Abstract
Stem cell quiescence has been hypothesized to suppress the rate at which genetic mutations accumulate within tissues by reducing the number of divisions a cell undergoes. However, recent studies have suggested that stem cells in the small intestine are rapidly dividing. This observation raises the issue of whether replication related errors are an important contributor to the accumulation of genetic damage and, if so, how genomic integrity is maintained within the small intestine. Here, reporter-marked small intestinal epithelial cells, resulting from mini-chromosome maintenance protein 2 (Mcm2) gene driven Cre-mediated recombination, are shown to be retained at the +1 position within the crypt and to contribute to the intestinal epithelia over long periods. Additionally, we show that the rate of cycling of +1 position Mcm2-expressing stem cells is heterogeneous with cycling times ranging between 1 and 4 days. Further, this heterogeneity depends on the p53 signaling pathway and could provide the basis for retention and expansion, through niche succession and crypt fission, of genetically intact stem cells. This somatic selection process would require active cellular replication.
Keywords: Intestinal stem cells, Mini-chromosome maintenance 2, Replication licensing factor, Somatic selection, Quiescence, DNA damage response
Introduction
A long standing assumption is that somatic stem cells exhibit the property of relative quiescence, dividing only intermittently, where this quiescence is presumed to protect stem cells from replication-related genetic damage [1]. However, recent BrdU labeling studies of Lgr5+ intestinal crypt stem cells (ISCs) have shown that approximately 80% of these cells divide within a 24-hour labeling period [2]. This observation suggests that most Lgr5+ cells cycle continuously. In support of this observation, in Lgr5-Cre marking studies, tracts of lacZ-marked progeny frequently form continuous crypt to villi units with no evidence that the progeny from a marked cell are interrupted by progeny from adjacent unmarked cells. Such crypt to villi units have been considered a hallmark of reporter-marked ISCs [2–6]. These observations are contradictory to the notion that genetic integrity is maintained in somatic stem cells of the small intestinal crypt through stem cell quiescence.
Despite these observations, other recent studies have shown that mice in which components of the replication machinery are compromised exhibit markedly accelerated rates of cancer, consistent with a contribution of replication-related damage to genomic instability. Mini-chromosome maintenance proteins (Mcm’s) are components of the complex of proteins that license origins of DNA replication for use during S-phase [7]. Mice that are homozygous for an Mcm4 hypomorphic mutation on a C3H genetic background show high rates of breast cancer [8]. Similarly, homozygous mice carrying an insertion of IRES-CreERT2 3′ to the Mcm2 coding sequence, which reduces Mcm2 expression to approximately one-third of normal levels, exhibit an early cancer phenotype with 100% penetrance of thymoma and additional tumor types on a 129/Sv genetic background [9].
As Mcm’s must assemble onto the chromatin early during the G-1 phase of the cell cycle, their expression marks not only actively dividing cells but also cells that are competent to divide but quiescent [10]. In heterozygous Mcm2-CreERT2 mice, tamoxifen-induced Cre-mediated recombination has been shown to mark a population of ISCs [9]. Here, we have utilized a variety of approaches to examine the rate of cycling of these cells under different conditions. We show that they are heterogeneous in their rate of cycling, where this heterogeneity is lost in the context of p53 mutation. These observations suggest that cell cycle heterogeneity in ISCs is not a consequence of programmed quiescence but rather reflects the activity of the DNA damage response pathway. This activity may play a key role in maintaining genetic integrity of stem cells by facilitating somatic selection for genetically intact cells, a mechanism that requires ongoing cell division.
Materials and Methods
The methodologies used are modified from those described previously [9, 10] as indicated in the text and Supporting Information, “Methods” Section. Briefly, for Cre-dependent reporter marking experiments, 6–8-week-old F1 heterozygotes carrying the Mcm2CreERT2 and reporter transgenes were injected on 3–5 consecutive days with 1 µg/g body weight of hydroxytamoxifen (Sigma-Aldrich, St Louis, MO). Treated mice were then rested between 4 weeks and 11 months in the absence of additional treatments. In some cases, mice were then administered a series of three s.c. injections of 10 µg insulin-like growth factor 1 (IGF1) at daily intervals where the final injection was given approximately 3 hours prior to examination of tissues.
For nucleoside analog labeling experiments, 3-month-old p53 knockout mice and wild-type controls on the BALB/c background were administered 0.5 mM IdU, 0.5 mM CldU, or mixtures in their drinking water or by injection as indicated in the text.
Results
Intermittent Production of Progeny from Mcm2-CreERT2-Marked Intestinal ISCs
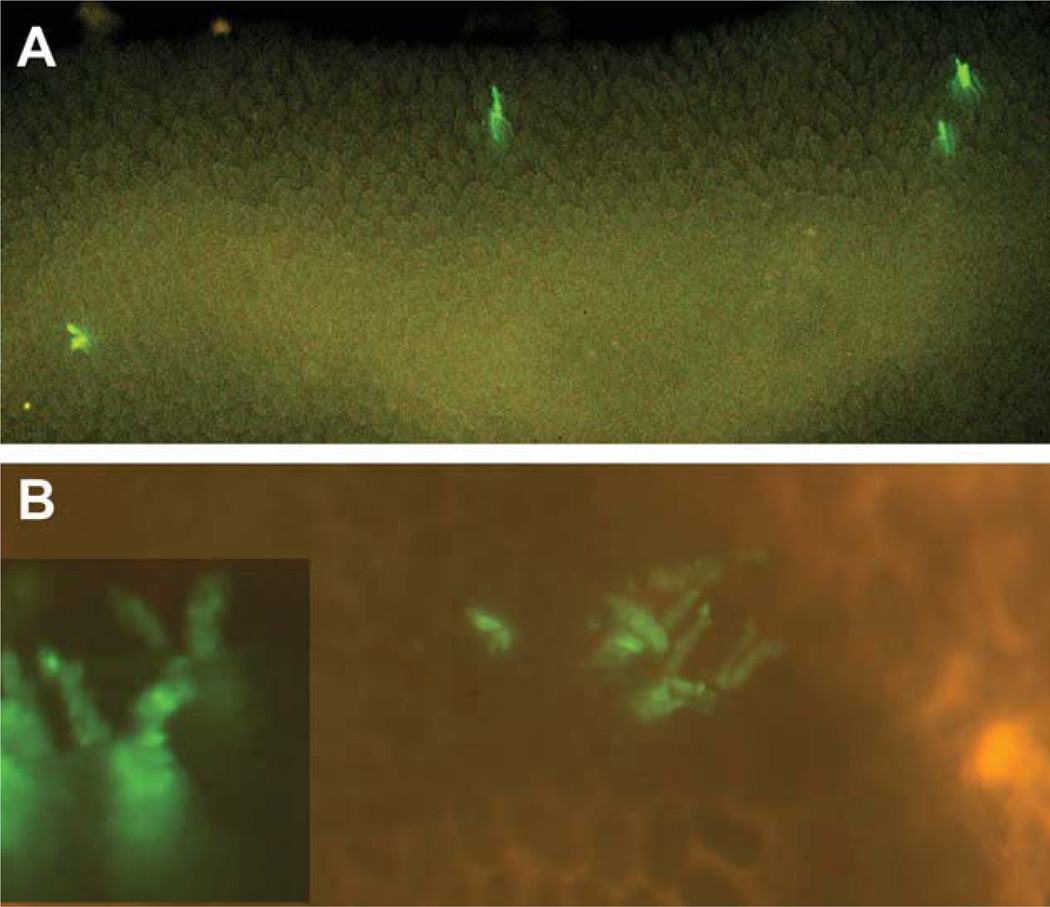
Mice heterozygous for both the Mcm2-CreERT2 transgene and a Cre-dependent reporter that expresses enhanced green fluorescent protein (EGFP) following Cre-mediated recombination (Z/EG) [11] were treated with tamoxifen at approximately 6–8 weeks of age and rested for between 1–3 months prior to examination of the small intestine. Except as indicated below, all results are from the lower third (ileum) of the small intestine. Tracts of EGFP-marked cells that are continuous from the crypt through the villus are found (e.g., Fig. 1C), similar to those described previously [2]. Additionally, EGFP-marked cells contribute to multiple lineages including enterocytes, goblet cells, and enteroendocrine cells (see Supporting Information, Section 1). These results are consistent with the observation that Mcm2 is expressed in essentially all cells within the base of the crypt except Paneth cells, including the Lgr5+ crypt basal columnar cells (Supporting Information, Section 1). However, EGFP+ tracts that are discontinuous, exhibiting breaks in the runs of EGFP+ cells that are reflected in two or three adjacent villi, are also present at high frequency (Fig. 1B). Additionally, short v-like formations and small spots, consisting of one or a few EGFP+ cells are found within the crypts and dissection of individual-marked crypts confirms this interpretation (Fig. 1D–1F).
