Abstract
There is limited information on the role of penicillin-binding proteins (PBPs) in the resistance of Acinetobacter baumannii to β-lactams. This study presents an analysis of the allelic variations of PBP genes in A. baumannii isolates. Twenty-six A. baumannii clinical isolates (susceptible or resistant to carbapenems) from three teaching hospitals in Spain were included. The antimicrobial susceptibility profile, clonal pattern, and genomic species identification were also evaluated. Based on the six complete genomes of A. baumannii, the PBP genes were identified, and primers were designed for each gene. The nucleotide sequences of the genes identified that encode PBPs and the corresponding amino acid sequences were compared with those of ATCC 17978. Seven PBP genes and one monofunctional transglycosylase (MGT) gene were identified in the six genomes, encoding (i) four high-molecular-mass proteins (two of class A, PBP1a [ponA] and PBP1b [mrcB], and two of class B, PBP2 [pbpA or mrdA] and PBP3 [ftsI]), (ii) three low-molecular-mass proteins (two of type 5, PBP5/6 [dacC] and PBP6b [dacD], and one of type 7 (PBP7/8 [pbpG]), and (iii) a monofunctional enzyme (MtgA [mtgA]). Hot spot mutation regions were observed, although most of the allelic changes found translated into silent mutations. The amino acid consensus sequences corresponding to the PBP genes in the genomes and the clinical isolates were highly conserved. The changes found in amino acid sequences were associated with concrete clonal patterns but were not directly related to susceptibility or resistance to β-lactams. An insertion sequence disrupting the gene encoding PBP6b was identified in an endemic carbapenem-resistant clone in one of the participant hospitals.
INTRODUCTION
Acinetobacter baumannii is an opportunistic nosocomial pathogen responsible for a variety of serious infections, especially in intensive care units (ICU) (23, 27). Its ability to survive on dry surfaces (6, 21, 41) and to acquire antimicrobial resistance, as well as its suitability for genetic exchange, has contributed to the longevity of this microorganism and its success in causing epidemic outbreaks (19). Carbapenems are currently one of the few options for the treatment of infections caused by multidrug-resistant A. baumannii (15). However, since the early 1990s, the frequency of outbreaks caused by carbapenem-resistant A. baumannii isolates has increased worldwide (5, 16), becoming a significant public health concern (40).
The mechanisms underlying resistance to carbapenems in A. baumannii include (i) the production of β-lactamases, particularly acquired carbapenem-hydrolyzing class D β-lactamases (CHDLs) (7, 31), metallo-β-lactamases (30), and, in rare cases, class A carbapenemases (32), (ii) outer membrane impermeability, associated with the loss or decreased expression of porins (25), and probably (iii) the overproduction of efflux pumps (20). However, there is limited information on the role of penicillin-binding proteins (PBPs) in this phenotype in A. baumannii (12, 13, 26).
PBPs are a family of enzymes that share an evolutionary origin. These enzymes catalyze the synthesis of peptidoglycan, the primary component of the bacterial cell wall, and are also associated with cell morphogenesis and the cell division complex (33). PBPs have been classified into two groups, the high-molecular-mass (HMM) and low-molecular-mass (LMM) PBPs, according to their apparent molecular weights on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels, their amino acid sequences, and their enzymatic and cellular functions (4, 14, 17, 33). The HMM PBPs can be divided into class A (transpeptidase/glycosyltransferase activities) (2, 3) and class B (transpeptidase activity, elongase activity, or divisome) (10, 29) PBPs, depending on the structure of their N-terminal domains (33). The LMM PBPs, or class C PBPs, are d,d-carboxypeptidases and/or endopeptidases involved in cell separation or peptidoglycan maturation or recycling (14). The monofunctional transglycosylases (MGTs), a group of enzymes present in some bacteria, have a single glycosyltransferase domain similar to those of class A PBPs, and their function is still unknown (35).
This study aimed to determine the nucleotide sequences of the genes encoding PBPs in A. baumannii and to analyze their allelic variations in isolates susceptible or resistant to β-lactams.
(This work was presented in part as an oral presentation at the 8th International Symposium on the Biology of Acinetobacter in Rome, Italy, 2010.)
MATERIALS AND METHODS
Bacterial isolates.
