sequence (coincidentally I is the corresponding amino acid at this site in rat NKCC2). In control experiments (not shown) we have confirmed that the mutation L913I has no detectable effect on the dimerization properties of NKCC1. The chimeras aa750-912, aa913-998 and aa999-1203 were obtained by ligating suitable PCR products between the restriction sites BamHI (at G750) and BspEI, BspEI and HindIII (at K998), and HindIII and XhoI (located at the end of the NKCC1 3′UTR), respectively. Additional replacements of amino acid regions between residues 750 and 912 were obtained via PCR-mediated gene splicing by overlap extension (23) to generate a BamHI/BspE1 fragment that was then ligated into the NKCC1 clone incorporating the BspE1 site. Mutations of single residues or pairs of residues were made with the Quikchange kit. To the extent possible chimeric boundries were located in regions of high homology between NKCC1 and NKCC2 so as to obtain seamless or near-seamless junctions (the rat NKCC1 and NKCC2 C-termini are 56% identical). All chimeras and mutants were sequenced to confirm that they were correct.
Cell culture and transient transfection
HEK293 cells were cultured in Dulbecco's Modified Essential Medium supplemented with 2 mM glutamine, 100 μg/ml each of penicillin and streptomycin (all from Biofluids), and 10% heat inactivated fetal bovine serum (GibcoBRL). Cells were grown in 10 cm plastic dishes in a humidified incubator at 37°C and 5% CO2, and subcultured every 3-4 days. Subconfluent (∼80%) HEK293 cells were transiently transfected overnight (19-24h) with expression vectors using FuGENE 6 (Roche) according to the manufacturer's instructions.
Membrane Preparations and chemical cross-linking
Crude membranes from transiently transfected HEK-293 cells were prepared as previously described (19). In the cross-linking studies (see ref. (19) for details) crude membranes (1 mg protein/ml) were solubilized in 0.3% Triton X-100 and centrifuged at 100000×g for 15 min, then the covalent cross-linker DTSSP was added at a concentration of 1 mM. After 30 min the cross-linking reaction was terminated and the samples were analyzed by Western blotting using the antibody α-wNT(r).
Co-immunoprecipitation assay
The co-immunoprecipitation assay is described in detail in reference (19). Briefly, HEK293 cells in 6 cm dishes were transiently transfected with nttNKCC1 and full length NKCC1, or the chimeras indicated, at a plasmid:plasmid ratio of 8:1 (3 μg total DNA). The following day the cells were solubilized in PBS containing 0.3% Triton X-100, centrifuged at 100000×g for 30 min to remove insoluble material, and immunoprecipitated using the antibody α-wNT(r) which had been preconjugated to protein G beads (Pierce). In preliminary experiments (not shown) we have confirmed that the amount of cell lysate (185 μl) used in these experiments was more than sufficient to saturate the beads. Following incubation with the antibody the beads were eluted with PBS containing 0.3% Triton X-100 and 0.1% SDS, and the eluate was probed with the antibody α-wCT(g) via Western blotting. As discussed in more detail in Results, we have previously shown that treatment with 0.1% SDS is sufficient to break dimer pairs (13, 19). Subsequent elution of the beads with SDS-PAGE sample buffer (containing 2% SDS and 100 mM DTT) was found to release considerable bound full length NKCC1 (not shown) indicating that 0.1% SDS did not break the binding of the precipitating antibody, α-wNT(r), to the NKCC1 N-terminus. In general the level of expression of all chimeras and point mutants was comparable to that of wild-type NKCC1 (not shown).
SDS-PAGE and Western Blotting
SDS-PAGE and Western blotting were carried out using 4-12% Tris-glycine gels (Bio-Rad) as previously described (19). The positions of Multi Mark colored standards (Invitrogen) are indicated on the blots.
86Rb flux assay
The 86Rb flux assay was performed as previously described (24).
Data Presentation
All experiments were carried out 3 or more times (unless otherwise noted) with similar findings. Quantitative results are expressed as means ± S.E.M.
Results
C-terminal amino acids are responsible for NKCC1 dimerization
In previous work from our laboratory (13;19) we have shown that NKCC1 exists as a homodimer in the plasma membrane that is stable after membrane solubilization in mild non-ionic detergents but is disrupted by low concentrations of SDS (< 0.1%). We also showed that this dimerization interaction occurs between the 50 kDa cytosolic C-termini of the dimer subunits (19). Our initial studies were carried out with the chemical cross-linking reagent DTSSP (see Methods). In these experiments (13) we demonstrated that after cross-linking NKCC1 migrated on SDS-PAGE gels at a molecular weight that was twice its monomeric size indicating its status as a dimer. A similar result was found with an N-terminally truncated NKCC1 (nttNKCC1) demonstrating that the N-terminus of NKCC1 does not play a significant role in its dimerization. However, deletion of the NKCC1 C-terminus resulted in a protein that failed to cross-link with DTSSP and thus apparently failed to dimerize (19).
To study NKCC1 dimerization in more detail we subsequently developed a novel co-immunoprecipitation assay (19). Briefly stated (see Methods for details), in this assay we transiently co-express full-length NKCC1 and nttNKCC1 in HEK293 cells, immunoprecipitate from solubilized cells with the antibody α-wNT(r) raised against the NKCC1 N-terminus, then treat the immunoprecipitated material with 0.1% SDS to specifically break dimer pairs. Since the sequence against which the antibody α-wNT(r) was raised is missing from nttNKCC1 (see Methods), this truncated protein should only appear in the immunoprecipitate as a component of full-length NKCC1/nttNKCC1 dimer pairs from which it will be released by 0.1% SDS. Thus by modifying the C-terminus of the full length protein and probing for nttNKCC1 in the SDS-eluted material we are able to evaluate the effects of these modifications on dimer formation. Using this assay we demonstrated that when the C-terminus of NKCC1 was replaced with the C-terminus of its close homologue NKCC2 the resulting protein was unable to immunoprecipitate nttNKCC1 ((19); see also below). However, this NKCC1/NKCC2 chimera was able to form homodimers (as demonstrated by cross-linking studies with DTSSP) and was fully functional (19). Taken together with our other results these observations confirmed that an interaction between the C-termini of the two dimer subunits is responsible for NKCC1 dimerization and indicated that the C-termini of NKCC1 and NKCC2 are not capable of this interaction with each other.
