Abstract
Staphylococcus aureus is exposed to multiple antimicrobial compounds, including oxidative burst products and antibiotics. The various mechanisms and regulatory pathways governing susceptibility or resistance are complex and only superficially understood. Bacillus subtilis recently has been shown to control disulfide stress responses by the thioredoxin-related YjbH protein, which binds to the transcriptional regulator Spx and controls its degradation via the proteasome-like ClpXP protease. We show that the S. aureus YjbH homolog has a role in susceptibility to the disulfide stress-inducing agent diamide that is similar to that in B. subtilis, and we demonstrate that the four cysteine residues in YjbH are required for this activity. In addition, the inactivation of YjbH led to moderate resistance to oxacillin and other β-lactam antibiotics, and this phenotypic change was associated with higher penicillin-binding protein 4 levels and increased peptidoglycan cross-linking. Of note, the impact of YjbH on β-lactam susceptibility still was observed when the four cysteines of YjbH were mutated, indicating that the roles of YjbH in disulfide stress and β-lactam resistance rely on different types of interactions. These data suggest that the ClpXP adaptor YjbH has more target proteins than previously thought, and that oxidative burst and β-lactam resistance mechanisms of S. aureus are closely linked.
INTRODUCTION
Staphylococcus aureus is a major human pathogen causing a wide spectrum of diseases that range from mild skin infections to life-threatening septicemia, pneumonia, and toxic-shock syndrome (27). β-Lactam antibiotics such as oxacillin are among to the most effective drugs against S. aureus infections, but their usefulness is continuously decreasing because β-lactam-resistant strains, particularly methicillin-resistant S. aureus (MRSA), are spreading in hospitals and, more recently, in the community at large, alarming international health authorities (11). Resistance is based on β-lactamases (penicillin-resistant S. aureus) or on an alternative peptidoglycan-biosynthetic enzyme, the penicillin-binding protein 2a (MRSA) (5, 14). In addition, mutations in a variety of proteins affecting cell wall biosynthesis and turnover can affect β-lactam susceptibility, leading either to hypersensitivity or to moderate levels of resistance (3, 5, 9, 17, 24, 36). In many cases it remains unclear how the mutated proteins affect staphylococcal β-lactam resistance, and the underlying mechanisms remain only superficially understood.
During the process of infection, S. aureus is engulfed by phagocytic cells such as neutrophils and macrophages, and the bacteria are exposed to the microbicidal activity of these cells. One of the most powerful antimicrobial mechanisms is based on the production of reactive oxygen species (ROS) or nitrogen species (RNS) by phagocyte NADPH oxidase and nitric oxide synthase, respectively (19, 29). In addition, ROS also are generated during incomplete electron transfer in the bacterial respiratory chain (21). Therefore, even nonpathogenic bacteria such as Bacillus subtilis require an ROS detoxification mechanism (40). ROS can interact with numerous targets, including lipids, DNA, and proteins (38). Protein cysteine residues are particularly sensitive to ROS-induced thiol oxidation and concomitant disulfide bond formation within the reducing environment of the cytoplasm (20, 26). S. aureus has developed several strategies to resist oxidative stresses, including thiol-specific low-molecular-weight thiol redox systems such as the dicysteine protein thioredoxin (37), bacillithiol (30), and coenzyme A (13).
B. subtilis encodes a thioredoxin-related protein, YjbH, that controls a wide range of genes involved in disulfide stress or other types of stress. YjbH is inactivated by oxidizing reagents, leading to the disruption of its binding to the oxidative burst-specific transcriptional regulator Spx (25). As a consequence, mutants lacking yjbH exhibit enhanced sensitivity to the nitrosating agent sodium nitroprusside (35), reduced sensitivity to the thiol oxidant diamide, and the down- or upregulation of distinct transcripts that are part of the Spx regulon (25). The cytoplasmic concentration of Spx is adjusted by controlled proteolysis via the ClpXP protease. YjbH represents a ClpXP adaptor protein that directs the susceptibility of Spx to degradation (18).
We identified a yjbH homolog in S. aureus and characterized a defined deletion mutant. YjbH turned out to control the S. aureus susceptibility to disulfide stress as described before for B. subtilis (25). In addition, the deletion of yjbH resulted in moderate resistance to β-lactam antibiotics and in increased peptidoglycan (PGN) cross-linking in S. aureus. While the four cysteine residues of S. aureus YjbH were essential for its ability to affect susceptibility to disulfide stress, they were dispensable for the effect on β-lactam susceptibility, indicating that the two phenotypes rely on different types of interactions of YjbH.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The S. aureus SA113 (ATCC 35556) strains and plasmids used in this study are listed in Table 1. Escherichia coli strain TOP10 was used in cloning experiments. Unless otherwise noted, bacteria were grown in BM broth (1% tryptone, 0.5% yeast extract, 0.5% NaCl, 0.1% K2HPO4, 0.1% glucose) supplemented with appropriate antibiotics at a concentration of 100 μg/ml ampicillin or 10 μg/ml chloramphenicol.