A total of 26 nonduplicate A. baumannii clinical isolates presenting different carbapenem susceptibility profiles were collected in three teaching hospitals in Spain: the University Hospital Marqués de Valdecilla, Santander (n = 12), the Hospital Clínic, Barcelona (n = 12), and the University Hospital Virgen Macarena, Seville (n = 2) (Table 1). The two isolates from the third hospital have been described previously (12). These isolates were representative of the most prevalent clones in each institution. Presumptive identification of the isolates as A. baumannii was carried out by amplifying the complete open reading frame of the blaOXA-51-like gene, which is considered chromosomally intrinsic to A. baumannii (31), using primers OXA-69A and OXA-69B as described previously (18). Both primers were also used to detect the presence of ISAba1 upstream of the blaOXA-51-like gene (18). Amplified rRNA gene restriction analysis (ARDRA), using the CfoI, AluI, MboI, RsaI, and MspI enzymes, was carried out as described previously (39) to confirm the genomic species identification of A. baumannii. The reference strain A. baumannii RUH-134 (11) was included as a control for both genomic identification and PCR amplification of PBP genes.
Table 1.
Genetic characterization of the 26 A. baumannii clinical isolates
| Strain | Hospitala/city | PFGE clone | MIC (μg/ml) of the following drugb: |
Oxacillinase type(s) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IPM | MEM | FEP | CAZ | CRO | CTX | ATM | AMK | GEN | MIN | CIP | TGC | CST | ||||
| HUMV-823 | HUMV/Santander | A | >32 | >32 | >32 | >32 | >32 | >32 | >16 | >16 | >8 | 8 | >2 | 1 | 0.5 | OXA-51-like + ISAba1 |
| HUMV-1175 | HUMV/Santander | A1 | >32 | >32 | >32 | >32 | >32 | >32 | >16 | >16 | >8 | 8 | >2 | 1 | 0.5 | OXA-51-like + ISAba1 |
| HUMV-3743 | HUMV/Santander | A2 | >32 | >32 | >32 | >32 | >32 | >32 | >16 | >16 | >8 | 8 | >2 | 1 | 0.5 | OXA-51-like + ISAba1 |
| HUMV-1102 | HUMV/Santander | B | 2 | 2 | 8 | 16 | >32 | >32 | 16 | ≤8 | ≤4 | 4 | >2 | 1 | 1 | OXA-51-like |
| HUMV-2790 | HUMV/Santander | B | 1 | 1 | 8 | >32 | >32 | >32 | >16 | ≤8 | ≤4 | 4 | >2 | 1 | 1 | OXA-51-like |
| HUMV-1319 | HUMV/Santander | C | >32 | >32 | >32 | 16 | >32 | >32 | >16 | >16 | >8 | 2 | >2 | 1 | 0.5 | OXA-51-like, OXA-24 |
| HUMV-5118 | HUMV/Santander | C | >32 | >32 | >32 | >32 | >32 | >32 | >16 | >16 | >8 | 1 | >2 | 1 | 0.5 | OXA-51-like, OXA-24 |
| HUMV-2471 | HUMV/Santander | D | >32 | >32 | 8 | ≤8 | 4 | 4 | 8 | >16 | >8 | 2 | >2 | 0.25 | 0.25 | OXA-51-like, OXA-24 |
| HUMV-4066 | HUMV/Santander | D | >32 | >32 | >32 | ≤8 | 32 | 32 | >16 | >16 | >8 | 2 | >2 | 0.25 | 0.5 | OXA-51-like, OXA-24 |
| HUMV-6457 | HUMV/Santander | D | >32 | >32 | >32 | 16 | 8 | 8 | 16 | ≤8 | ≤4 | 2 | >2 | 0.25 | 0.5 | OXA-51-like, OXA-24 |
| HUMV-2120 | HUMV/Santander | E | 0.25 | 0.25 | 4 | ≤8 | 8 | 8 | 16 | ≤8 | ≤4 | 1 | ≤0.125 | 0.5 | 0.5 | OXA-51-like |
| HUMV-4674 | HUMV/Santander | F | 0.5 | 0.5 | 8 | ≤8 | 8 | 8 | 16 | ≤8 | ≤4 | 0.5 | 1 | 2 | 0.5 | OXA-51-like |
| HC-360 | HC/Barcelona | G | 1 | 2 | 32 | 32 | >32 | >32 | >16 | >16 | >8 | 2 | >2 | 1 | 0.25 | OXA-51-like |
| HC-3581 | HC/Barcelona | G | 1 | 2 | 32 | 32 | >32 | >32 | >16 | >16 | >8 | 2 | >2 | 1 | 0.25 | OXA-51-like |
| HC-4249 | HC/Barcelona | G | >32 | >32 | >32 | 32 | >32 | >32 | >16 | 16 | >8 | <0.5 | >2 | 0.5 | 0.5 | OXA-51-like, OXA-24 |
| HC-4275 | HC/Barcelona | G | >32 | >32 | >32 | 32 | >32 | >32 | >16 | >16 | >8 | 1 | >2 | 1 | 0.125 | OXA-51-like + ISAba1, OXA-24 |