In what follows we have systematically replaced sequence in the C-terminus of NKCC1 with the corresponding sequence of NKCC2 in order to localize and identify the amino acids involved in NKCC1 dimerization.
NKCC1/NKCC2 chimeras
Fig. 1 shows the results of the above co-immunoprecipitation assay for a series of NKCC1/NKCC2 chimeras. In Fig. 1A we illustrate the C-terminal sequence of each of these chimeras schematically, indicating NKCC1 sequence in white and NKCC2 sequence in black. Each chimera is named according to the NKCC1 sequence that has been replaced with NKCC2 sequence (see Figure 1 caption). In control experiments (not shown) we have verified that these chimeras, and all other NKCC1/NKCC2 chimeras studied here, are expressed and complex-glycosylated at levels comparable to wild-type NKCC1 and form homodimers with themselves (as determined via crosslinking studies employing DTSSP). In Fig. 1B we show the results of a typical experiment where we have cotransfected HEK-293 cells with nttNKCC1 and each of the full-length proteins described in Fig. 1A. In each case the material eluted by 0.1% SDS from the α-wNT(r) immunoprecipitate was probed by Western blotting with the antibody α-wCT(g) against the NKCC1 C-terminus, which recognizes both nttNKCC1 and wild-type NKCC1. As previously shown, nttNKCC1 is co-immunoprecipitated with wild-type NKCC1 but a much reduced band is seen with the chimera aa750-1203 in which the complete NKCC1 C-terminus is replaced by that of NKCC2. It is also clear from Fig. 1B that nttNKCC1 is readily co-immunoprecipitated with aa913-998 and aa999-1203 but that the co-precipitated nttNKCC1 signal with aa750-912 is much weaker. The result seen with aa999-1203 confirms that of a complimentary experiment shown in Fig. 6 of ref. (19) where we replaced amino acids 999-1203 in nttNKCC1 with NKCC2 sequence and co-immunoprecipitated with wild-type NKCC1.
Figure 1. Co-immunoprecipiation of nttNKCC1 with various NKCC1/NKCC2 chimeras.
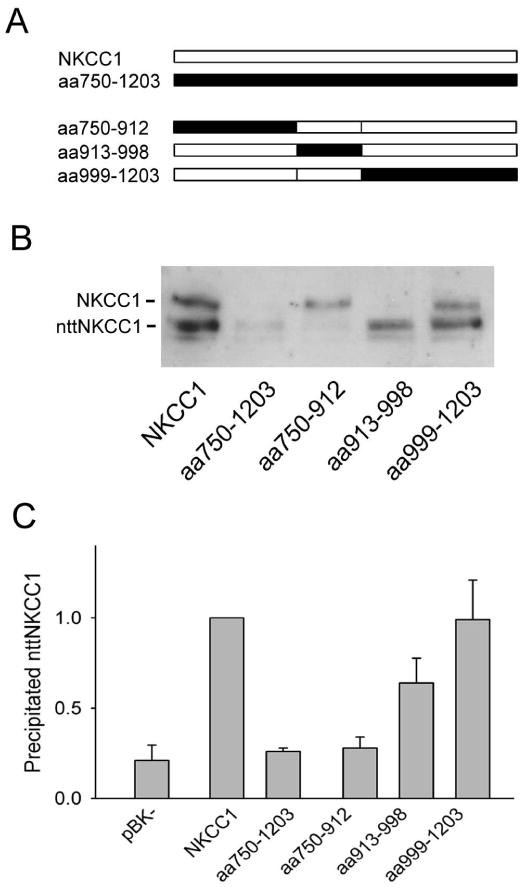
A. Schematic representations of the C-terminal sequence (amino acids 750-1203) of each of the NKCC1/NKCC2 chimeras studied in panels B and C. NKCC1 sequence is shown in white and NKCC2 sequence in black. Chimeras are named according to the NKCC1 sequence replaced by the corresponding sequence of NKCC2; thus, for example, the chimera aa750-1203 has the complete NKCC1 C-terminus replaced by that of NKCC2. B. HEK293 cells were transiently transfected with nttNKCC1 and the chimera indicated. The next day cells were solubilized and immunoprecipitated with α-wNT(r), the precipitate was eluted with 0.1% SDS, and the eluate was probed via Western blotting with the antibody α-wCT(g). See Methods for details. The figure shows the results of a typical Western blot with the positions of NKCC1 and nttNKCC1 indicated. C. Quantitated results of nttNKCC1 co-immunoprecipitation with the NKCC1/NKCC2 chimeras indicated. The results have been normalized to the nttNKCC1 signal observed with NKCC1 on the same day. The background signal seen when the full length clones are replaced with the empty vector pBK− is also shown. Each data point represents the mean ± S.E. for 3 or more independent experiments.
The experiments shown in Fig. 1B have been quantitated in Fig. 1C. These quantitated results show that the nttNKCC1 signals seen with aa750-1203 and aa750-912 are not significantly different from that seen when the empty vector pBK− is used in place of NKCC1 and thus represent a background signal. These experiments are therefore consistent with the hypothesis that there are amino acids required for the dimerization of NKCC1 occurring between residues 750-912 that cannot be effectively replaced by the corresponding amino acids of NKCC2. We also note that the nttNKCC1 signal seen with the chimera aa913-998 is significantly less than that seen with wild-type NKCC1 indicating a weakened dimerization interation. Thus these latter amino acids apparently also play a role, albeit not an essential one, in NKCC1 dimerization.
In order to further localize the amino acids involved in NKCC1 dimerization between residues 750-912 we systematically subdivided this region into the successive pairs of additional chimeras illustrated in Fig. 2A and assayed them for their ability to dimerize with nttNKCC1 (Fig. 2B). In each case one member of the pair was found to yield an nttNKCC1 signal that was not significantly different from that observed with aa750-1203 indicating that it contained important amino acids for dimerization that could not be replaced by the corresponding residues of NKCC2. This sequence was then subdivided to produce the next pair of chimeras, and so on. In this way we were able to localize the essential amino acids for nttNKCC1 co-immunoprecipitation to residues 806-814 (Fig. 2B). We also note that a number of other chimeras (aa842-912, aa825-841, aa815-825) display positive but weakened nttNKCC1 signals relative to wild-type NKCC1 indicating that these amino acids also play some role in the strength of the dimerization interaction.