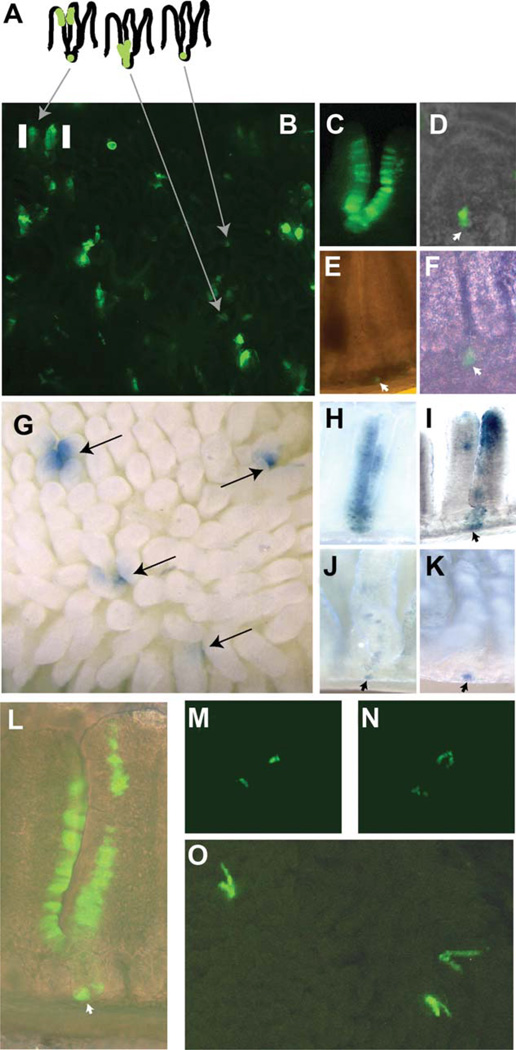
Figure 1.
Mcm2-CreERT2-induced reporter expression profiles in small intestine. The schematic in panel (A) represents an interpretation of various enhanced green fluorescent protein (EGFP)-marked clusters of cells in panel (B) which is an image of EGFP fluorescence in a living explant of the small intestine of an Mcm2Cre-ERT2;Tg(ACTB-Bgeo/GFP)21Lbe (commonly referred to as Z/EG) mouse at approximately 1 month following treatment with tamoxifen. Panels (C–F) are images of EGFP expressing cells in individual crypts and associated villi at between 1 and 3 months following tamoxifen treatment and prepared by mild formalin fixation and manual dissection. Panel (G) is an image of the small intestine from an Mcm2Cre-ERT2;R26R mouse at approximately 10 weeks following tamoxifen treatment and stained for β-galactosidase activity. Panels (H–K) show images of β-galactosidase stained cells in individual crypts and associated villi following manual dissection. Panel (L) shows a manually dissected crypt containing EGFP expressing cells at 10 months following tamoxifen treatment. Panels (M, N) show regions of EGFP expression in living explants immediately following isolation [panel (M)] or after 12 hours in culture [panel (N)] where the same location is shown. Panel (O) is an image of the small intestine of an Mcm2Cre-ERT2;Z/EG mouse at approximately 1 month following treatment with tamoxifen where during the final 3 days prior to isolation the mouse received daily injections of insulin-like growth factor 1 (IGF1). The images in panels (B, C, E, M, N, O) were taken using a fluorescence dissecting stereomicroscope at magnifications between ×20 and ×40. The images in panels (G–K) were taken using a dissecting stereomicroscope and bright-field illumination at magnifications between ×20 and ×40. The images in panels (D, F, L) were taken using an inverted fluorescence microscope at magnifications ranging between ×40 and ×200. Additional images of the crypt shown in panel (D) are presented in the Supporting Information, Section 1, Figure S2.
To determine if the apparent difference in the distribution of reporter-marked cells following Mcm2-CreERT2 marking in the present study versus the continuous tracts of marked cells observed following Lgr5-CreERT2 marking in prior studies [2] results from use of a different Cre-dependent reporter line, mice carrying the Mcm2-CreERT2 transgene and the ROSA26-Cre-dependent lacZ reporter [12] used in the prior studies were established and a tracing study similar to that described above was performed. In this experiment, mice were rested for approximately 10 weeks following tamoxifen treatment. Although occasional continuous crypt to villi tracts are found (Fig. 1H), results from this study again demonstrate a high frequency (greater than half) of interrupted tracts of reporter-marked cells similar to those observed using the EGFP reporter strain (Fig. 1G, 1I–1K).
As Mcm2 is expected to be expressed in all proliferation competent cells, including both stem cells and proliferative progenitors, one possibility to account for the interrupted tracts of reporter-marked cells is that, even 10 weeks following tamoxifen treatment, a fraction of the Mcm2-marked cells do not mark stem cells but rather long-lived differentiated cells or progenitors. To test this possibility, Mcm2-CreERT2-marked cells were examined at 11 months following treatment with tamoxifen using the Z/EG reporter. Even with the extended chase period, many crypts are found in which discontinuous tracts of EGFP expressing cells are present supporting the notion that these tracts result from reporter-marked ISCs (e.g., Fig. 1L).
Another interpretation of the discontinuous tracts of reporter expressing cells is that they result from intermittently cycling stem cells located near to the base of the crypts as shown schematically in Figure 1A. Each crypt is thought to contain multiple stem cells only one of which is expected to be marked at lower levels of recombination. In this case, if the marked stem cell fails to cycle continuously, progeny from unmarked cells would also occupy the epithelia producing the apparent gaps in the reporter expressing tracts. Under situations where multiple stem cells within a crypt are marked, as might occur at high levels of Cre-mediated recombination or following niche succession, gaps in the reporter tracts would not be apparent. Consistent with this interpretation, many of the continuous runs of reporter-marked cells derive from crypts in which a high proportion of the cells within the crypt express the reporter suggesting that these crypts contain multiple-marked stem cells (e.g., Fig. 1C, 1H).
To examine the possibility that Mcm2-CreERT2-marked cells present at 1 month or longer cycle intermittently we, first, tested the ability of single or small clusters of EGFP+ cells located at the base of the crypts to give rise to progeny in short-term explants in vitro. Figure 1M shows a region containing two such clusters immediately following isolation. The same location is shown in Figure 1D, 12 hours later demonstrating a transition of the EGFP+ spots to multicell v-like structures. This observation demonstrates that many of the isolated EGFP+ cells remain capable of entering the cell cycle.
To further examine whether intermittent stem cell cycling is responsible for the observed patterns of EGFP expressing cells, we sought to modulate cell cycling rates in vivo using IGF1 administration. Here, Mcm2-CreERT2; Z/EG mice in which a subset of stem cells had been marked by tamoxifen injections and rested for over 1 month were treated for three consecutive days with 10 µg of IGF1/injection/mouse. Three hours following the final IGF1 injection, small intestine was examined for the pattern of EGFP expression (Fig. 1O). Unlike control mice which had not received IGF1 injections, nearly all tracts of EGFP+ cells in IGF1-treated mice are continuous, consistent with an uninterrupted production of progeny from EGFP-marked stem cells. The ability of the discontinuous tracts of EGFP expressing epithelial cells found in untreated mice to convert to continuous tracts of EGFP+ cells, considered a hallmark of progeny resulting from marked ISCs, strongly supports that by 1 month following tramoxifen treatment most tracts of EGFP+ cells marked by Mcm2-CreERT2 result from marked ISCs. These observations and additional studies below support an intermittent pattern of cycling of many Mcm2-CreERT2-marked ISCs.