| HC-60 | HC/Barcelona | H | 1 | 4 | 32 | ≤8 | 16 | 16 | >16 | 16 | >8 | <0.5 | 1 | 1 | 0.125 | OXA-51-like |
| HC-3343 | HC/Barcelona | H | 1 | 4 | 32 | ≤8 | 16 | 16 | >16 | 16 | >8 | <0.5 | 1 | 1 | 0.125 | OXA-51-like |
| HC-4256 | HC/Barcelona | H | >32 | >32 | 16 | ≤8 | 32 | 32 | >16 | ≤8 | >8 | <0.5 | 1 | 0.5 | 0.25 | OXA-51-like, OXA-24 |
| HC-3202 | HC/Barcelona | H | >32 | >32 | 32 | 32 | >32 | >32 | >16 | >16 | >8 | 2 | >2 | 1 | 0.25 | OXA-51-like, OXA-24 |
| HC-771 | HC/Barcelona | I | 1 | 4 | 16 | >32 | >32 | >32 | >16 | 16 | >8 | 8 | >2 | 0.5 | 0.25 | OXA-51-like |
| HC-769 | HC/Barcelona | I | 1 | 8 | 16 | >32 | >32 | >32 | >16 | >16 | >8 | 8 | >2 | 1 | 0.25 | OXA-51-like |
| HC-181 | HC/Barcelona | I | >32 | >32 | 16 | >32 | >32 | >32 | >16 | >16 | >8 | 8 | >2 | 0.5 | 0.25 | OXA-51-like + ISAba1 |
| HC-1959 | HC/Barcelona | I | >32 | >32 | >32 | >32 | >32 | >32 | >16 | >16 | 8 | 8 | >2 | 0.5 | 0.25 | OXA-51-like + ISAba1 |
| HUS-31 | HUVM/Seville | J | 0.25 | 0.5 | 32 | 16 | >32 | >32 | >16 | >16 | >8 | 2 | >2 | 2 | 0.25 | OXA-51-like |
| HUS-457 | HUVM/Seville | K | 4 | 4 | 4 | 16 | 32 | 32 | >16 | 16 | >8 | <0.5 | >2 | 0.5 | 0.125 | OXA-51-like |
HUMV, Hospital Universitario Marqués de Valdecilla; HC, Hospital Clinic; HUVM, Hospital Universitario Virgen Macarena.
IPM, imipenem; MEM, meropenem; FEP, cefepime; CAZ, ceftazidime; CRO, ceftriaxone; CTX, cefotaxime; ATM, aztreonam; AMK, amikacin; GEN, gentamicin; MIN, minocycline; CIP, ciprofloxacin; TGC, tigecycline; CST, colistin.
Testing of susceptibility to antimicrobial drugs.
Tigecycline and colistin MICs were determined at the three participating centers by broth microdilution according to Clinical and Laboratory Standards Institute (CLSI) guidelines (8). The MICs of imipenem, meropenem, cefepime, ceftazidime, ceftriaxone, cefotaxime, aztreonam, amikacin, gentamicin, minocycline, and ciprofloxacin were also determined by microdilution for the isolates from the Hospital Clínic and the University Hospital Virgen Macarena, but for the isolates from the University Hospital Marqués de Valdecilla, the MICs of these drugs were determined with Etest strips according to the manufacturer's (AB bioMérieux, Solna, Sweden) recommendations,. The results for tigecycline were interpreted according to the U.S. Food and Drug Administration (FDA) breakpoints for Enterobacteriaceae (for susceptibility, ≤2 μg/ml; for intermediacy, 4 μg/ml; for resistance, ≥8 μg/ml), and the results for the other antimicrobial agents tested were interpreted according to the CLSI breakpoints (9). Pseudomonas aeruginosa ATCC 27853 and Escherichia coli ATCC 25922 were used as quality control strains.
Molecular typing by PFGE.
The clonal relationships of the A. baumannii isolates were determined by pulsed-field gel electrophoresis (PFGE) using the ApaI restriction enzyme (Roche Diagnostics, Indianapolis, IN), as described elsewhere (34). The restriction fragments were separated on 1% (wt/vol) agarose gels in 0.5% Tris-borate-EDTA (TBE) buffer in a CHEF-DR (contour-clamped homogeneous electric field–dynamically regulated) III Mapper electrophoresis system (Bio-Rad Laboratories, Richmond, CA) for 19 h at 14°C using a pulse ramping rate changing from 5 to 20 s at 6 V/cm. DNA fingerprints were interpreted as recommended by Tenover et al. (37). The reference strains A. baumannii RUH-875 and RUH-134 (11), representatives of major international clones I and II, respectively, were also used as comparators.