Figure 2. Localization of important amino acids for NKCC1 dimerization.
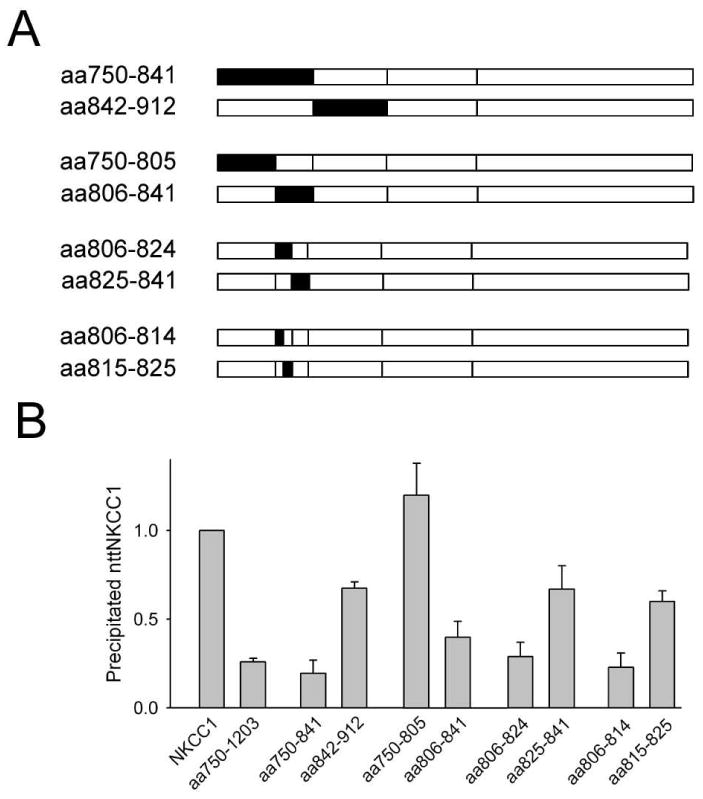
A. Schematic representations of the C-terminal sequences of the NKCC1/NKCC2 chimeras studied in panel B (see Fig. 1 caption for details). B. Co-immunoprecipitation of nttNKCC1 with the chimeras indicated was determined and quantitated as described in the caption of Fig. 1. Each data point represents the mean ± S.E. for 3 or more independent experiments.
The functions of some of the chimeric proteins studied in Figs. 1 and 2 are explored in Fig. 3. Here we show the bumetanide-sensitive (i.e., NKCC-specific) component of 86Rb influx in transiently transfected HEK293 cells. Transfection with NKCC1 itself yields a ∼6-fold increase in 86Rb uptake over the basal level observed with the empty vector pBK−. All of the other constructs studied in Fig. 3 show some level of specific functional activity. Interestingly, however, while replacing the entire NKCC1 C-terminus with that of NKCC2 (chimera aa750-1203) has little effect on functional activity, many of the replacements of smaller regions of NKCC1 C-terminal sequence markedly reduce bumetanide-sensitive 86Rb uptake. Thus these results suggest that multiple interactions within the NKCC1 C-terminus may play significant roles in the function of this protein. Further exploration of this apparently complex role of the NKCC1 C-terminus in its functional expression is beyond the scope of the present paper.
Figure 3. Bumetanide-sensitive 86Rb fluxes via NKCC1/NKCC2 chimeras.
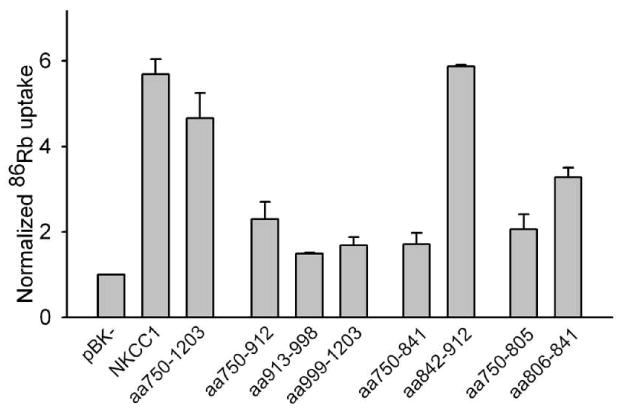
86Rb fluxes were measured as previously described (24) in HEK293 cells transiently transfected with the NKCC1 constructs indicted. Flux was measured in the presence and absence of 250 μM bumetanide. For each chimera the bumetanide-sensitive component of 86Rb flux was calculated and normalized to that measured in cells transfected with pBK− on the same day. For aa750-912, aa913-998 and aa999-1203 n=2, for all others n≥3. The results for NKCC1, aa750-1203 and aa999-1203 were taken from Fig. 7 of ref. (19) and are included for comparison purposes.
As already mentioned, all of the chimeras studied in Figs. 1 and 2 are normally expressed and processed by the HEK293 cells and are able to form homodimers. Inspection of the results of Figs. 1-3 indicates that there is no apparent relationship between the functional activities of these chimeras and their abilities to dimerize with nttNKCC1; for example, the chimeras aa913-998 and aa842-912 co-precipitate similar amounts of nttNKCC1 but the 86Rb transport activity of the former is relatively low and that of the latter relatively high. Thus taken together these results support the view that the differences illustrated in Figs. 1 and 2 do indeed reflect the abilities of these chimeras to interact with nttNKCC1 and not non-specific differences related to their handling and processing by the HEK293 cells.