Mcm2-CreERT2-Marked Intestinal ISCs Are Located at the +1 Position of the Base of the Crypt
The intermittent cycling of Mcm2-CreERT2-marked stem cells makes it possible to define the location of these cells within the crypt since cells that have not cycled recently, or cycled only very recently, should reflect their position. Careful dissection of over 20 crypts containing single or small clusters of reporter-marked cells was performed and in each case it was possible to identify at least one wedge-shaped-marked cell at the extreme base (+1 position) of the crypt using either the EGFP- or lacZ-based reporters. Several examples are shown in Figure 1D–1F and 1I–1K. No other location within the crypts consistently exhibited a marked cell.
Cycling Times of +1 Position Mcm2 Expressing Cells
To examine the cycling rates of +1 position Mcm2 expressing cells directly, a modification of a double labeling method was utilized in which different ratios of IdU and CldU were administered in discrete steps over a period of several days (ratiometric labeling). It is expected that half of the DNA, one strand of each chromosome, present in a replicating nucleus will reflect the mixture of IdU:CldU that was present on the most recent day on which it divided. Further, to the extent that the ratio of IdU to CldU within different regions of DNA within a nucleus can be measured, each nucleus will also contain a record of any other divisions it underwent during the labeling period (see Supporting Information, Section 2).
To test this approach, wild-type 129/Sv mice were administered drinking water containing varying ratios of IdU and CldU as follows: d1, 100% IdU, d2, 75% IdU:25% CldU, d3, 50% IdU:50% CldU; d4, 25% IdU:75% CldU; and d5, 100% CldU. Following labeling, paraffin sections of the small intestine were prepared and assayed for incorporation of IdU (red) and CldU (green) as described previously [10]. A low magnification image taken using compound fluorescence microscopy is shown in Figure 2A. As expected, nuclei near the tips of the villi are red and become progressively green at positions closer to the crypt. However, within the crypts, nuclei appear yellow suggesting a continued presence of IdU from more slowly dividing cells. Crypts were examined using confocal microscopy and optical sectioning at intervals of 0.5 µm as shown in Figure 2B–2F. There are two nuclei within the basal portion of the crypt shown in Figure 2 that have incorporated IdU and CldU (arrows). Further, subdomains within the nucleus on the right, which exhibit differing ratios of CldU:IdU, are identifiable (also see higher magnification in panel F). Three to four different color domains that are contiguous in the z-axis can be identified in each of the three serial sections (panels C–E and Supporting Information, Section 2) in this nucleus. These observations suggest that this nucleus synthesized DNA during three or four intervals of the 5-day labeling period consistent with a cycling time of approximately 48 hours or less. In contrast, the nucleus on the left exhibits a predominant orange color domain consistent with the presence of DNA synthesized on day 4 of the experiment and showing that this cell has remained quiescent for a period of 4 days (although quantitative analysis in Supporting Information, Section 2, suggests that an additional round of DNA synthesis had initiated during the final labeling interval on day 5). In these experiments, examples of nuclei that had color domains consistent with cell cycling intervals ranging between 1 and 4 days were found. These observations suggest that although a subset of +1 position cells cycle rapidly, as often as once per day, many others cycle more slowly. Further, there is a significant degree of heterogeneity in the rates at which these cells cycle even within the same crypt.
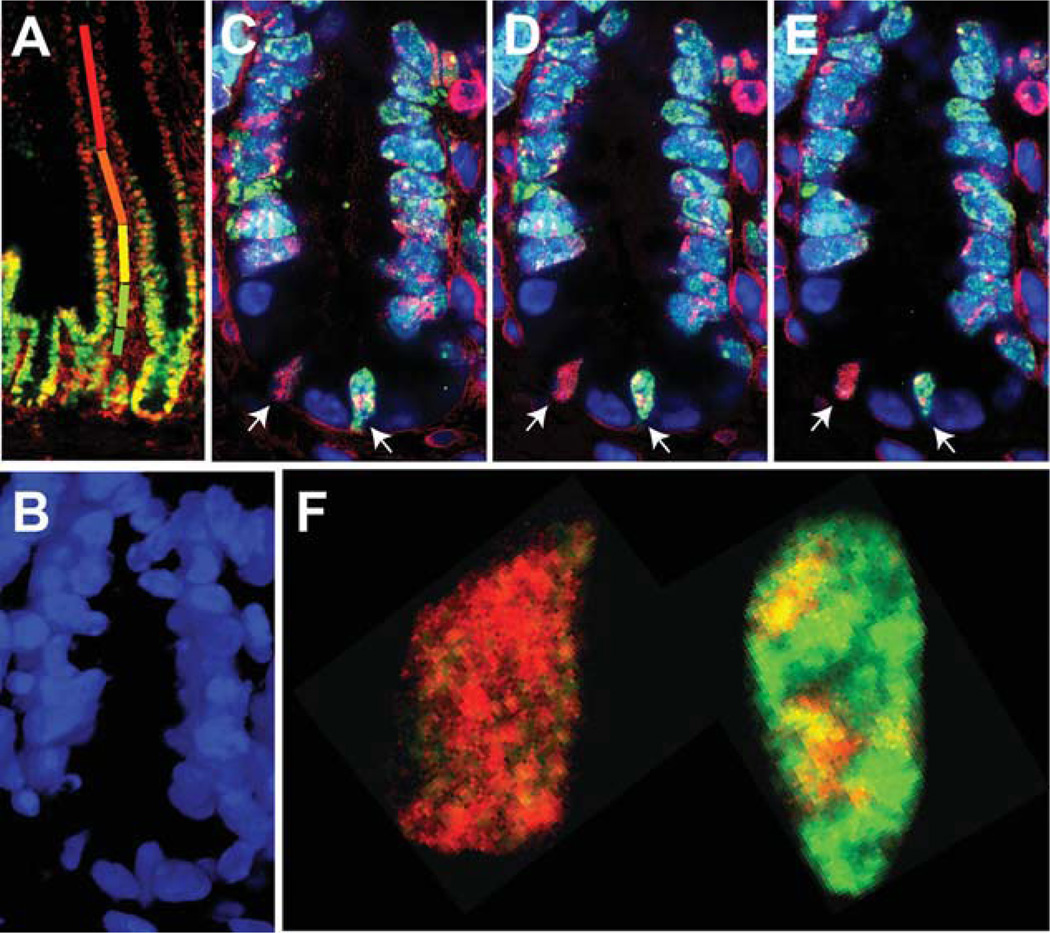
Figure 2.
Five-step ratiometric labeling to define days on which replication occurred for most cells within the small intestinal crypt. A wild-type 129/Sv mouse was administered drinking water containing varying ratios of IdU and CldU as follows: d1, 100% IdU, d2, 75% IdU:25% CldU, d3, 50% IdU:50% CldU; d4, 25% IdU:75% CldU; and d5, 100% CldU. Following labeling, 7-µm thick paraffin sections were prepared of the small intestine and assayed for incorporation of IdU and CldU using methods described previously [10]. A low magnification image taken at ×40 magnification using a compound fluorescence microscope is shown in panel (A). Different colored bars represent the different color transitions between nuclei near the tips of the villi, which are red, and become progressively more green at positions closer to the crypt. Within the crypts, however, nuclei appear more yellow, suggesting a continued presence of IdU, which would result from more slowly dividing cells. Examination of crypts using confocal microscopy at ×680 magnification and optical sectioning at intervals of 0.5 µm is shown in panels (B–E), where panel (B) is a three-dimensional reconstruction of DAPI stained nuclei over 6.0 µm and panels (C–E) are adjacent optical sections from within this interval stained for IdU (red) and CldU (green). There are two nuclei with the morphology of crypt basal columnar cells within the basal portion of this crypt each of which has incorporated IdU and CldU. Further, subdomains within the nucleus on the right which exhibit differing ratios of CldU:IdU are identifiable. Contiguous domains of three different hues can be identified in each of the three serial sections in this nucleus. These observations are consistent with this nucleus having undergone DNA synthesis on three or four different days during which different ratios of IdU to CldU were incorporated. In contrast, the nucleus on the left exhibits a predominant orange color domain demonstrating the presence of DNA synthesized on day 2 of the experiment and suggesting that this cells has remained quiescent for a period of at least 4 days (however, see Supporting Information, Section 2). Panel (F) shows a higher magnification of one optical section from each of these nuclei.