Identification of PBP genes.
The genes encoding PBPs were identified on the basis of the six complete genomes of A. baumannii that had been deposited in GenBank by the time this study was started. The following organisms were considered: A. baumannii strains AB0057 (accession no. NC_011586), ATCC 17978 (accession no. NC_009085), SDF (accession no. NC_010400), AYE (accession no. NC_010410), ACICU (accession no. NC_010611), and AB307-0294 (accession no. NC_011595). Consensus sequences were obtained for each PBP gene identified, and a series of primers for PCR amplification and sequencing was designed, as listed in Table S1 in the supplemental material. The A. baumannii genomes deposited in GenBank after the beginning of this study were also considered for comparison and analysis of the PBP genes. These genomes include those of A. baumannii strains AB056 (accession no. NZ_ADGZ01000571), AB058 (accession no. NZ_ADHA01000108), AB059 (accession no. NZ_ADHB01000264), AB900 (accession no. NZ_ABXK01000007), and ATCC 19606 (accession no. NZ_ACQB00000000).
Analysis of the PBP genes in A. baumannii clinical isolates.
Genomic DNA from clinical isolates was extracted with InstaGene Matrix (Bio-Rad Laboratories, Hercules, CA) according to the manufacturer's recommendations. PBP genes were amplified by PCR under the following conditions: 95°C for 5 min, followed by 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 to 3 min, according to the amplicon size, and finally 72°C for 7 min. PCR products were analyzed on 1% (wt/vol) agarose gels stained with ethidium bromide. PCR products were purified by using a High Pure PCR product purification kit (Roche Diagnostics, Mannheim, Germany). Bidirectional DNA sequencing was performed by Macrogen Inc. (Seoul, South Korea). Each PBP gene was named and classified on the basis of homology with other PBP sequences from different microorganisms deposited in GenBank, as well as by the recognition of conserved motifs. All PBP sequences were compared with those of A. baumannii ATCC 17978 (1).
Nucleotide sequence accession numbers.
The nucleotide sequences of the PBP genes of the A. baumannii clinical isolates in this study have been deposited in the GenBank nucleotide sequence database and were assigned the following accession numbers, according to each PBP/MTG gene and strain: for ponA, JF746077 to JF746102; for mrcB, JF746103 to JF746128; for pbpA, JF745973 to JF745998; for ftsI, JF745999 to JF746024; for dacC, JF746025 to JF746050; for dacD, JF746051 to JF746074; for pbpG, JF746129 to JF746154; and for mtgA, JF745947 to JF745972.
RESULTS
PBP genes in A. baumannii.
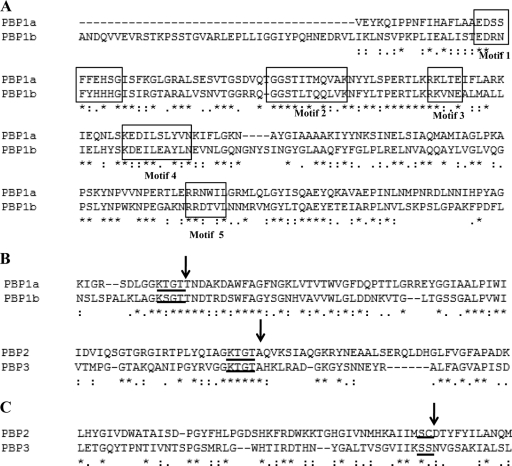
Seven PBP genes and one MTG gene were identified in the six A. baumannii genomes analyzed in this study (Table 2). Four HMM PBPs were found: PBP1a (encoded by ponA), PBP1b (mrcB), PBP2 (pbpA or mrdA), and PBP3 (ftsI). According to their N-terminal domains, they were assigned to class A (PBP1a and PBP1b) or class B (PBP2 and PBP3). Sequence alignment revealed the five conserved motifs [EDXXFXXHXG, GXSTXX(M/Q)QXXK, RKXXE, KXXIXXYXN, and RXXXXL (where X is any amino acid)] of the glycosyltransferase N-terminal domain in both HMM class A PBPs (Fig. 1A). In the C-terminal penicillin-binding (PB) domain, with transpeptidase activity, the residue following motif 3 [K(T/S)GT] was a threonine in class A PBPs (KTGTT for PBP1a and KSGTT for PBP1b) and an alanine in class B PBPs (KTGTA), as expected (Fig. 1B). The HMM class A PBPs, PBP1a and PBP1b, are the major transglycosylases-transpeptidases in A. baumannii and are probably involved in the elongation of non-cross-linked glycan chains of peptidoglycan.