Table 1.
Bacterial strains and plasmids used in this study
| Strain | Plasmid | Function | Restriction sites used for cloning | Primer used for PCR amplification or site-directed mutagenesisa |
|---|---|---|---|---|
| SA113 | pMADΔyjbH | Allelic replacement of yjbH | SacI/BamHI | 5′GAAGCAATCGGAGCTCAATGGATCGA |
| Upstream fragment | 5′CGATACTACGGATCCTGAGTTAGAGTTCG | |||
| NcoI/XbaI | 5′AATTCATGCCATGGAAGAAGCAGGTG | |||
| Downstream fragment | 5′CCAGATGGCGATTTCTAGAAATCTAAAATGCC | |||
| SA113ΔyjbH | pRB474yjbH | Complementation of mutant strain with wild-type copy of S. aureus yjbH | PstI/XbaI | 5′GAGCGCTTAAGATTAACTGCAGATCATATGGTG |
| 5′CTCGTTACATCTAGAGTGTACGAGGTC | ||||
| SA113ΔyjbH | pRB474yjbHBs | Complementation of mutant strain with wild-type copy of B. subtilisyjbH | HindIII/BamHI | 5′ATCAAGCTTAAAGGAGGTATCATCATGACAAACTATCAGCATGAGCTATAC |
| 5′CATATGGATCCCTGCGGCTATTTTTCACATG | ||||
| SA113 | pRB474 | Control wild-type strain with empty vector | ||
| SA113ΔyjbH | pRB474 | Control mutant strain with empty vector | ||
| SA113ΔyjbH | pRB474yjbHC2 (C114G, C116G) | Complementation of mutant strain with mutated yjbH | 5′CAATACCTGCATTTTGAATACCGTCACCAATCATTGATTCTG | |
| 5′CAGAATCAATGATTGGTGACGGTATTCAAAATGCAGGTATTG | ||||
| SA113ΔyjbH | pRB474yjbHC3a (C30G, C114G, C116G) | Complementation of mutant strain with mutated yjbH | 5′GATTGCTGATAATTTGAAGCCATCGGAGCTAAATGGATC | |
| 5′GATCCATTTAGCTCCGATGGCTTCAAATTATCAGCAATC | ||||
| SA113ΔyjbH | pRB474yjbHC3b (C64G, C114G, C116G) | Complementation of mutant strain with mutated yjbH | 5′GGATGTACTTTGAGCTTGGCCTTTCGTTAATACTTTTAACG | |
| 5′CGTTAAAAGTATTAACGAAAGGCCAAGCTCAAAGTACATCC | ||||
| SA113ΔyjbH | pRB474yjbHC4 (C30G, C64G, C114G, C116G) | Complementation of mutant strain with mutated yjbH | 5′GATTGCTGATAATTTGAAGCCATCGGAGCTAAATGGATC | |
| 5′GATCCATTTAGCTCCGATGGCTTCAAATTATCAGCAATC |
Restriction sites in primers used for cloning or mutated nucleotides in primers used for site-directed mutagenesis are indicated in boldface.
Construction and plasmid complementation of yjbH mutant.
The yjbH gene of SA113 was replaced by introducing a tetracycline (tet) resistance cassette originally derived from pT181 (22). Upstream (0.76 kb) and downstream (0.91 kb) DNA fragments flanking the yjbH gene were amplified by PCR and inserted into pMAD (1) along with the 1.7-kb tet fragment using appropriate restriction enzymes. Subsequently, the resulting plasmid pMADΔyjbH was transferred into wild-type SA113, and gene replacement was performed by incubating the temperature-sensitive plasmids at 42°C according to standard procedures (1). The integration of the tet cassette in the resulting mutant ΔyjbH was verified by PCR. The yjbH gene was cloned for complementation experiments in the E. coli- and Staphylococcus-specific shuttle plasmid pRB474 (8) at the PstI and XbaI restriction sites to generate plasmid pRB474yjbH. The gene was inserted downstream of the constitutive plasmid-carried vegII promoter (8) to ensure its expression in S. aureus. Primers and resulting plasmids are given in Table 1.
Site-directed mutagenesis of yjbH.
Selected cysteines within the YjbH protein were replaced with glycines by site-directed mutagenesis using the QuickChange kit (Stratagene). Plasmid pRB474yjbH was used as a template for the first round of mutagenesis, yielding plasmid pRB474yjbHC2 containing mutations C114G and C116G. The latter then served as a template for the second round of mutagenesis for exchanging a third cysteine at position 30 or 64, leading to plasmid pRB474yjbHC3a or pRB474yjbHC3b, respectively. Plasmid pRB474yjbHC3b was used in the last round of mutagenesis to exchange all four cysteines, yielding plasmid pRB474C4. Primers and resulting plasmids are given in Table 1.