Heterogeneity in +1 Position Mcm2 Expressing Cell Cycling Rates Is Dependent on p53
To determine if heterogeneity in +1 position-Mcm2+ cell cycling rates is affected by DNA damage response pathways we utilized ratiometric labeling to compare cycling rates of +1 position cells in p53 null [13] or wt animals of the same strain (BALB/c). In this experiment, however, the labeling scheme was inverted, relative to that used in Figure 2, and the number of discrete CldU to IdU ratios was reduced to four such that mice received 100% CldU on day 1 (green), 67% CldU:33% IdU on day 2 (yellow), 33% CldU;67% IdU on day 3 (orange), and 100% IdU on day 4 (red). Longitudinal sections showing crypts from wt and p53 null mice are shown in Figure 3A–3F, where panels A, B, and E are wt and panels C, D, and F are p53 null. Transverse sections are shown in panels H–Q, where panels H–J and N–O are wt and K–M and P–Q are p53 null. Consistent with the observations for wt animals on the 129/Sv strain, a range of cycle times between 1 and 4 days was observed for +1 position Mcm2+ cells in the wt BALB/c crypts. For example, in panels A and B arrows indicate Mcm2+ cell nuclei, panel A, which exhibit various different colors resulting from IdU and CldU incorporation that are predominately of a single hue ranging from red through green, panel B, and demonstrating that the most recent division had occurred between 1 and 4 days prior to the end of the labeling period in different nuclei. Panel E shows a similar result at higher resolution using confocal microscopy. Panel G shows a distribution of the minimum length of the cell cycle estimated for 56 + 1 position Mcm2+ cell nuclei from p53 wild-type mice (blue bars). Transverse sections in which all of the +1 position Mcm2+ cell nuclei within a crypt can be identified show similar results (e.g., the innermost ring of Mcm2+ nuclei, in Fig. 3I, show a range of hues resulting from incorporation of CldU and IdU, Fig. 3J, that vary between red, yellow, and green consistent with the most recent division occurring in these cells at times ranging between 1 and 4 days prior to the end of the labeling period).
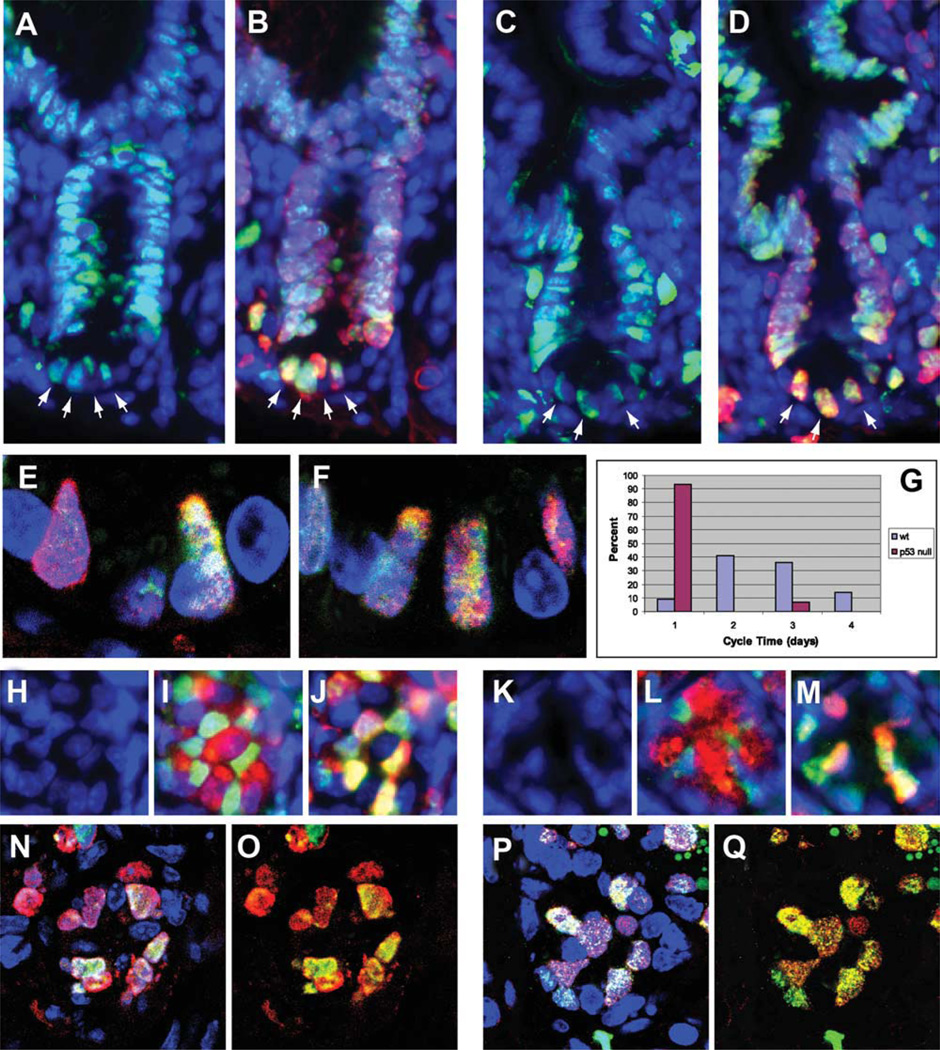
Figure 3.
Use of ratiometric labeling to estimate cycle times of +1 position Mcm2+ cells in wild-type and p53 null mice. Four step ratiometric labeling was utilized in wt and p53 null mice where mice received 100% CldU on day 1, 67% CldU:33% IdU on day 2, 33% CldU:67% IdU on day 3, and 100% IdU on day 4. Images are from 7-µm paraffin sections where longitudinal sections are shown in panels (A–F) and transverse sections are shown in panels (H–Q). Images in panels (A–D) were taken at ×200 magnification using a compound fluorescence microscope. Panels (A) and (B) are images from wild-type mice stained for Mcm2 (green) and DAPI (blue) in panel (A) and IdU (red), CldU (green), and DAPI (blue) in panel (B). Panels (C, D) are images from p53 null mice stained for stained for Mcm2 (green) and DAPI (blue) in panel (C) and IdU (red), CldU (green) and DAPI (blue) in panel (D). Images in panels (E, F) are 0.5-um optical sections taken at ×680 magnification using confocal microscopy. Panel (E) is from a wild-type mouse stained for IdU (red), CldU (green) and DAPI (blue). Panel (F) is from a p53 null mouse stained for IdU (red), CldU (green) and DAPI (blue). Panel (G) is a plot of the minimal estimated interval between cell divisions of +1 position Mcm2+ cells determined for each of 56 nuclei from wt mice (blue bars) and 30 nuclei from p53 null mice (red bars) as determined from the largest and, if present, second largest color domains present in ratiometrically labeled sections stained for IdU and CldU. The percent of the nuclei showing cycling intervals ranging between 1 and 4 days are plotted. Images in panels (H–M) are from transverse sections taken at ×200 magnification using a compound fluorescence microscope and show individual crypts from wt (panels [H–J]) and p53 null mice. Panels (H, K) show DAPI stained nuclei (blue); panels (I, J) show staining for Mcm2 (green), lysozyme (red), and DAPI (blue); and panels (J, M) show staining for IdU (red), CldU (green), and DAPI (blue). Images in panels (N–Q) are 0.5-um thick optical sections taken in the transverse plane by confocal microscopy where panels (N, O) are from a wt mouse crypt and panels (P, Q) are from a p53 null mouse crypt. In these panels staining is for DAPI (blue), IdU (red), and CldU (green).