Table 2.
The penicillin-binding proteins of A. baumannii
| PBP (gene) | PBP classificationa | Molecular function | Possible physiological functionb | % identityc with: |
|
|---|---|---|---|---|---|
| Pseudomonas aeruginosa | Escherichia coli | ||||
| PBP1a (ponA) | HMM class A (subclass A1) | Transglycosylase and transpeptidase | Peptidoglycan synthesis | 43 | 36 |
| PBP1b (mrcB) | HMM class A (subclass A2) | Transglycosylase and transpeptidase | Peptidoglycan synthesis | 42 | 32 |
| MtgA (mtgA) | Monofunctional enzymes (MGTs) | Monofunctional transglycosylase | Unknown | 34 | 32 |
| PBP2 (mrdA, also called pbpA) | HMM class B (subclass B2) | Transpeptidase | Cell elongation | 45 | 39 |
| PBP3 (ftsI) | HMM class B (subclass B3) | Transpeptidase | Septum formation (cell division) | 39 | 39 |
| PBP5/6 (dacC) | LMM class C (type 5) | d-Ala-d-Ala-carboxypeptidase | Unknown | 48 | 40 |
| PBP6b (dacD) | LMM class C (type 5) | d-Ala-d-Ala-carboxypeptidase | Unknown | 31 | |
| PBP7/8 (pbpG) | LMM class C (type 7) | Endopeptidase | Unknown | 38 | 34 |
HMM, high molecular mass; LMM, low molecular mass.
Enzymatic activities and functions predicted by sequence homology analysis.
Amino acid sequence homology with the PBP and MGT proteins from the genomes of P. aeruginosa strain PA7 (accession no. NC_009656), E. coli O157:H7 strain EDL933 (accession no. NC_002655.2), and A. baumannii strain SDF (accession no. NC_010400). Analysis was performed using CLUSTAL W2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/zurlx).
Fig. 1.
Identification of the conserved motifs in the consensus sequences encoded by the HMM PBP genes of A. baumannii genomes used for the classification of PBPs, according to the work of Sauvage et al. (33). (A) Localization of the five characteristic conserved motifs of the N-terminal domain (boxed) in both HMM class A PBPs, PBP1a and PBP1b. (B) Arrows indicate differences in motif 3 of the C-terminal domain (underlined) between the HMM class A and class B PBPs. (C) Arrows indicate the motif 2 residues in the third position of the active site of the HMM class B PBPs, used to divide these PBPs into subclasses B2 and B3. The first two residues of the active site are underlined.
The presence of an aspartic acid residue at the third position of motif 2 (SXD) in the active site of PBP2 (pbpA) and the presence of an asparagine residue at the same position in PBP3 confirmed the classification of these PBPs into subclasses B2 and B3, respectively (Fig. 1C). Both PBP2 and PBP3 are monofunctional transpeptidases. PBP2 is a member of subclass B2, a group of proteins involved in cell elongation. PBP3, as a member of subclass B3, is probably associated with cell division. The fstI gene was localized in the A. baumannii genomes in an operon with the mraW, mraY, ftsL, murE, and murF genes, which are associated with the divisome. MtgA (encoded by mtgA) is a monofunctional transglycosylase with a glycosyltransferase domain similar to those of PBP1a and PBP1b.
Three LMM PBPs have been found in A. baumannii: PBP5/6 (encoded by dacC), PBP6b (dacD), and PBP7/8 (pbpG). These LMM PBPs are associated with cell separation and the maturation or recycling of peptidoglycan. Analysis of the genes encoding PBP5 and PBP6 in A. baumannii genomes showed that they are identical at the nucleotide and amino acid levels. Thus, this PBP is referred to as PBP5/6 (dacC). This PBP and PBP6b (dacD) were classified as class C type 5 PBPs, and both are presumably d-Ala-d-Ala-carboxypeptidases. PBP7/8 (pbpG) is a class C type 7 PBP with putative endopeptidase activity. No class C type 4 PBP genes seem to be present in A. baumannii.
Hot spot mutations in PBP genes and β-lactam resistance.