Cloning of the yjbH gene from B. subtilis.
To complement ΔyjbH with the yjbH gene of B. subtilis (accession number BSU11550), genomic DNA was extracted from B. subtilis strain 168 using the NucleoSpin tissue kit (Macherey-Nagel). The gene was PCR amplified with Phusion hot start high-fidelity DNA polymerase (New England BioLabs) and subcloned into the shuttle expression vector pRB474 at the BamHI and HindIII restriction sites. The resulting plasmid and primers used are listed in Table 1.
Phenotypic characterization.
The inhibition of bacterial growth upon treatment with diamide [diazenedicarboxylic acid bis(N,N-dimethylamide)] (Sigma-Aldrich) was performed by diluting bacteria from overnight cultures to an optical density (OD) of 0.1 at 578 nm in BM medium containing serial dilutions of the thiol oxidant agent. Ninety percent inhibitory concentrations (IC90) were determined photometrically after 24 h of incubation at 37°C by continued shaking in 24-well plates.
Growth differences between wild-type and ΔyjbH strains under 1,920 different cultivation conditions were analyzed in phenotypic microarrays by Biolog Inc. (Hayward, CA) according to the manufacturer's protocols (6). Briefly, the strains were tested for phenotypic changes in microplates (PM01 to PM08) that examined catabolic pathways, including carbon, nitrogen, phosphorus, and sulfur sources. In addition, plates PM09 to PM20 measured sensitivities to salt and pH stresses and to a variety of antibiotics, antimetabolites, and other inhibitors. As a reporter for active metabolism, the reduction of tetrazolium violet was used (6).
Susceptibility of bacterial strains to different β-lactam and glycopeptide antibiotics was determined by an elipsometry test (Etest) by following the recommendations of the manufacturer (bioMérieux). Briefly, overnight cultures were diluted to an OD of 0.1 at 578 nm in phosphate-buffered saline (PBS) and subsequently plated on BM agar plates using a cotton swab. Etest strips were laid on top of the agar, and the MIC was determined after 24 h of incubation at 37°C.
Extraction of membrane proteins and labeling of penicillin-binding proteins (PBPs) using bocillin-FL.
To isolate membrane proteins, strains SA113, ΔyjbH, and ΔyjbH(pRByjbH) were grown until mid-exponential phase in tryptic soy broth (Difco). The pellet was resuspended in 1/250 of the initial culture volume of buffer A (50 mM KPO4 buffer, pH 7.4, 10 mM MgCl2) and lysed for 40 min with 100 μg/ml lysostaphin (Sigma-Aldrich), at room temperature with stirring, in the presence of 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml DNase, and RNase. A second lysis step was carried out using a French press three times at 800 lb/in2. The resulting lysate was centrifuged at 7,000 × g for 30 min at 4°C. The supernatant was centrifuged at 80,000 rpm in a Beckman Ultracentrifuge TL100 using a TLA 100.3 rotor for 1 h at 4°C, and the membrane pellet was washed with buffer B (50 mM potassium phosphate buffer, pH 7.4, 10 mM MgCl2, 20% glycerol) and resuspended in the same buffer. Total membrane proteins were quantified using the bicinchoninic acid (BCA) kit assay (Pierce) according to the manufacturer's procedures, diluted to 10 mg/ml in buffer B, quickly frozen with liquid N2, and stored at −80°C. Subsequently, the membrane proteins (100 μg) were labeled with 100 μM bocillin-FL (Molecular Probes) for 10 min at 30°C. The reaction was stopped by adding 5-fold-concentrated SDS-PAGE sample buffer (500 mM dithiothreitol [DTT]; 10% SDS; 250 mM Tris-HCl, pH 6.8; 30% glycerol; 0.02% bromophenol blue). Labeled membrane proteins (20 μg) were separated on a 7.5% SDS-PAGE gel and detected using a 473-nm laser of a Fuji FLA-5100 reader. The quantification of the intensity of the fluorescent bands was done using Image J software.
Analysis of mutanolysin-treated PGN by high-performance liquid chromatography (HPLC).