In contrast, CldU/IdU stained domains from similar images of +1 position Mcm2+ cell nuclei in the p53 null crypts showed, with very few exceptions (which may reflect the miss-identification of +1 position Mcm2+ cells), largely red hues in lower magnification images (e.g., Fig. 3D, 3M) consistent with division within 1 day prior to end of the labeling period and multiple different color domains in high-resolution images (e.g., Fig. 3F, 3P, 3Q) that would result from four or more cell divisions during the 4-day labeling period. Panel G (red bars) shows a distribution of the length of the cell cycle estimated for 30 + 1 position nuclei from p53 null mice. These results are consistent with a cycle time of once per day or faster for the +1 position Mcm2+ cells of p53 null mice.
The increased rate of cycling of +1 position Mcm2+ cells in p53 null mice observed using the ratiometric labeling approach is sufficiently large, relative to wt mice, that it should be detectable as an increase in the fraction of cells that incorporate nucleoside analogue over short pulses. To determine if this is the case, mice were administered a bolus of CldU and assayed for incorporation in the small intestinal crypts 2 hours later. Wild-type and p53 null mice that had been treated over a period of 3 days with daily injections of 10 µg of IGF1 were also included in the experiment where the final IGF1 injection was given 1 hour prior to CldU administration. Nuclei were stained for incorporation of CldU (green) and Mcm2 (red) as shown in Figure 4A–4D and the proportion of Mcm2+ nuclei that also stained for CldU incorporation as a function of the position of the nuclei from the base of the crypt is plotted in Figure 4E. Consistent with the result of the ratiometric labeling experiment, the proportion of Mcm2+ nuclei at the +1 position that incorporate CldU during a 2-hour pulse is approximately 15% in wild-type animals and doubled to approximately 30% in p53 null mice. IGF1 treatment also affects the proportion of +1 position Mcm2+ nuclei that incorporate CldU, where the proportion is approximately 44% in wt mice and 52% in p53 null mice (Fig. 4E).
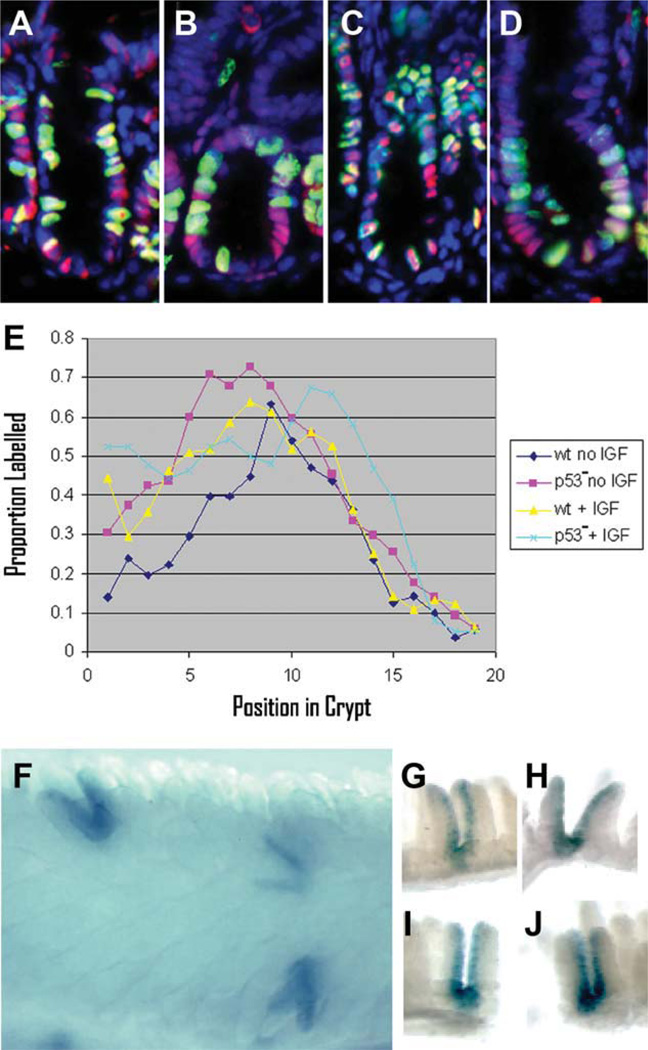
Figure 4.
Pulse labeling and Mcm2-CreERT2-mediated marking studies confirm an increased rate of cycling in the stem cells of p53 null mice relative to wild type. In panels (A–E) wt or p53 null mice were treated with daily IGF1 injections for 3 days and compared to mice of the same genotypes that had not been treated with IGF1. A pulse of CldU was given 2 hours prior to harvesting the small intestine. Panels (A–D) show 7-µm-thick longitudinal paraffin sections stained for Mcm2 (red), CldU (green), and DAPI (blue) where panel (A) is wt, no IGF1; panel (B) is p53 null, no IGF1; panel (C) is wt, IGF1 treated; and panel (D) is p53 null, IGF1 treated. Panel (E) shows of the proportion of Mcm2 stained nuclei that are also stained for CldU (y-axis) plotted as a function of position of the nuclei from the base of the crypt (x-axis) from 50 or more crypts for each of the different experimental conditions as indicated in the box. Panel (F) is a bright-field image of a whole mount preparation of small intestine stained for β-galactosidase activity from a p53 null, Mcm2-CreERT2 mouse carrying a Cre-dependent lacZ reporter at approximately 1 month following tamoxifen treatment. Panels (G–J) are individual β-galactosidase expressing crypts prepared by microdissection. Abbreviation: IGF, insulin-like growth factor 1.
The effect of the p53 null mutation on the production of Mcm2CreERT2-dependent reporter-marked progeny was also determined. If stem cells are producing progeny continuously in p53 null mice, uninterrupted tracts of marked progeny should be observed, similar to the effect of IGF1 treatment. For this experiment, trigenic mice that were wild-type or homozygous for the p53 null allele and also carried the Mcm2CreERT2 transgene and the Cre-dependent lacZ reporter [12] were established (Z/EG could not be used because it is carried on the same chromosome as p53). Mice were treated with tamoxifen as before and either 4 or 10 weeks following tamoxifen treatment in different experiments, small intestines were recovered and stained for expression of lacZ, Figure 4F. Similar results were obtained at each time point. Additionally, the proportion of marked crypts expressing lacZ was similar to levels observed for wt mice. As shown previously (Fig. 1G–1K), the majority of crypts from mice that are wild type for p53 expression show a discontinuous pattern of lacZ expression. However, in p53 null mice essentially all crypts containing a marked stem cell exhibited continuous tracts of lacZ expressing progeny extending from the crypt through the tips of the villi. These observations support the conclusion that, in wild-type animals, heterogeneity in stem cell cycling rates accounts for the subset of crypts exhibiting discontinuous tracts of reporter-marked cells and that this heterogeneity is dependent on the p53-mediated DNA damage response.
Expansion of Subsets of Reporter-Marked Cells over Long Periods of Time
ISCs are capable of dividing symmetrically to give rise to two daughter stem cells and expansion of subsets of stem cells through niche succession, in which progeny from a single stem cell populate all of the stem cell niches within a crypt, and crypt fission, in which new crypts are formed by splitting of an existing crypt, have been demonstrated [14, 15]. DNA damage response-dependent heterogeneity in the rate of cycling of ISCs might be expected to affect the contribution of subsets of stem cells through these processes. Here, we have observed that in wild-type mice, at long periods following tamoxifen treatment, regions of small intestine are found in which there are high densities of EGFP-marked crypts even under tamoxifen treatment regimens that result in relatively low overall levels of marked cells (e.g., Fig. 5A, 5B). This distribution is different from that seen at shorter intervals following tamoxifen treatment, where there is a more uniform distribution of marked crypts (e.g., Fig. 1B).