Point mutations of NKCC1 residues 806-814
The chimera aa806-814 contains only 9 NKCC2 residues, five of which are in fact identical to the corresponding residues in NKCC1 (Fig. 4A). In Fig. 4B we show the effects of the individual amino acid differences in residues 806-814 between NKCC1 and NKCC2 on dimerization. Each of the mutations V806S, G812C, and H813E have significant effects on the ability of the resulting full length NKCC1 point mutant to co-immunoprecipitate nttNKCC1, and the signal seen with G812C is not statistically significantly different from that found with aa750-1203, i.e., from background levels (see above). The effects of additional point mutations of G812 and H813 are shown in Fig. 4C. None of the nttNKCC1 signals seen with G812S, G812T, GH-SE, GH-CE or GH-CC are significantly different from that seen with aa750-1203 demonstrating that even relatively conservative mutations of G812 result in proteins that have a dramatically reduced ability to dimerize with nttNKCC1. In additional studies (not shown) we also generated and characterized the full length mutants G812A and G812P, but cross-linking studies with DTSSP indicated that both these proteins were significantly aggregated (not shown) and thus misprocessed in the HEK293 cells. We also found that G812P was not complex-glycosylated indicating that it was retained in the endoplasmic reticulum. Thus co-immunoprecipitation studies with these mutants could not be interpreted.
Figure 4. Effects of point mutations of NKCC1 amino acids 806-814 on dimerization.
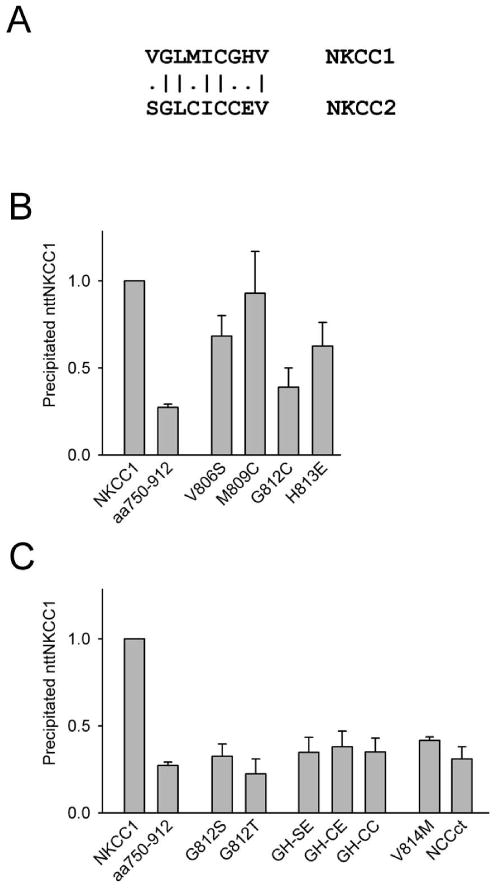
A. Comparison of NKCC1 amino acids 806-814 to the corresponding residues of NKCC2. B and C. Co-immunoprecipitation of nttNKCC1 with the mutants indicated was determined and quantitated as described in the caption of Fig. 1. In the mutants GH-SE, GH-CE and GH-CC both G812 and H813 were mutated as indicated. NCCct, a chimera in which the C-terminus of NKCC1 was replaced with that of NCC, has been described previously (19). Each data point represents the mean ± S.E. for 3 or more independent experiments except M809C, G812T and GH-CE for which n=2.
We show two additional results in Fig. 4C, the co-immunoprecipitation of nttNKCC1 with the point mutant V814M and with the chimera NCCct in which the C-terminus of NKCC1 was replaced by that of rat NCC (19). V814 is conserved in NKCC1, NKCC2 and NCC. The corresponding mutation in human NCC (V677M) has been detected in a patient with Gitelman's Syndrome (25) an inherited renal tubular disorder linked to mutations in NCC (SLC12A3). To our knowledge the functional consequences of this mutation in human NCC has not been explored but it is clear from Fig. 4C that the corresponding mutation in NKCC1 leads to a markedly reduced dimer interaction with nttNKCC1. We include the result with NCCct because the residues around G812 in NKCC1 are conserved in NCC (the amino acids corresponding to residues 806-814 of NKCC1 are LSLMICGHV in NCC). We note, however, that there is no significant co-immunoprecipitation of nttNKCC1 with NCCct consistent with the hypothesis that, although G812 and the surrounding amino acids play a central role in the dimer interaction, other regions in the NKCC1 C-terminus also are involved in dimerization in a significant way.
In Fig. 5 we show the bumetanide-sensitive component of 86Rb transport into HEK293 cells transiently transfected with the point mutants studied in Fig. 4. All of these point mutants increase this NKCC-specific component of 86Rb flux to ∼3 or more times basal levels indicating that they are all well-expressed, properly processed and functional in the HEK293 cells. Thus the inability of many of these mutants to co-immunoprecipitate nttNKCC1 is not related to their mishandling in the HEK293 cells. Interestingly the mutation V812M associated with Gitelman's Syndrome does not result in lack of function of NKCC1. While more direct studies with NCC are obviously required to clarify these results we speculate that the apparent defect in NCC associated with the Gitelman's Syndrome mutation V677M may be related to problems with dimerization (the patient with this mutation was also found to have the mutation R209W in the other NCC allele (25)).
Figure 5. Bumetanide-sensitive 86Rb fluxes via NKCC1 point mutants.
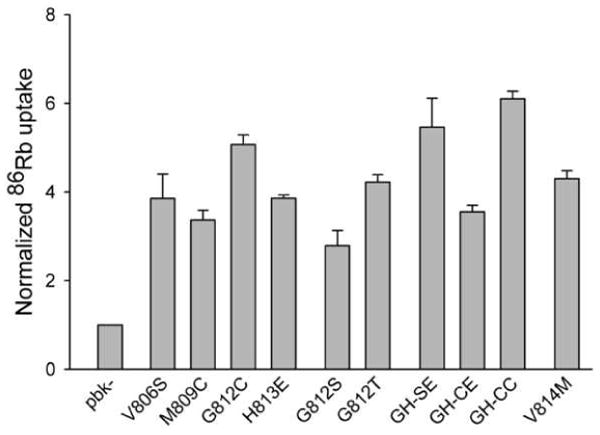
Bumetanide-sensitive 86Rb fluxes were measured and normalized as described in the caption of Fig. 3. Each data point represents the mean ± S.E. for 3 or more independent experiments except GH-CE for which n=2.