To analyze the degree of PGN cross-linking in SA113 wild-type and ΔyjbH strains, PGN was isolated from exponentially grown bacteria as described before (4, 12). Lyophilized PGN was incubated with hydrofluoric acid (HF) at 4°C for 48 h to remove wall teichoic acid, washed twice with Tris-HCl buffer (50 mM, pH 6.8), and subsequently washed with distilled water until neutral pH was reached. Mutanolysin (62.5 U; Sigma-Aldrich) was used to digest approximately 2 mg of pure PGN in 125 μl sodium phosphate buffer (12.5 mM, pH 5.5) for 16 h at 37°C. After heat inactivation for 5 min at 90°C, insoluble material was removed by centrifugation. Soluble muropeptides were dried, resuspended in distilled water, and reduced with sodium borohydride. An excess of borohydride was neutralized by using 20% phosphoric acid. Analysis was carried out on an Beckman Coulter System Gold with a Prontosil 120-3-C18 AQ column (250 by 4.6 mm, 3 μm diameter; Bischoff Chromatography) using a linear gradient of 10 mM sodium phosphate buffer (pH 2.5) to 30% methanol for 150 min. Muropeptides were detected at 205 nm.
RESULTS AND DISCUSSION
Both S. aureus and B. subtilis encode YjbH homologs, but the two proteins differ with respect to number and location of cysteine residues.
S. aureus was found to encode a YjbH homolog (accession number Q99V89) sharing about 30% sequence identity with the B. subtilis YjbH. The yjbH gene is located in a similar genetic environment in a putative operon with the yjbI gene in both species (25). However, the S. aureus YjbH homolog is shorter in length (216 versus 299 amino acids), the number of cysteines is lower (four versus seven), and the cluster of histidine residues at the N terminus is lacking compared to the B. subtilis protein (Fig. 1). The YjbH proteins share similarity with thioredoxin-like proteins (25) containing characteristic cysteine pairs within the active site (32). While the B. subtilis protein has a CxxC motif close to the N terminus, the S. aureus YjbH contains a CxC motif in the middle of the protein resembling the cysteine pair of the disulfide isomerase YphP of B. subtilis (15). Despite their different locations in the B. subtilis and S. aureus YjbH proteins, the two motifs may have similar roles in the redox status-dependent functions of YjbH. The alignment of the YjbH proteins of S. aureus and B. subtilis with those of Bacillus cereus, Listeria monocytogenes, Lactobacillus acidophilus, and Staphylococcus epidermidis revealed four or more cysteines in all proteins, albeit without allocation to conserved sequence motifs (Fig. 1). Only the cysteine at position 30 of the S. aureus YjbH is consistently conserved, which hints to a particularly important role of this residue.
Fig. 1.
Alignment of S. aureus YjbH with homologs from other bacteria of the phylum Firmicutes. Proteins from the following species were compared (Swiss-Prot accession numbers are given in parentheses): Bsu, Bacillus subtilis (O31606); Bce, Bacillus cereus (Q813V3); Lmo, Listeria monocytogenes (C1L1N5); Lac, Lactobacillus acidophilus (Q5FLA1); Sep, Staphylococcus epidermidis (Q8CT67); and Sau, Staphylococcus aureus (Q99V89). Amino acid positions conserved in all or in the majority of the compared proteins are boxed in black or gray, respectively. Cysteine residues in the S. aureus YjbH are underlined.
Increased resistance to disulfide stress upon yjbH deletion.
To determine whether the inactivation of yjbH has an effect on environmental stress in S. aureus similar to that in B. subtilis, yjbH was deleted from the penicillin-sensitive S. aureus SA113 by homologous recombination. In contrast to recent findings for B. subtilis (25), the resulting S. aureus mutant (ΔyjbH) had no growth defect. Moreover, ΔyjbH grew as well as the SA113 wild-type strain in the presence of sodium nitroprusside, indicating that YjbH plays no major role in S. aureus growth under laboratory conditions or during nitrosative stress (data not shown). However, as reported for B. subtilis (25), S. aureus ΔyjbH also exhibited increased tolerance to the disulfide stress-inducing azo compound diamide (2.4-fold) (Table 2), indicating that the protein has a similar function in disulfide stress in the two species. IC90 values for SA113 wild-type and ΔyjbH strains were 1.9 and 4.5 mM, respectively. The complementation of ΔyjbH with a plasmid-encoded copy of yjbH restored wild-type diamide susceptibility. The expression of the yjbH gene from B. subtilis strain 168 in ΔyjbH complemented the mutant phenotype in a manner similar to that of the S. aureus gene, indicating that the two proteins have similar functions and can replace each other (Table 2).
Table 2.
IC90s of diamide and MICs of oxacillin against S. aureus strains
| Strain | IC90s of diamidea (mM) | MIC of oxacillina (μg/ml) |
|---|---|---|
| Wild type | 1.88 ± 0.042 | 0.11 ± 0.008b |
| ΔyjbH | 4.51 ± 0.043 | 0.28 ± 0.026b |
| ΔyjbH(pRB474yjbH) | 2.25 ± 0.099 | 0.11 ± 0.008 |
| ΔyjbH(pRB474yjbHBs) | 2.34 ± 0.013 | 0.09 ± 0.000 |
| ΔyjbH(pRB474yjbHC2) | 4.37 ± 0.025 | 0.08 ± 0.010 |
| ΔyjbH(pRB474yjbHC3a) | 4.62 ± 0.078 | 0.11 ± 0.010 |
| ΔyjbH(pRB474yjbHC3b) | 4.44 ± 0.019 | 0.13 ± 0.000 |
| ΔyjbH(pRB474yjbHC4) | 4.47 ± 0.050 | 0.13 ± 0.000 |
Data represent means ± standard errors of the means from at least three independent experiments.