Figure 5.
EGFP expression profiles in small intestine of Mcm2-CreERT2:Z/EG mice at approximately 10 months following tamoxifen treatment. Panel (A) is an image taken using a fluorescence dissecting microscope at approximately ×20 magnification of EGFP fluorescence in scattered individual crypts of living explant from the small intestine of an Mcm2Cre-ERT2;Z/EG mouse at approximately 10 months following treatment with tamoxifen. Panel (B) shows an image taken at approximately ×30 magnification of clusters of EGFP fluorescence-marked crypts which are also found in these mice at approximately 10 months following tamoxifen treatment, where the inset in this figure is a higher magnification of a similar cluster.
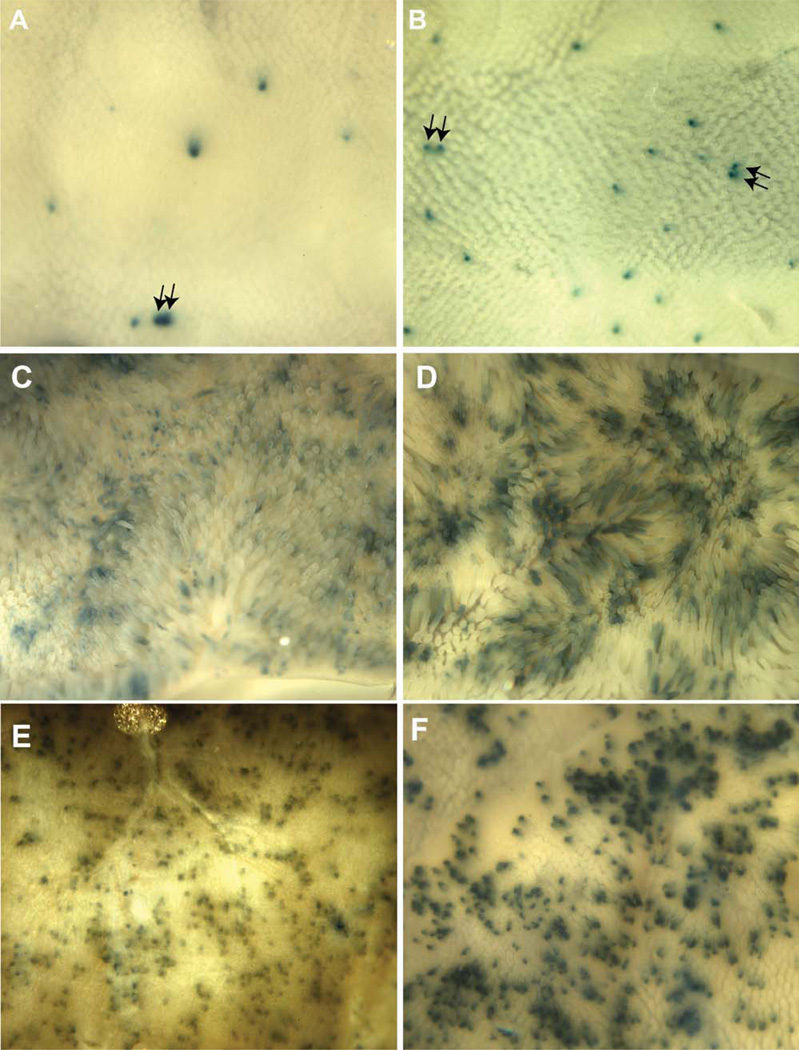
If the expansion of a subset of stem cells into multicrypt domains is affected by p53-dependent heterogeneity in their cycling rates, p53 null mice might be expected to exhibit differences in the extent to which reporter-marked crypts undergo crypt succession or expand into multicrypt domains over time. To test this possibility, p53 wild-type or null mice were marked using Mcm2-CreERT2 and the lacZ reporter as previously. Because of the limited lifespan of mice lacking p53 activity, mice were assayed at a relatively short time, approximately 10 weeks, following tamoxifen treatment. Dissected crypts were used to estimate the proportion of crypts in which the majority versus only a portion of the base of the crypt was stained as a measure of crypt succession. Approximately 28% (n = 25) of crypts from wt mice showed complete staining whereas approximately twice as many, 57% (n = 21), were completely stained in crypts from p53 null mice (these values may somewhat overestimate succession rates due to staining artifacts). Small intestine was also examined for the presence of adjacent reporter-marked crypts as an index of crypt fission. Within the lower third of the intestine (ileum), the region studied in the experiments described in previous sections pairs of adjacent-marked crypts were identified at a frequency of 2.4% ± 0.85% and 6.7% ± 0.07% of total-marked crypts in wt and p53 null mice, respectively (Fig. 6A, 6B). The upper small intestine (duodenum), which shows higher rates of crypt fission, was also examined in this experiment (Fig. 6C–6F). In this region, a higher proportion of crypts are marked overall and many adjacent-marked crypts are found in both wt and p53 null mice. However, the size of the marked multicrypt domains generally appears larger in p53 null relative to wt mice.
Figure 6.
Distribution of Mcm2-CreERT2-marked β-galactosidase expressing crypts in the ileum and duodenum of wt and p53 null mice. Wild-type and p53 null mice carrying the Mcm2-CreERT2 and R26R transgenes were treated with tamoxifen and 10 weeks following tamoxifen treatment small intestine was recovered, fixed, and stained for β-galactosidase activity. Panels (A, B) are images taken from the outside of the ileum to facilitate identification of stained crypts and these and similar images were used to identify adjacent-marked crypts (arrows in panels [A, B]) as discussed in the text. Panels (C–F) are similar preparations from the duodenum where (C) and (D) are images taken from the inside to show villi and (E) and (F) are images taken from the outside to show the distribution of marked crypts. All images were taken under bright-field illumination at a magnification of approximately ×12.5.
Discussion
The relative quiescence of somatic stem cells compared with proliferative progenitors has been considered to contribute to genome stability within tissues because a reduced rate of cycling would, in principle, reduce the acquisition of replication related genetic errors [1, 16]. Studies demonstrating the key role that DNA damage response and repair proteins [17], and more recently DNA replication proteins [8, 9], play in cancer and aging support the notion that the accumulation of replication-related genetic errors is detrimental. Further, a number of studies support that somatic stem cells in many tissues cycle slowly (reviewed for the hematopoietic, hair follicle, and intestinal crypt systems [16] and neural stem cells [10]). However, other studies have raised the possibility that quiescent stem cells constitute a specific subset of stem cells that are not responsible for tissue maintenance but, rather, a reserve that functions only following tissue damage [17]. The intestinal crypt is of particular interest in that, although different studies have suggested different locations for ISCs and different rates of cycling, in all studies the rate of cycling within these stem cells is far more rapid than inferred for other tissues.
Here, we have utilized tamoxifen induction of Cre-recombinase activity driven from the Mcm2 gene to mark cells within the intestinal crypt. Mcm2 is expressed in replication competent cells and is expected to allow marking of both actively dividing stem/progenitor cells and, if present, quiescent stem cells [9, 10, 18]. Following tamoxifen treatment, mice were resting for periods of between 1 and 11 months and assayed for expression of reporter-marked progeny. These studies demonstrate that reporter-marked cells capable of contributing to multiple cell lineages of the intestinal epithelia remain within the crypt for at least 11 months. This result is consistent with the observation that Mcm2 is expressed in all cells within the base of the crypt except Paneth cells, which will include the Lgr5 expressing crypt basal columnar cells (Supporting Information, Section 1), and + 4 position Bmi1 expressing cells (Supporting Information, Section 4) each of which has been shown to exhibit ISC cell properties in prior studies [2, 6].