Additional complimentary studies
In order to further clarify and confirm the above results we carried out several additional studies. In Fig. 6 we examine the results of some complimentary experiments to those illustrated in Fig. 4. Here we employed the protein nttGH-SE in which the mutations of G812 and H813 to S and E, respectively, are made in nttNKCC1 rather than in NKCC1. We first show that nttGH-SE does not co-immunoprecipitate with wild-type NKCC1. This result confirms that of the complimentary experiment in Fig. 4C where we showed that nttNKCC1 does not co-precipitate with GH-SE. We next show that nttGH-SE does co-immunoprecipitate with GH-SE. This result is consistent with our general finding stated above that our various NKCC1/NKCC2 chimeras form homodimers irrespective of whether they display significant dimerization with nttNKCC1. We note, however, that the co-immunoprecipitated nttGH-SE signal seen with GH-SE in Fig. 6 is significantly lower than the nttNKCC1 signal seen with wild-type NKCC1, indicating a somewhat weaker dimer interaction between the modified proteins. Finally we show that nttGH-SE does not co-immunoprecipitate with aa750-1203. These two clones have the same amino acids at positions 812 and 813 (S and E, respectively) but the remaining C-terminal amino acids of nttGH-SE are those of NKCC1 while the C-terminal amino acids of aa750-1203 are those of NKCC2. Thus this result again confirms that residues in the NKCC1 C-terminus other than those at and near position 812 also play significant roles in the dimerization interaction.
Figure 6. Complimentary co-immunoprecipitation studies.
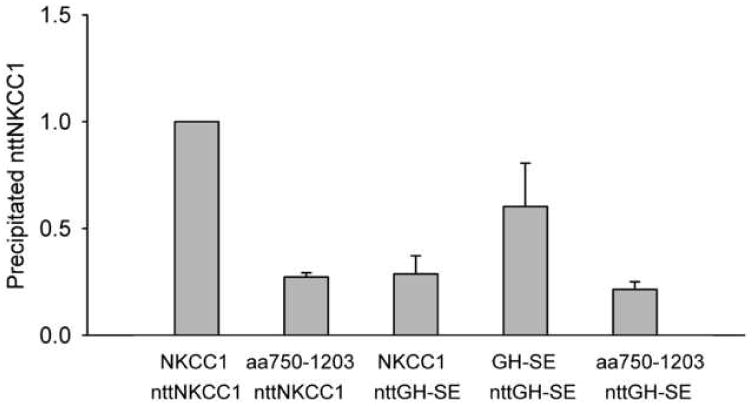
Co-immunoprecipitation of nttNKCC1 and nttGH-SE (nttNKCC1 with the residues G812 and H813 mutated to S and E, respectively) with the full length chimeras and mutants indicated was determined and quantitated as described in the caption of Fig. 1. Each data point represents the mean ± S.E. for 3 or more independent experiments.
To further explore the above observation that the dimerization interaction between GH-SE and nttGH-SE is apparently weaker than that between wild-type NKCC1 and nttNKCC1 (Fig. 6) we carried out the cross-linking studies illustrated in Fig. 7. In these experiments membranes from HEK293 cells transiently transfected with the (single) proteins indicted were solubilized in 0.3% Triton X-100 then treated with the covalent crosslinker DTSSP and analyzed by Western blotting. In the case of wild-type NKCC1 we typically find that virtually all of the monomeric protein is lost following DTSSP treatment and appears in a higher molecular weight band that migrates at twice its monomeric size. This observation formed the basis of our original demonstration that NKCC1 exists as a dimer in the plasma membrane that is stable in mild detergent solution (13). In the case of the mutants GH-SE, G812S and G812T, however, we find that significant amounts of monomer typically remain after DTSSP treatment and that, in fact, little crosslinked dimer is seen for G812T. In the case of GH-SE this result is consistent with our observation of a weakened interaction between GH-SE and nttGH-SE in Fig. 6. Our interpretation of these results is that the dimerization interactions of GH-SE and G812S are weaker than that of wild-type NKCC1 and thus are somewhat destabilized in 0.3% Triton X-100. The dimerization of G812T would appear to be even weaker still.
Figure 7. Chemical crosslinking of NKCC1 and NKCC1 mutants.
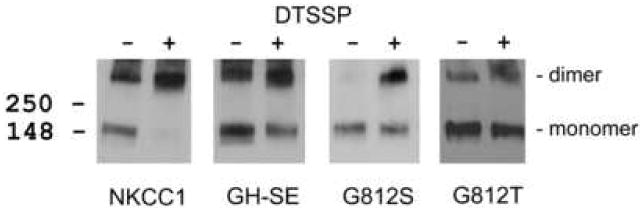
Membranes from HEK293 cells transiently transfected with NKCC1 or the mutants indicated were prepared, solubilized in 0.3 Triton X-100, treated with (+) or without (−) the crosslinker DTSSP as indicated, and probed by Western blotting as described in Methods. No disulfide reducing agent was added to the SDS-PAGE sample buffer. The positions of the NKCC1 monomer and dimer are indicated.
Finally, in order verify the participation of G812 and its surrounding amino acids in NKCC1 dimerization using a rather different methodology, we examined the effect of the sense peptide LMISGHVHMG and the scrambled peptide GIHMLGVSMH on the NKCC1 dimer in detergent solution (Fig. 8). This sense peptide represents NKCC1 amino acids 808-817 (centered on G812 and H813) with the modification C811S to prevent the formation of interpeptide disulfide bonds. In these experiments we solubilized membranes from HEK293 cells transiently transfected with wild-type NKCC1 in 0.3% Triton X-100. We then incubated these solubilized membranes with the sense or scrambled peptides in the presence of an additional 0.01% SDS added to weaken the dimerization interaction and thereby enhance competition of the peptide for the dimer interface. One hour later the crosslinker DTSSP was added and amount of dimer was determined by Western blotting. In these studies we found that significantly less dimer was seen in the presence of the sense peptide than in the presence of the scrambled peptide (p = 0.03, paired t-test). This observation suggests that the sense peptide is able to compete with NKCC1 dimer partners for the dimer interface consistent with the hypothesis that the sequence around G812 forms a part of this interface.