Strains contained the empty control vector pRB474.
The B. subtilis YjbH is known to act as an adaptor protein for the proteasome-like ClpXP protease complex that is responsible for protein quality control and controlled turnover of key proteins, such as transcription factors (18, 25). Such adaptor proteins control the degradation of dedicated target proteins by, e.g., exposing ClpXP-susceptible epitopes in target proteins (23). So far, only the transcriptional regulator Spx has been identified as a target protein for YjbH in B. subtilis (23). Spx also is present in S. aureus and has been shown to have a function in controlling the expression of oxidative stress response genes similar to that in B. subtilis (31). Our finding that YjbH inactivation in S. aureus has an effect on resistance to diamide that is similar to that in B. subtilis (25) and that these changes can be reverted to wild-type properties by complementation with the B. subtilis yjbH gene suggests that YjbH also controls Spx levels in S. aureus.
Lack of yjbH leads to moderate β-lactam resistance.
Diamide is a synthetic compound that does not occur in nature. To elucidate under which environmental conditions YjbH affects the fitness of S. aureus, the behaviors of SA113 wild-type and ΔyjbH strains in phenotypic microarrays (6) were compared. Among the 1,920 different tested conditions, only a few revealed differences in the growth behavior of the two strains. Eventually, only the growth difference observed in the presence of β-lactam antibiotics was reproducible (Tables 2 and 3 ). The lack of yjbH led to a 2.0- to 2.5-fold increase in the MIC values of various β-lactam antibiotics compared to those of the SA113 wild type, with the largest observed difference being for oxacillin (Table 3). In contrast, there was no significant difference in the MICs of the glycopeptide antibiotic vancomycin. The wild-type β-lactam sensitivity of the YjbH mutant could be restored by complementation with a wild-type copy of yjbH from S. aureus or B. subtilis, confirming that the phenotype was caused by the targeted yjbH deletion and that the B. subtilis yjbH can replace the S. aureus gene (Table 2).
Table 3.
MICs for five different β-lactams and the glycopeptide antibiotic vancomycin
| Strain | MICa (μg/ml) |
|||||
|---|---|---|---|---|---|---|
| Oxacillin | Cefuroxime | Cefotaxime | Ceftazidime | Penicillin G | Vancomycin | |
| Wild type (pRB474) | 0.11 ± 0.008 | 0.44 ± 0.035 | 0.60 ± 0.091 | 4.00 ± 0.408 | 0.02 ± 0.004 | 1.30 ± 0.122 |
| ΔyjbH (pRB474) | 0.28 ± 0.026 | 1.00 ± 0.000 | 1.25 ± 0.100 | 8.50 ± 0.500 | 0.04 ± 0.003 | 1.55 ± 0.122 |
| Factor (ΔyjbH/wild type)b | 2.5 | 2.3 | 2.1 | 2.1 | 2 | 1.2 |
Data represent means ± standard errors of the means from at least three independent experiments.
Fold increase between mutant and wild-type strains.
PBP4 is upregulated in ΔyjbH.
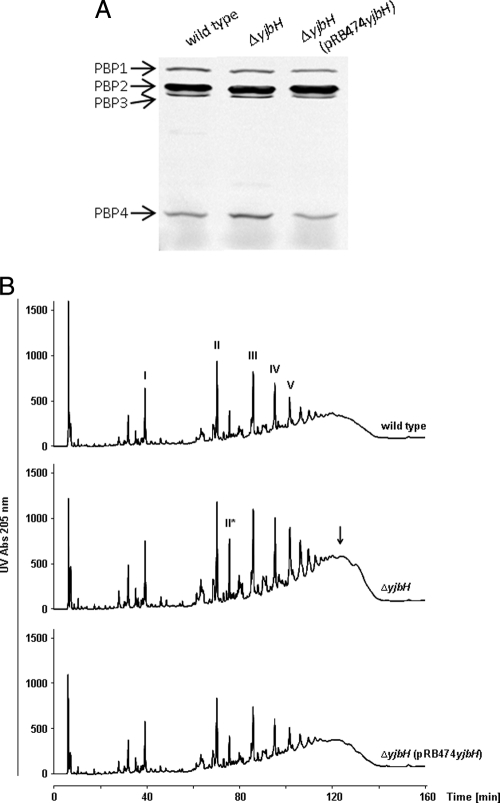
It is likely that YjbH controls the degradation of more than one protein, as shown for other bacterial proteinase adaptor proteins (23), and may direct the turnover of proteins involved in β-lactam tolerance. Staphylococcal susceptibility to β-lactams is governed by several cell wall-biosynthetic proteins, including the penicillin-binding proteins PBP2 and PBP4 (24, 28, 33). To determine if YjbH affects the abundance and activity of these components, penicillin-binding proteins were visualized in wild-type and ΔyjbH strains using bocillin-FL, a fluorescent derivative of penicillin V and a substrate analog for PBPs (2). While yjbH inactivation seemed to have no influence on PBP1, PBP2, and PBP3, a notable, ca. 60% increase in the amount of PBP4 was found in ΔyjbH compared to the level in the wild type, as calculated from the ratio of PBP4 to PBP1 (Fig. 2A). The complementation of ΔyjbH with a plasmid-encoded copy of yjbH restored the wild-type PBP4 level.