Although the long-term contribution of Mcm2-CreERT2-marked cells to multiple cell types of the intestinal epithelia demonstrates that intestinal stem cells are marked by this approach, one apparent difference is observed between these cells and those identified in prior studies. Specifically, prior studies emphasized the contribution of stem cells marked using Lgr5-CreERT2 [2] or Bmi-CreERT2 [6] to progeny that are present in continuous runs from the crypt to the ends of the villi. In contrast, many of the cells marked by Mcm2-CreERT2 produce tracts of marked cells that do not completely mark the epithelia including interrupted runs of marked cells, short runs of marked cells that extend from the crypt through only a portion of the villi and single-marked cells or small clusters of marked cells near to the base of the crypts. Mcm2-CreERT2 marking using either EGFP-based or lacZ-based reporters gives similar patterns of marked progeny; where the lacZ-based reporter is the same as was used in prior studies [2, 6]. Several additional lines of evidence support that Mcm2-CreERT2 reporter-marked cells, present in mice at times longer than 1 month following tamoxifen treatment, arise from marked ISCs. First, many of the single-marked cells near the base of the crypt divide in culture. Additionally, most of the single-marked cells and interrupted tracts of cells are converted to continuous crypt to villus tracts following treatment with IGF1 or in the absence of p53 activity, demonstrating that most cells marked by Mcm2-CreERT2 are capable of generating crypt to villus units with the hallmark appearance of those containing a marked stem cell.
We hypothesize that discontinuous tracts of Mcm2-CreERT2-marked cells result from stem cells that divide intermittently to produce progeny. When the level of recombination is low, and as each crypt contains multiple stem cells, crypts containing a marked stem cell will also contain unmarked stem cells. Many crypts containing a single-marked stem cell will ultimately undergo niche succession in which the marked cell is either eliminated or expanded to occupy multiple stem cell niches within the crypt [14, 15] (also see Supporting Information, Section 1). However, prior to this point, it is expected that reporter-marked progeny of a marked stem cell will be intermixed with unmarked cells. As relatively higher levels of Cre-mediated recombination were used in prior studies, the presence of multiple-marked stem cells within the crypts in those studies may in part account for apparent difference in the frequency of continuous or discontinuous tracts between those and the present study. However, two additional factors may also contribute. First, Mcm2 is shown to be expressed in all cells present within the crypt with the exception of Paneth cells (see Supporting Information, Section 1). This expression pattern is expected to include all Lgr5 expressing cells and, potentially, a larger population of cells with stem cell properties as well. Preliminary studies have also shown that Mcm2 expression occurs within Bmi1 expressing cells (Supporting Information, Section 4). Second, the Mcm2-CreERT2 allele itself may affect the rate of stem cell cycling. This transgene has been shown in prior studies to result in reduced expression of Mcm2 and elevated rates of cancer and stem cell deficiencies when present in a homozygous condition [9]. These phenotypes suggest that reduced Mcm2 expression results in constitutively elevated rates of genetic damage. Although these phenotypes are not seen in mice that are heterozygous for the transgene, it is nonetheless possible that the basal level of replication-related genetic damage is higher in Mcm2-CreERT2 heterozygous mice than in wild-type animals. Consistent with this possibility, recent studies have demonstrated in the absence of p53 activity even a single copy of the Mcm2-CreERT2 results in increased cancer incidence [19], suggesting that p53 activity may mask an elevated level of genetic damage in heterozygous Mcm2-CreERT2 mice.
Although the present studies demonstrate that Mcm2-CreERT2-marked cells participate in niche succession, the rate at which crypts become fully occupied by marked cells appears reduced relative to prior estimates. Here, we observe many crypts at 3 (e.g., Supporting Information, Figs. 2 and 3) or even 11 (Fig. 1L) months following tamoxifen treatment which continue to exhibit unmarked cells. In contrast, prior studies have suggested that, in mice, crypts become fully occupied by marked cells over a period of approximately 7–12 weeks [20]. However, the earlier studies, on which this estimate is largely based, utilized mutagen-induced somatic mutation to mark subsets of cells and there is a concern that the mutagen treatment may have increased the rate of niche succession. As discussed above, more recent studies using CreERT2 marking have been performed under conditions where a higher proportion of crypts initially contain multiple-marked stem cells and which could obscure the presence of a subset of mixed crypts. Alternatively, it is also possible that the Mcm2-CreERT2 transgene influences the rate of niche succession through an effect on stem cell cycling rates.
The intermittent production of progeny and relatively slow rate of niche succession from marked cells has permitted localization of the position of the Mcm2-CreERT2-marked stem cells within the crypt to the +1 position. This localization is based on the finding (by microdissection using fluorescence stereomicroscopy in which labeled cells were unlikely to have been missed) of a reporter-marked stem cell at the basal most (+1) position of crypts containing one or more marked cells in all of well over 20 dissected crypts. Although cases are found in which marked cells are also present at higher locations in the crypt, their presence at locations other than the +1 position was variable. This interpretation is also consistent with whole mount and sectioned images reported in prior studies for both Mcm2-CreERT2- [9] and Lgr5-CreERT2-marked cells [2, 21]. Based on this localization, multiple different assays were used to directly estimate the cycling time these cells. Further, the use of the ratiometric labeling technique has made it possible to measure heterogeneity in the rate of cycling between different +1 position cells within the same crypt. Results from this method show that the rate of cycling of +1 position Mcm2+ cells is heterogeneous and varies from 24 hours or less to as long as 4 days. This result directly confirms that many +1 position Mcm2 expressing cells cycle intermittently. Further, as the mice used in these studies did not carry the Mcm2-CreERT2 transgene, heterogeneity in the rate of cycling measured by this approach cannot be attributed to trangene-dependent elevated levels of genetic damage.
The role of the p53-mediated DNA damage response in establishing cell cycle heterogeneity has also been assessed using multiple assays including ratiometric labeling, the pattern of reporter expression in progeny cell tracts, and short-term pulse labeling at the +1 Mcm2+ cell position. Each of these assays supports the conclusion that +1 position Mcm2+ cells cycle more rapidly in p53 mutant mice, where both the ratiometric labeling assay and the loss of an intermittent pattern in tracts of cells in Mcm2-CreERT2 marking studies suggest that this increased cycling rate is a consequence of loss of the heterogeneity observed in p53 wt mice.
The observation that p53 mediates heterogeneity in the rate of ISC cycling has implications for maintenance of genomic integrity of the tissue. ISCs are known to undergo symmetric divisions to give rise to two daughter stem cells. Further, although crypts contain approximately four stem cells, a variety of tracing studies have shown that over time crypts become clonogenic such that all of the stem cells within the crypt are derived from the same cell lineage [22]. Additionally, fission of crypts can lead to expansion of progeny from a specific stem cell lineage within the tissue as a whole [14, 15, 20, 23]. It is expected that cells that cycle more rapidly would compete more effectively in these processes and the role that mutations leading to a growth advantage may play in accelerating niche succession and progression toward tumorogenesis has been described [22]. Here, +1-position Mcm2+ cells in p53 null mice are shown to cycle more rapidly, on average, than in wild-type mice. However, the increased average rate of cycling is largely a consequence of reducing the degree of heterogeneity in cycling times observed in wild-type mice, resulting in a situation where all cells cycle rapidly. We hypothesize that the p53-mediated heterogeneity observed in wild-type animals is a consequence of a delay in cycling due to DNA damage repair events. This implies that over half of the +1 position Mcm2+ stem cells undergo at least a transient delay in cycling due to the DNA damage response with each cell cycle. Even a transient p53-mediated suppression of cycling of genetically damaged cells could provide a basis for selection against these potentially damaged cells and a higher likelihood that cells not undergoing a damage response will expand through niche succession and/or fission. This mechanism provides an explanation, in addition to stochastic events or mutations allowing a growth advantage, to account for the high rate of niche succession within the intestine. Further, rather than the expansion of crypts containing mutated cells, the consequence of such an ongoing selection process would be enhanced genetic integrity within the tissue as a whole. The observation that deletion of p53 impairs the depletion of chromosomal-instable intestinal stem cells in aging telomere-dysfunctional mice [24] is consistent with this proposed role of the DNA damage response.