Figure 8. Effect of the sense peptide LMISGHVHMG on the NKCC1 dimer.
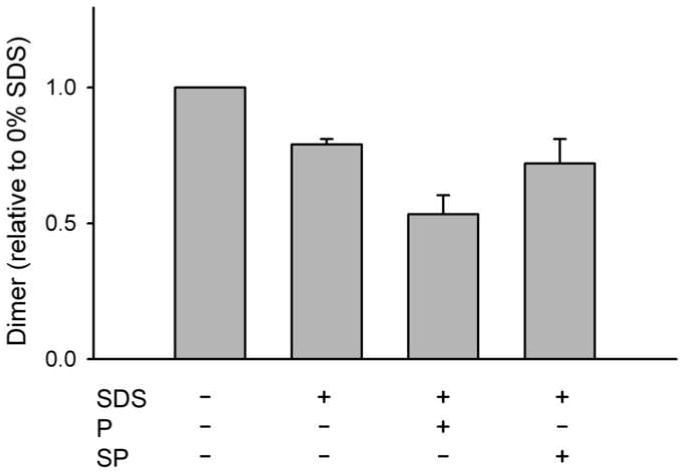
Membranes from HEK293 cells transiently transfected with NKCC1 were prepared, solubilized in 0.3 Triton X-100 in the presence or absence of 0.01% SDS and 1 mM of the sense peptide LMISGHVHMG (P) or the scrambled peptide GIHMLGVSMH (SP) as indicated, then treated with the crosslinker DTSSP (1 mM). Peptides were added 1 hour before the crosslinker. The resulting material was probed by Western blotting to determine the amount of crosslinked dimer in each sample. Results were normalized to the amount of dimer observed in the absence of both SDS and peptide. The results of 3 independent experiments were averaged to produce the figure.
Concluding Remarks
In the experiments reported here we have made use of a novel co-immunoprecipitation assay (19) to study the dimerization interaction of NKCC1. This assay takes advantage of the known properties of the NKCC1 dimer (13), namely its stability in Triton-X100 and its disruption by low concentrations of SDS, to specifically and sensitively probe the dimerization interaction. Our starting point for the present studies was our previous observation (19) that the chimeric protein aa750-1203, in which the cytosolic C-terminus of NKCC1 had been replaced with that of NKCC2, was unable to co-immunoprecipitate nttNKCC1. Thus there are amino acids in the NKCC1 C-terminus that play a central role in dimerization that cannot be replaced by the corresponding residues of NKCC2. Our results presented here from additional NKCC1/NKCC2 chimeras and point mutants indicate that the substitutions of a number of residues in the C-terminus of NKCC1 with the corresponding sequence from NKCC2 result in weakened dimerization with nttNKCC1, indicating that various regions of the NKCC1 C-terminus play a role in this interaction. Most interestingly, however, we find that the replacement of a single NKCC1 residue, G812, with cysteine, the corresponding amino acid in NKCC2, essentially recapitulates the inability of aa750-1203 to co-immunoprecipitate nttNKCC1. In addition to this effect on heterodimer formation we also find that mutations at G812 can weaken the corresponding homodimer interactions.
It is also interesting to note that, although the substitution of NKCC1 C-terminal residues with those of NKCC2 can result in proteins that no longer show significant heterodimerization with nttNKCC1, these proteins nevertheless typically still homodimerize. This observation puts significant constraints on the possible configurations of the NKCC1 dimer interface since the effect of these mutations must be to modify this interface in some sort of complimentary way that preserves homodimer formation while interfering with heterodimerization with wild-type. In general, a simple distortion of the dimer interface as a result of mutation would not be expected to have this complimentary effect. In Fig. 9 we present a very schematic model for the NKCC1 dimer interface that can account for our observations and which incorporates a known mechanism for protein dimer and oligomer assembly known as “domain swapping” (26). We want to emphasize at this point that we are not proposing this as an actual physical model for NKCC1 dimerization. Our data do not address the physical form of the dimer interface and other models may be equally capable of accounting for our observations. Our purpose here is to simply demonstrate that our results can be accounted for using an established mechanism for dimer formation. The basic idea of dimer formation via domain swapping is illustrated in Fig. 9A (in these models we imagine that we are looking up at the C-termini (CT) of two interacting NKCC1 molecules from a vantage point below the plasma membrane). Here domain ‘a’ of each dimer partner interacts with a complimentary domain‘c’ of the other partner in a lock and key-type fashion. It has been shown that a number of protein dimers form via such complimentary domain interactions (26) and it has been suggested that dimers of this type might have evolved from stable monomers in which the interaction between ‘a’ and ‘c’ was originally intramolecular. Thus the resulting dimer would have evolved via the swapping of intramolecular for intermolecular domain interactions. In Fig. 9B we illustrate, again very schematically, how a mutation in such a dimer interface could lead to a protein (mutant 1) that no longer can dimerize with the wild-type protein (because the space between a and c has changed) but can nevertheless form homodimers with essentially the same interaction strength as wild-type. In Fig. 9C we illustrate how a mutation could lead to a protein that can no longer dimerize with wild-type and also has a reduced homodimer interaction because of a mutation-induced distortion in domain ‘a’. In a model of this type we would envision G812 at a position where its mutation could induce the types of changes illustrated in mutants 1 and 2.
Figure 9. Schematic representation of the mechanism of dimer formation by “domain swapping”.