Fig. 2.
yjbH inactivation leads to increased PBP4 expression and more highly cross-linked muropeptides than those for the wild type and complemented mutant. (A) Visualization of penicillin-binding proteins with bocillin-FL. (B) HPLC analysis of mutanolysin-digested PGN of SA113 wild-type, ΔyjbH, and ΔyjbH(pRB474yjbH) strains. The polymerization state of muropeptides is indicated by numbers (monomer, I; dimer, II; trimer, III; etc.). A macrocyclic muropeptide dimer with two oligoglycine bridges is marked in the chromatogram of ΔyjbH by II*. An additional arrow points to highly cross-linked muropeptide species in ΔyjbH.
These results are in accordance with previous findings that mutations in pbp4 and other genes, such as pbp2 or fmtA, that cause reduced PGN cross-linking also lead to increased β-lactam susceptibility in S. aureus (24, 28, 33). Conversely, the inactivation of the putative PGN hydrolase LytH has resulted in increased β-lactam tolerance (16, 17). Thus, YjbH may reduce β-lactam susceptibility by increasing PBP4-dependent PGN cross-linking.
yjbH inactivation results in increased PGN cross-linking.
To compare the muropeptide cross-linking patterns of SA113 wild-type, ΔyjbH, and complemented mutant strains, PGN of the three strains was hydrolyzed with mutanolysin, which cleaves the PGN sugar strands and generates muropeptides with different degrees of cross-linking, ranging from monomers to higher oligomers (12). The amounts of muropeptides with higher degrees of cross-linking were more abundant in ΔyjbH, and this tendency increased with the oligomerization state (Fig. 2B, arrow). Moreover, an unusual peak previously shown to represent a macrocyclic muropeptide dimer with two oligoglycine bridges (7) accumulated substantially in PGN of ΔyjbH compared to that in the wild-type strain and complemented strain ΔyjbH(pRB474yjbH). Thus, the increase in S. aureus β-lactam resistance upon the deletion of yjbH is associated with increased PGN cross-linking, most probably as a consequence of increased amounts of PBP4.
The cysteine residues in YjbH are crucial for its role in disulfide stress but not for that in β-lactam resistance.
YjbH controls Spx degradation in B. subtilis in response to disulfide stress, and the cysteine residues in the B. subtilis YjbH are thought to play a crucial role in this process (18). To study if the S. aureus YjbH functions in a similar way, its four cysteine residues were successively replaced with glycine residues, and the ability of the resulting genes to complement ΔyjbH were analyzed. The replacement of C114 and C116 forming the CxC cysteine pair abolished the ability of plasmid-encoded yjbH to restore wild-type-level diamide susceptibility (Table 2), indicating that these cysteine residues are indeed essential for the role of YjbH in disulfide stress. The fact that most of the cysteine residues are not conserved in the various YjbH orthologs (Fig. 1) raises the possibility that they are less important for determining the active conformation of YjbH, but they are important for disrupting its structure when they become oxidized. Future in vitro studies will be necessary to evaluate this hypothesis.
Of note, the replacement of C114 and C116 or of any of the additional four cysteine residues in YjbH did not affect the ability of yjbH to complement ΔyjbH in the β-lactam resistance assay (Table 2). Thus, this second activity of YjbH was independent of the presence of the cysteine residues and may depend on interaction with other target proteins affecting PBP4 expression. Taken together, the roles of S. aureus YjbH in diamide and β-lactam resistance appear to rely on different types of interactions, only the first of which may involve disulfide bridge formation.
Concluding comments.