The 26 clinical isolates were identified as A. baumannii by ARDRA, and all isolates carried a blaOXA-51 allele (Table 1). PFGE profile analysis revealed that the 26 isolates were categorized into 11 different clones. All A. baumannii isolates were susceptible to colistin and tigecycline, with MICs ranging from 0.25 to 1 μg/ml and 0.5 to 1 μg/ml, respectively. Minocycline also showed good coverage against the A. baumannii isolates tested (73.1% susceptibility; MIC at which 50% of isolates were inhibited [MIC50], 2 μg/ml). High resistance rates were observed for expanded-spectrum cephalosporins (MIC50, ≥32 μg/ml), cefepime (MIC50, ≥32 μg/ml), and aztreonam (MIC50, >16 μg/ml). The MICs of imipenem and meropenem ranged from 0.25 to >32 μg/ml, and for 53% of the isolates, the MICs of both these antimicrobial agents were >32 μg/ml. The majority of the isolates were resistant to ciprofloxacin (MIC50, >2 μg/ml) and aminoglycosides (MIC50s, >8 μg/ml for gentamicin and >16 μg/ml for amikacin). All the A. baumannii isolates resistant to carbapenems (n = 14) carried the plasmid-borne carbapenemase gene blaOXA-24 or the insertion sequence ISAba1 upstream of the chromosomal blaOXA-51-like gene and/or AmpC (data not shown).
Analysis of the nucleotide sequences of the seven PBP genes and one MGT gene in the A. baumannii clinical isolates revealed specific hot spot mutation regions in all genes. However, most of the allelic variations observed were silent mutations (data not shown). The main changes in the different PBP genes relative to the sequences of strain ATCC 17978 are shown in Table 3. Although a few mutations were found in the amino acid sequences of the PBPs, none of these mutations could be related to β-lactam resistance. The observed changes in the amino acid sequence were associated with specific clonal patterns, and in general, the same mutations were observed in organisms from the different hospitals as well as in the 10 genomes already sequenced (Table 4). Interestingly, comparison of the amino acid consensus sequence corresponding to each PBP gene in the A. baumannii genomes deposited in GenBank with the corresponding amino acid consensus sequence of the 26 A. baumannii clinical isolates showed that these genes were highly conserved (identity, 99.6% to 100.0%).
Table 3.
Point mutations observed in the PBP genes of A. baumannii strains susceptible and resistant to carbapenems
| Strain origina and susceptibilityb | Point mutation(s)c (clonal pattern[s]) |
|||||||
|---|---|---|---|---|---|---|---|---|
| PBP1a (ponA) | PBP1b (mrdA) | PBP2 (pbpA) | PBP3 (fstl) | PBP5/6 (dacC) | PBP6b (dacD) | PBP7/8 (pbpG) | MtgA (mtgA) | |
| HUMV | ||||||||
| Imipenem susceptible | L147I (B), T636A (E) | P112S (B) | P665A (E) | 0d | N329S (B), T374V (B, E), N296D (B, E), N307S (B, E) | P28S (B), T188P (B) | T45S (B, F), A84T (B) | F18L (B, E, F), T49P (B, E, F), I54V (E, F), N179S (B, E, F) |
| Imipenem resistant | A244T (C), S382N (C), T636A (C, D) | P112S (A), P764S (C) | V509I (C), E110Q (D) | G523V (C) | N329S (A, C, D), T374V (A), N296D (A), N307S (A) | P28S (A), A277T (D), V350I (D), S429N (D) | T45S (A, C, D), A84T (A) | F18L (A, C, D), T49P (A, C, D), Q100E (C), N179S (A, C, D) |
| HC | ||||||||
| Imipenem susceptible | T38A (H), L147I (I), A244T (G), S382N (G), T636A (G) | P112S (I), P764S (G) | V509I (G) | G523V (G), H370Y (I) | N329S (I) | P28S (I), T188P (G) | T39I (I), T45S (G, H, I), A84T (I) | F18L (G, H, I), T49P (G, I), Q100E (G), N179S (G, H, I) |
| Imipenem resistant | L147I (I), A244T (G, H), S382N (G, H), T636A (G, H) | P112S (I), P764S (G, H) | V509I (G) | G523V (G), H370Y (I) | N329S (I) | P28S (I), T188P (G, H) | T39I (I), T45S (G, H, I), A84T (I) | F18L (G, H, I), T49P (G, I), Q100E (G, H), N179S (G, H, I) |
| HUVM (imipenem susceptible) | A244T (J), S382N (J), T636A (J) | P764S (J) | V509I (J), E110Q (K) | G523V (J) | 0d | A277T (K), V350I (K), S429N (K) | T45S (J, K) | F18L (J, K), T49P (J, K), Q100E (J), N179S (J, K) |
HUMV, Hospital Universitario Marqués de Valdecilla; HC, Hospital Clinic; HUVM, Hospital Universitario Virgen Macarena.