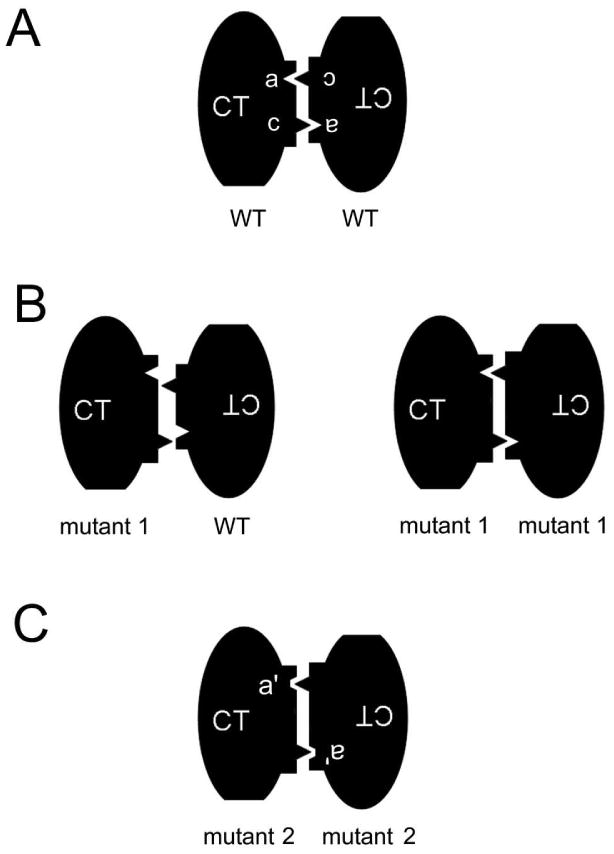
See text for details.
Interestingly Warmuth et al. (27) have recently reported the crystal structure of the recombinant C-terminus of an SLC12 homologue, MaCCC, from the archaeon Methanosarcina acetivorans. The crystallized C-terminus has a mixed α/β fold made up of 7 α helical and 10 β strand regions forming a compact trapezoidal structure. They found that this protein formed dimers in solution and that its crystal structure suggested a relatively small hydrophilic dimer interface in which residues on α helices 1 and 2 in one subunit were in proximity to residues located in the loop connecting β strands 5 and 6 in the second subunit, and vice versa. Thus this structure corresponds rather well to the schematic model in Figure 9A with, for example, the relevant residues on α helices 1 and 2 represented by domain ‘a’ and those in the loop connecting β strands 5 and 6 by the domain ‘b’ or vice versa. Alignments of the sequences of NKCC1 and NKCC2 with that of MaCCC place G812 in β strand 2 which lies between α helices 1 and 2 (27) and thus close to the dimer interface and in a position that could potentially alter the positions of the residues in α helices 1 and 2 that participate in dimerization. Although these results from MaCCC are consistent with the concepts illustrated in Figure 9 we should emphasize that the sequence conservation between the C-termini of MaCCC and NKCC1 is rather low (∼12% identity) so comparisons should be treated with some caution. Additional work will be required to explore the significance of the MaCCC C-terminal crystal structure and its dimer interface to NKCC1 and its mammalian homologues.
In conclusion we have demonstrated that the residue G812 plays a central role in the configuration of the dimer interface of NKCC1. Although at first glance it seems odd that the mutation of a single residue can result in a protein that no longer can dimerize with wild-type NKCC1 but nevertheless can still form homodimers, we illustrate that this phenomenon is in fact consistent with dimer formation via the mechanism of domain swapping and that these ideas are further supported by the recently reported crystal structure of the MaCCC C-terminus.
Acknowledgments
This research was supported by the Intramural Research Program of the National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda MD.
Abbreviations
- NKCC
Na+-K+-2Cl− cotransporter
- NCC
Na+-Cl− cotransporter
- KCC
K+-Cl− cotransporter
- nttNKCC1
N-terminally truncated NKCC1
- DTSSP
3,3′-Dithiobis(sulfosuccinimidylpropionate)
References
- 1.Xu JC, Lytle C, Zhu TT, Payne JA, Benz E, Jr, Forbush B., III Molecular cloning and functional expression of the bumetanide-sensitive Na-K-Cl cotransporter. Proc Natl Acad Sci U S A. 1994;91:2201–2205. doi: 10.1073/pnas.91.6.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gamba G, Miyanoshita A, Lombardi M, Lytton J, Lee WS, Hediger MA, Hebert SC. Molecular cloning, primary structure, and characterization of two members of the mammalian electroneutral sodium-(potassium)-chloride cotransporter family expressed in kidney. J Biol Chem. 1994;269:17713–17722. [PubMed] [Google Scholar]
- 3.Payne JA, Xu JC, Haas M, Lytle CY, Ward D, Forbush B., III Primary structure, functional expression, and chromosomal localization of the bumetanide-sensitive Na-K-Cl cotransporter in human colon. J Biol Chem. 1995;270:17977–17985. doi: 10.1074/jbc.270.30.17977. [DOI] [PubMed] [Google Scholar]
- 4.Gamba G. Molecular physiology and pathophysiology of electroneutral cation-chloride cotransporters. Physiol Rev. 2005;85:423–493. doi: 10.1152/physrev.00011.2004. [DOI] [PubMed] [Google Scholar]
- 5.Delpire E, Mount DB. Human and murine phenotypes associated with defects in cation-chloride cotransport. Annu Rev Physiol. 2002;64:803–843. doi: 10.1146/annurev.physiol.64.081501.155847. [DOI] [PubMed] [Google Scholar]
- 6.Kaneko H, Putzier I, Frings S, Kaupp UB, Gensch T. Chloride accumulation in mammalian olfactory sensory neurons. J Neurosci. 2004;24:7931–7938. doi: 10.1523/JNEUROSCI.2115-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gavrikov KE, Nilson JE, Dmitriev AV, Zucker CL, Mangel SC. Dendritic compartmentalization of chloride cotransporters underlies directional responses of starburst amacrine cells in retina. Proc Natl Acad Sci U S A. 2006;103:18793–18798. doi: 10.1073/pnas.0604551103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Price TJ, Cervero F, Gold MS, Hammond DL, Prescott SA. Chloride regulation in the pain pathway. Brain Res Rev. 2009;60:149–170. doi: 10.1016/j.brainresrev.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kahle KT, Staley KJ, Nahed BV, Gamba G, Hebert SC, Lifton RP, Mount DB. Roles of the cation-chloride cotransporters in neurological disease. Nat Clin Pract Neurol. 2008;4:490–503. doi: 10.1038/ncpneuro0883. [DOI] [PubMed] [Google Scholar]