We confirm a role of YjbH in disulfide stress regulation in S. aureus that is similar to that in B. subtilis and furthermore demonstrate that YjbH affects the expression and activity of a virtually unrelated protein, S. aureus PBP4, thereby altering PGN cross-linking and susceptibility to β-lactam antibiotics. It remains to be analyzed if YjbH influences the abundance of PBP4 in a direct way or indirectly by affecting regulators of pbp4 expression. Interestingly, the B. subtilis YjbH could revert the β-lactam resistance phenotype of S. aureus ΔyjbH to the wild-type level, suggesting that this second activity also is relevant in B. subtilis. It may be related to the recently described implication of YjbH point mutations in low-level S. aureus glycopeptide resistance, which involved additional mutations in the regulatory proteins Stp1 and VraS (34). Of note, S. aureus and Listeria monocytogenes yjbH transposon mutants have been shown to be affected in desiccation resistance (10) and hemolytic activity (39), respectively, suggesting that YjbH has even more functions in Gram-positive cell integrity and stress responses. The investigation of the biological significance of the ClpXP adaptor protein YjbH is a matter of ongoing research to identify ligand proteins in addition to Spx and to understand the entire scheme by which YjbH affects β-lactam susceptibility.
ACKNOWLEDGMENTS
We thank Daniel Kühner for excellent technical assistance.
This work was supported by German Research Foundation grants TR-SFB34 to A.P. and SFB766 to A.P., G.X., and U. B. and Fundação para a Ciência e Tecnologia grant PTDC/BIA-MIC/099151/2008 to M.G.P.
Footnotes
Published ahead of print on 26 September 2011.
REFERENCES
- 1. Arnaud M., Chastanet A., Debarbouille M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70:6887–6891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Atilano M. L., et al. 2010. Teichoic acids are temporal and spatial regulators of peptidoglycan cross-linking in Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 107:18991–18996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Banerjee R., Gretes M., Harlem C., Basuino L., Chambers H. F. 2010. A mecA-negative strain of methicillin-resistant Staphylococcus aureus with high-level beta-lactam resistance contains mutations in three genes. Antimicrob. Agents Chemother. 54:4900–4902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bera A., Herbert S., Jakob A., Vollmer W., Götz F. 2005. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol. Microbiol. 55:778–787 [DOI] [PubMed] [Google Scholar]
- 5. Berger-Bächi B., Rohrer S. 2002. Factors influencing methicillin resistance in staphylococci. Arch. Microbiol. 178:165–171 [DOI] [PubMed] [Google Scholar]
- 6. Bochner B. R. 2009. Global phenotypic characterization of bacteria. FEMS Microbiol. Rev. 33:191–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boneca I. G., Xu N., Gage D. A., de Jonge B. L., Tomasz A. 1997. Structural characterization of an abnormally cross-linked muropeptide dimer that is accumulated in the peptidoglycan of methicillin- and cefotaxime-resistant mutants of Staphylococcus aureus. J. Biol. Chem. 272:29053–29059 [DOI] [PubMed] [Google Scholar]
- 8. Brückner R. 1992. A series of shuttle vectors for Bacillus subtilis and Escherichia coli. Gene 122:187–192 [DOI] [PubMed] [Google Scholar]
- 9. Campbell J., et al. 2011. Synthetic lethal compound combinations reveal a fundamental connection between wall teichoic acid and peptidoglycan biosyntheses in Staphylococcus aureus. ACS Chem. Biol. 6:106–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chaibenjawong P., Foster S. J. Desiccation tolerance in Staphylococcus aureus. Arch. Microbiol. 193:125–135 [DOI] [PubMed] [Google Scholar]
- 11. Chambers H. F., Deleo F. R. 2009. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 7:629–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Jonge B. L., Chang Y. S., Gage D., Tomasz A. 1992. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain. The role of penicillin binding protein 2A. J. Biol. Chem. 267:11248–11254 [PubMed] [Google Scholar]
- 13. del Cardayre S. B., Stock K. P., Newton G. L., Fahey R. C., Davies J. E. 1998. Coenzyme A disulfide reductase, the primary low molecular weight disulfide reductase from Staphylococcus aureus. Purification and characterization of the native enzyme. J. Biol. Chem. 273:5744–5751 [DOI] [PubMed] [Google Scholar]