Summary and Conclusion
The present study demonstrates that the major limitation to the rate of cell cycling of +1 position ISCs results from the activity of the p53-mediated DNA damage response and, at least during the routine maintenance of the small intestine, there is apparently no specific program by which quiescence is maintained within most +1 position stem cells apart from this response. The extent to which the apparent quiescence of stem cells present in other tissues also reflects the activity of the DNA damage response is unclear. However, p53 has been shown to influence the rate of cell cycling within a variety of tissue types. For example, cell cycle rates within the neurogenic region of the subventricular zone of the brain are increased in p53 null mice [25] and decreased in mice expressing a hyperdominant mutation in p53 [26]. The rate of cycling of neural stem cells within this region has been estimated to be approximately 15 days [10, 27] in wild-type animals; however, in p53 null animals this rate is reduced to approximately 3 days (Pruitt et al., unpublished observations). Similarly, p53 has been implicated in regulating the rate of proliferation of hematopoietic [28] and mesenchymal [29] stem cells. These observations suggest that, to a significant degree, the apparent quiescence of somatic stem cells in a variety of tissues is mediated by the DNA damage response.
Finally, we note that, to the extent that growth stimulatory signals suppress the degree of heterogeneity afforded by p53-dependent suppression of stem cell cycling, such signals would interfere with selection for genetically intact cells within a tissue. In the present study, IGF1 treatment is shown to override p53-mediated cell cycle heterogeneity in the small intestine of wild-type mice resulting in cycling rates that are similar to, or higher than, those in 53 null mice and which show little heterogeneity within the population. Based on the present studies, one predicted consequence of sustained growth stimulatory signals (e.g., constitutive IGF1 overexpression) is suppression of the differential proliferation meditated by p53 due to DNA damage events and a resulting accumulation of genetically damaged stem cells.
Supplementary Material
Acknowledgments
We thank Peter Demant for providing Trp53 mutant mice. The work was supported in part by NIH grant (CA130995), Ellison Medical Foundation Senior Scholar award, and a grant from NYStem to S.C.P. The RPCI DLAR assisted in maintenance of mice and was supported in part by a Comprehensive Cancer Center Support Grant (CA016056).
Footnotes
Author contributions: S.C.P.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing; A.F.: collection and/or assembly of data; A.K.: collection and/or assembly of data.
Disclosure of Potential Conflicts of Interest
S.C.P. the founder and owner of the Buffalo Molecular Target Labs. LLC. The other authors have no financial interests to disclose.
See www.StemCells.com for supporting information available online.
References
- 1.Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255:197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- 2.Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 3.Bjerknes M, Cheng H. Clonal analysis of mouse intestinal epithelial progenitors. Gastroenterology. 1999;116:7–14. doi: 10.1016/s0016-5085(99)70222-2. [DOI] [PubMed] [Google Scholar]
- 4.Bjerknes M, Cheng H. Multipotential stem cells in adult mouse gastric epithelium. Am J Physiol Gastrointest Liver Physiol. 2002;283:G767–G777. doi: 10.1152/ajpgi.00415.2001. [DOI] [PubMed] [Google Scholar]
- 5.Barker N, van de Wetering M, Clevers H. The intestinal stem cell. Genes Dev. 2008;22:1856–1864. doi: 10.1101/gad.1674008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40:915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blow JJ, Dutta A. Preventing re-replication of chromosomal DNA. Nat Rev. 2005;6:476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shima N, Alcaraz A, Liachko I, et al. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat Genet. 2007;39:93–98. doi: 10.1038/ng1936. [DOI] [PubMed] [Google Scholar]
- 9.Pruitt SC, Bailey KJ, Freeland A. Reduced Mcm2 expression results in severe stem/progenitor cell deficiency and cancer. Stem Cells. 2007;25:3121–3132. doi: 10.1634/stemcells.2007-0483. [DOI] [PubMed] [Google Scholar]
- 10.Maslov AY, Barone TA, Plunkett RJ, et al. Neural stem cell detection, characterization and age related changes in the sub-ventricular zone of mice. J Neurosci. 2004;24:1726–1733. doi: 10.1523/JNEUROSCI.4608-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Novak A, Guo C, Yang W, et al. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- 12.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 13.Donehower LA, Harvey M, Slagle BL, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 14.Graham TA, Wright NA. Investigating the fixation and spread of mutations in the gastrointestinal epithelium. Future Oncol. 2008;4:825–839. doi: 10.2217/14796694.4.6.825. [DOI] [PubMed] [Google Scholar]
- 15.Humphries A, Wright NA. Colonic crypt organization and tumorigenesis. Nat Rev Cancer. 2008;8:415–424. doi: 10.1038/nrc2392. [DOI] [PubMed] [Google Scholar]
- 16.Fuchs E. The tortoise and the hair: Slow-cycling cells in the stem cell race. Cell. 2009;137:811–819. doi: 10.1016/j.cell.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raaijmakers MH, Scadden DT. Divided within: Heterogeneity within adult stem cell pools. Cell. 2008;135:1006–1008. doi: 10.1016/j.cell.2008.11.034. [DOI] [PubMed] [Google Scholar]
- 18.Maslov AY, Bailey KJ, Mielnicki LM, et al. Stem/progenitor cell-specific enhanced green fluorescent protein expression driven by the endogenous Mcm2 promoter. Stem Cells. 2007;25:132–138. doi: 10.1634/stemcells.2006-0032. [DOI] [PubMed] [Google Scholar]
- 19.Kunnev D, Rusiniak ME, Kudla A, et al. DNA damage response and tumorigenesis in Mcm2-deficient mice. Oncogene. 2010 doi: 10.1038/onc.2010.125. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park HS, Goodlad RA, Wright NA. Crypt fission in the small intestine and colon. A mechanism for the emergence of G6PD locus-mutated crypts after treatment with mutagens. Am J Pathol. 1995;147:1416–1427. [PMC free article] [PubMed] [Google Scholar]
- 21.Haegebarth A, Clevers Wnt signaling, lgr5, and stem cells in the intestine and skin. H. Am J Pathol. 2009;174:715–721. doi: 10.2353/ajpath.2009.080758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leedham SJ, Wright NA. Expansion of a mutated clone: from stem cell to tumour. J Clin Pathol. 2008;61:164–171. doi: 10.1136/jcp.2006.044610. [DOI] [PubMed] [Google Scholar]
- 23.Totafurno J, Bjerknes M, Cheng H. The crypt cycle. Crypt and villus production in the adult intestinal epithelium. Biophys J. 1987;52:279–294. doi: 10.1016/S0006-3495(87)83215-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Begus-Nahrmann Y, Lechel A, Obenauf AC, et al. p53 deletion impairs clearance of chromosomal-instable stem cells in aging telomere-dysfunctional mice. Nat Genet. 2009;41:1138–1143. doi: 10.1038/ng.426. [DOI] [PubMed] [Google Scholar]
- 25.Meletis K, Wirta V, Hede SM, et al. p53 suppresses the self-renewal of adult neural stem cells. Development. 2006;133:363–369. doi: 10.1242/dev.02208. [DOI] [PubMed] [Google Scholar]
- 26.Medrano S, Burns-Cusato M, Atienza MB, et al. Regenerative capacity of neural precursors in the adult mammalian brain is under the control of p53. Neurobiol Aging. 2009;30:483–497. doi: 10.1016/j.neurobiolaging.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morshead CM, Reynolds BA, Craig CG, et al. Neural stem cells in the adult mammalian forebrain: A relatively quiescent subpopulation of subependymal cells. Neuron. 1994;13:1071–1082. doi: 10.1016/0896-6273(94)90046-9. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Elf SE, Asai T, et al. The p53 tumor suppressor protein is a critical regulator of hematopoietic stem cell behavior. Cell Cycle. 2009;8:3120–3124. doi: 10.4161/cc.8.19.9627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Armesilla-Diaz A, Elvira G, Silva A. p53 regulates the proliferation, differentiation and spontaneous transformation of mesenchymal stem cells. Exp Cell Res. 2009;315:3598–3610. doi: 10.1016/j.yexcr.2009.08.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.