- 10.Orlov SN, Tremblay J, Hamet P. NKCC1 and hypertension: a novel therapeutic target involved in the regulation of vascular tone and renal function. Curr Opin Nephrol Hypertens. 2010;19:163–168. doi: 10.1097/MNH.0b013e3283360a46. [DOI] [PubMed] [Google Scholar]
- 11.Dzhala VI, Kuchibhotla KV, Glykys JC, Kahle KT, Swiercz WB, Feng G, Kuner T, Augustine GJ, Bacskai BJ, Staley KJ. Progressive NKCC1-dependent neuronal chloride accumulation during neonatal seizures. J Neurosci. 2010;30:11745–11761. doi: 10.1523/JNEUROSCI.1769-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mejia-Gervacio S, Murray K, Lledo PM. NKCC1 controls GABAergic signaling and neuroblast migration in the postnatal forebrain. Neural Dev. 2011;6:4. doi: 10.1186/1749-8104-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore-Hoon ML, Turner RJ. The structural unit of the secretory Na+-K+-2Cl- cotransporter (NKCC1) is a homodimer. Biochemistry. 2000;39:3718–3724. doi: 10.1021/bi992301v. [DOI] [PubMed] [Google Scholar]
- 14.Starremans PG, Kersten FF, Van Den Heuvel LP, Knoers NV, Bindels RJ. Dimeric architecture of the human bumetanide-sensitive Na-K-Cl Co-transporter. J Am Soc Nephrol. 2003;14:3039–3046. doi: 10.1097/01.asn.0000097370.29737.5b. [DOI] [PubMed] [Google Scholar]
- 15.de Jong JC, Willems PH, Mooren FJ, Van Den Heuvel LP, Knoers NV, Bindels RJ. The structural unit of the thiazide-sensitive NaCl cotransporter is a homodimer. J Biol Chem. 2003;278:24302–24307. doi: 10.1074/jbc.M303101200. [DOI] [PubMed] [Google Scholar]
- 16.Casula S, Shmukler BE, Wilhelm S, Stuart-Tilley AK, Su W, Chernova MN, Brugnara C, Alper SL. A dominant negative mutant of the KCC1 K-Cl cotransporter: both N- and C-terminal cytoplasmic domains are required for K-Cl cotransport activity. J Biol Chem. 2001;276:41870–41878. doi: 10.1074/jbc.M107155200. [DOI] [PubMed] [Google Scholar]
- 17.Simard CF, Bergeron MJ, Cotton-Frenette R, Carpentier GA, Pelchat ME, Caron L, Isenring P. Homooligomeric and heterooligomeric associations between K-Cl cotransporter isoforms and between K-Cl and Na-K-Cl cotransporters. J Biol Chem. 2007 doi: 10.1074/jbc.M607811200. [DOI] [PubMed] [Google Scholar]
- 18.Uvarov P, Ludwig A, Markkanen M, Soni S, Hubner CA, Rivera C, Airaksinen MS. Coexpression and heteromerization of two neuronal K-Cl cotransporter isoforms in neonatal brain. J Biol Chem. 2009;284:13696–13704. doi: 10.1074/jbc.M807366200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parvin MN, Gerelsaikhan T, Turner RJ. Regions in the cytosolic C-terminus of the secretory Na(+)-K(+)-2Cl(−) cotransporter NKCC1 are required for its homodimerization. Biochemistry. 2007;46:9630–9637. doi: 10.1021/bi700881a. [DOI] [PubMed] [Google Scholar]
- 20.Kurihara K, Moore-Hoon ML, Saitoh M, Turner RJ. Characterization of a phosphorylation event resulting in upregulation of the salivary Na(+)-K(+)-2Cl(−) cotransporter. Am J Physiol. 1999;277:C1184–C1193. doi: 10.1152/ajpcell.1999.277.6.C1184. [DOI] [PubMed] [Google Scholar]
- 21.Evans RL, Park K, Turner RJ, Watson GE, Nguyen HV, Dennett MR, Hand AR, Flagella M, Shull GE, Melvin JE. Severe impairment of salivation in Na+/K+/2Cl- cotransporter (NKCC1)- deficient mice. J Biol Chem. 2000;275:26720–26726. doi: 10.1074/jbc.M003753200. [DOI] [PubMed] [Google Scholar]
- 22.Moore-Hoon ML, Turner RJ. Molecular and topological characterization of the rat parotid Na+-K+- 2Cl- cotransporter1. Biochim Biophys Acta. 1998;1373:261–269. doi: 10.1016/s0005-2736(98)00112-6. [DOI] [PubMed] [Google Scholar]
- 23.Vallejo AN, Pogulis RJ, Pease LR. Mutagenesis and synthesis of novel recombinant genes using PCR. In: Dieffenbach CW, D GS, editors. PCR primer, A laboratory manual. Cold Spring Harbor Press; Plainview, NY: 1995. pp. 603–612. [Google Scholar]
- 24.Dehaye JP, Nagy A, Premkumar A, Turner RJ. Identification of a functionally important conformation-sensitive region of the secretory Na+-K+-2Cl-cotransporter (NKCC1) J Biol Chem. 2003;278:11811–11817. doi: 10.1074/jbc.M213148200. [DOI] [PubMed] [Google Scholar]
- 25.Syren ML, Tedeschi S, Cesareo L, Bellantuono R, Colussi G, Procaccio M, Ali A, Domenici R, Malberti F, Sprocati M, Sacco M, Miglietti N, Edefonti A, Sereni F, Casari G, Coviello DA, Bettinelli A. Identification of fifteen novel mutations in the SLC12A3 gene encoding the Na-Cl Co-transporter in Italian patients with Gitelman syndrome. Hum Mutat. 2002;20:78. doi: 10.1002/humu.9045. [DOI] [PubMed] [Google Scholar]
- 26.Bennett MJ, Schlunegger MP, Eisenberg D. 3D domain swapping: a mechanism for oligomer assembly. Protein Sci. 1995;4:2455–2468. doi: 10.1002/pro.5560041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warmuth S, Zimmermann I, Dutzler R. X-ray structure of the C-terminal domain of a prokaryotic cation-chloride cotransporter. Structure. 2009;17:538–546. doi: 10.1016/j.str.2009.02.009. [DOI] [PubMed] [Google Scholar]