- 14. de Lencastre H., Oliveira D., Tomasz A. 2007. Antibiotic resistant Staphylococcus aureus: a paradigm of adaptive power. Curr. Opin. Microbiol. 10:428–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Derewenda U., et al. 2009. Structure and function of Bacillus subtilis YphP, a prokaryotic disulfide isomerase with a CXC catalytic motif. Biochemistry 48:8664–8671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fujimura T., Murakami K. 1997. Increase of methicillin resistance in Staphylococcus aureus caused by deletion of a gene whose product is homologous to lytic enzymes. J. Bacteriol. 179:6294–6301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fujimura T., Murakami K. 2008. Staphylococcus aureus clinical isolate with high-level methicillin resistance with an lytH mutation caused by IS1182 insertion. Antimicrob. Agents Chemother. 52:643–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Garg S. K., Kommineni S., Henslee L., Zhang Y., Zuber P. 2009. The YjbH protein of Bacillus subtilis enhances ClpXP-catalyzed proteolysis of Spx. J. Bacteriol. 191:1268–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hampton M. B., Kettle A. J., Winterbourn C. C. 1998. Inside the neutrophil phagosome: oxidants, myeloperoxidase, and bacterial killing. Blood 92:3007–3017 [PubMed] [Google Scholar]
- 20. Hochgräfe F., Mostertz J., Albrecht D., Hecker M. 2005. Fluorescence thiol modification assay: oxidatively modified proteins in Bacillus subtilis. Mol. Microbiol. 58:409–425 [DOI] [PubMed] [Google Scholar]
- 21. Imlay J. A. 2003. Pathways of oxidative damage. Annu. Rev. Microbiol. 57:395–418 [DOI] [PubMed] [Google Scholar]
- 22. Khan S. A., Carleton S. M., Novick R. P. 1981. Replication of plasmid pT181 DNA in vitro: requirement for a plasmid-encoded product. Proc. Natl. Acad. Sci. U. S. A. 78:4902–4906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kirstein J., Molière N., Dougan D. A., Turgay K. 2009. Adapting the machine: adaptor proteins for Hsp100/Clp and AAA+ proteases. Nat. Rev. Microbiol. 7:589–599 [DOI] [PubMed] [Google Scholar]
- 24. Komatsuzawa H., Ohta K., Labischinski H., Sugai M., Suginaka H. 1999. Characterization of fmtA, a gene that modulates the expression of methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 43:2121–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Larsson J. T., Rogstam A., von Wachenfeldt C. 2007. YjbH is a novel negative effector of the disulphide stress regulator, Spx, in Bacillus subtilis. Mol. Microbiol. 66:669–684 [DOI] [PubMed] [Google Scholar]
- 26. Leichert L. I., Scharf C., Hecker M. 2003. Global characterization of disulfide stress in Bacillus subtilis. J. Bacteriol. 185:1967–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lowy F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520–532 [DOI] [PubMed] [Google Scholar]
- 28. Memmi G., Filipe S. R., Pinho M. G., Fu Z., Cheung A. 2008. Staphylococcus aureus PBP4 is essential for beta-lactam resistance in community-acquired methicillin-resistant strains. Antimicrob. Agents Chemother. 52:3955–3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nathan C., Shiloh M. U. 2000. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. U. S. A. 97:8841–8848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Newton G. L., et al. 2009. Bacillithiol is an antioxidant thiol produced in bacilli. Nat. Chem. Biol. 5:625–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pamp S. J., Frees D., Engelmann S., Hecker M., Ingmer H. 2006. Spx is a global effector impacting stress tolerance and biofilm formation in Staphylococcus aureus. J. Bacteriol. 188:4861–4870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pan J. L., Bardwell J. C. 2006. The origami of thioredoxin-like folds. Protein Sci. 15:2217–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pinho M. G., de Lencastre H., Tomasz A. 2001. An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc. Natl. Acad. Sci. U. S. A. 98:10886–10891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Renzoni A., et al. 2011. Whole genome sequencing and complete genetic analysis reveals novel pathways to glycopeptide resistance in Staphylococcus aureus. PLoS One 6:e21577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rogstam A., Larsson J. T., Kjelgaard P., von Wachenfeldt C. 2007. Mechanisms of adaptation to nitrosative stress in Bacillus subtilis. J. Bacteriol. 189:3063–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Trotonda M. P., Xiong Y. Q., Memmi G., Bayer A. S., Cheung A. L. 2009. Role of mgrA and sarA in methicillin-resistant Staphylococcus aureus autolysis and resistance to cell wall-active antibiotics. J. Infect. Dis. 199:209–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Uziel O., Borovok I., Schreiber R., Cohen G., Aharonowitz Y. 2004. Transcriptional regulation of the Staphylococcus aureus thioredoxin and thioredoxin reductase genes in response to oxygen and disulfide stress. J. Bacteriol. 186:326–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zahrt T. C., Deretic V. 2002. Reactive nitrogen and oxygen intermediates and bacterial defenses: unusual adaptations in Mycobacterium tuberculosis. Antioxid. Redox Signal. 4:141–159 [DOI] [PubMed] [Google Scholar]
- 39. Zemansky J., et al. 2009. Development of a mariner-based transposon and identification of Listeria monocytogenes determinants, including the peptidyl-prolyl isomerase PrsA2, that contribute to its hemolytic phenotype. J. Bacteriol. 191:3950–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zuber P. 2009. Management of oxidative stress in Bacillus. Annu. Rev. Microbiol. 63:575–597 [DOI] [PubMed] [Google Scholar]