Imipenem susceptible (MIC, ≤4 μg/ml) or imipenem resistant (MIC, ≥16 μg/ml).
Amino acid substitution(s) relative to the genome of A. baumannii strain ATCC 17978.
No mutations relative to the genome of A. baumannii strain ATCC 17978 were observed.
Table 4.
Point mutations observed in the PBP genes of the 10 A. baumannii genomes and the reference strain RUH-134
| Strain | Point mutation(s) |
|||||||
|---|---|---|---|---|---|---|---|---|
| PBP1a (ponA) | PBP1b (mrdA) | PBP2 (pbpA) | PBP3 (fstl) | PBP5/6 (dacC) | PBP6b (dacD) | PBP7/8 (pbpG) | MtgA (mtgA) | |
| RUH-134 | L147I | 0a | 0 | 0 | N329S | P28S | A84T | F18L, T49P |
| ACICU | L147I | P112S | 0 | A346V, H370Y | 0 | P28S, K229Q | T45S, A84T | F18L, T49P, N179S |
| SDF | A224T, T636A | S274A, R590H, Q601E, R712H, Q765E | 0 | 0 | S5N | S418N | T45S, R218H | V8M, F18M, T49P, Q100E, N179S |
| AYE | T38A, A244T | N513H | P665A | 0 | 0 | 0 | T45S | F18L, T49P, Q100E, N179S |
| AB0057 | T38A, A244T | N513H | P665A | 0 | 0 | 0 | T45S | F18L, T49P, Q100E, N179S |
| AB307-0294 | T38A, A244T, A613T | N513H | P665A | 0 | N329S | 0 | T45S | F18L, T49P, Q100E, N179S |
| AB900 | T636A | 0 | E110Q | 0 | 0 | A277T, V350I, S429N | T45S | F18L, T49P, N179S |
| AB056 | T38A, A244T, T776A | 0 | P665A | 0 | 0 | 0 | T45S | F18L, T49P, Q100E, N179S |
| AB058 | T38A, A244T, T776A | 0 | P665A | V565L | 0 | 0 | T45S | F18L, T49P, Q100E, N179S |
| AB059 | T38A, A244T, T776A | 0 | P665A | 0 | 0 | 0 | T45S | F18L, T49P, Q100E, N179S |
| ATCC 19606 | V623I | 0 | 0 | 0 | 0 | Q143K | T45S | F18L, T49P, N179S |
0, no mutations relative to the A. baumannii ATCC 17978 genome were observed.
In two isolates (HUMV-1319 and HUMV-5118) of an endemic carbapenem-resistant clone (MIC, >32 μg/ml) from one of the participating centers, PCR amplification of the carboxypeptidase PBP6b gene showed a fragment of 2,410 bp instead of the expected 1,320 bp. Sequencing of this amplicon indicated the presence of an insertion sequence (IS) disrupting the PBP6b gene. This IS was identical to an IS30 family transposase found in the genome of A. baumannii strain ACICU and was similar to ISAba125, differing only in His222Tyr codons in the transposase gene. Two imperfect inverted repeats (IR-L [AAACTTGAAGTCGACA] and IR-R [TGTCGCACCTCATGTTT]) bordering the transposase gene were also identified. The IS was not found in the carO gene in these two isolates (data not shown).
DISCUSSION
PBPs are involved in the metabolism of peptidoglycan, an essential component of the bacterial cell wall (33). β-Lactams mimic the d-Ala-d-Ala dipeptide and act as suicide inhibitors binding covalently to the PBPs (45). Inhibition of PBPs causes instability in the cell wall, resulting in growth inhibition or lysis (42). In this study, seven PBP genes and one MGT gene were found in the genomes of several A. baumannii strains, encoding PBP1a, PBP1b, PBP2, PBP3, PBP5/6, PBP6b, PBP7/8, and MtgA. The amino acid sequences of these PBPs in the A. baumannii clinical isolates were compared with those in strain ATCC 17978. This strain was isolated in the early 1950s (1), prior to the development of the majority of the antimicrobial agents used in clinical practice. This comparison showed that PBP genes are highly conserved in all A. baumannii strains analyzed. The few point mutations observed could not be associated with carbapenem resistance, since they were found in both susceptible and resistant strains. Some point mutations were observed in specific isolates, especially in the genome of strain SDF (Table 4), a multisusceptible A. baumannii strain. It is most probable that these variations are associated with clonal patterns.
PBPs with low affinity for β-lactams had been described for Acinetobacter spp. (13, 26, 38). Gehrlein et al. (13) described seven PBPs in an A. baumannii clinical strain with apparent molecular sizes of 94, 84, 65, 61, 48, 40, and 24 kDa, based on phenotypic assays; the first six could be identified as PBP1a (94.74 kDa), PBP1b (88.26 kDa), PBP2 (74.42 kDa), PBP3 (67.66 kDa), PBP5/6 (48.84 kDa), and PBP6 (41.78 kDa), respectively, but the last PBP did not correlate with PBP7/8 (36.86 kDa). They also observed that imipenem could select in vitro for a resistant A. baumannii mutant showing a complex reorganization of PBPs. Because no alterations in outer membrane proteins (OMPs) or in β-lactamase production were detected in the imipenem-resistant mutant, the authors associated the alterations in PBP profiles with the observed imipenem resistance. In another study, Obara and Nakae (26) evaluated 12 imipenem-susceptible strains of Acinetobacter calcoaceticus and detected six PBP bands of 94 (PBPla), 92 (PBPlb), 86 (PBPlc), 74 (PBP2), 59 (PBP3), and 42 (PBP4) kDa. In vitro-selected mutants resistant to cefoxitin, cefoperazone, or ceftazidime showed reduced expression of porins, as well as alterations in PBP expression and/or affinity for β-lactams. In another study, an imipenem-resistant nosocomial strain of A. baumannii showing PBPs with low affinity for β-lactamase inhibitors, especially clavulanic acid, was reported (38). This study also showed that sulbactam bound to PBPs better than tazobactam, even in imipenem-resistant strains, which may explain the satisfactory in vitro activity of sulbactam against some multidrug-resistant A. baumannii isolates (22, 28).
Fernández-Cuenca et al. (12) used 12% SDS-PAGE gels marked with 125I-labeled ampicillin to evaluate the PBP profiles of two groups of A. baumannii isolates with imipenem and meropenem MICs of 0.25 to 2 μg/ml and 4 to 32 μg/ml, respectively. Isolates HUS-31 and HUS-457, included in the present study, are representative of these two groups, respectively. Isolates with carbapenem MICs of ≥4 μg/ml showed reduced expression of a 73.2-kDa PBP named PBP2 (which may correspond to the PBP2 identified in this study [74.42 kDa]), associated with the production of several β-lactamases, including oxacillinases (which has been confirmed in the present study), and, in some isolates, with the loss of a 22.5-kDa porin. We have not observed any mutations in PBP2 (or any other PBP) of strain HUS-457, suggesting that regulatory mechanisms could be involved in the reported decreased expression of the 73.2-kDa protein in this isolate.
An IS has been found to disrupt the dacD gene (encoding PBP6b) in two isolates of a carbapenem-resistant endemic clone. This IS is similar to ISAba125, which was previously reported to disrupt the carO gene (25), coding for a porin associated with resistance to carbapenems in A. baumannii. Although E. coli mutants lacking one or all of the LMM PBPs failed to substantially affect cell division or elongation (14), Moya et al. (24) have reported that in P. aeruginosa, inactivation of the dacB gene (encoding the nonessential PBP4, but absent in A. baumannii) is associated with a complex high-level β-lactam resistance, triggering ampC overproduction and the specific activation of the CreBC two-component regulator, which also activates the expression of β-lactamase in an Aeromonas PBP4-like mutant (36). Zamorano et al. (44) also showed that dacB inactivation produced significantly higher MICs of antipseudomonal penicillins and cephalosporins than ampD inactivation. We may speculate that the importance of inactivation of the PBP6 gene in carbapenem resistance might be marginal. It was observed in only 2 of the 14 carbapenem-resistant isolates we studied (both of which belonged to the same clone), in which resistance can be explained by the production of OXA-24. In fact, other OXA-24-producing isolates are also resistant to carbapenems (Table 1), even though they do not have an inactivated PBP6 gene. Unfortunately, attempts to silence the dacD gene in the carbapenem-susceptible A. baumannii strain ATCC 19606 (to verify the actual role of PBP6b in β-lactam resistance) were unsuccessful until now. Additional studies on this topic are warranted.
Recently, Yun et al. (43) evaluated proteome regulation in an imipenem-resistant A. baumannii strain under antibiotic stress conditions. They observed that the levels of RND family transporters (AdeABC and AdeJIK), the PBP genes ponA (PBP1a), ftsI (PBP3), and dacC (PBP5/6), and, noticeably, AmpC β-lactamase were increased in the presence of imipenem. In contrast, repression of the OMPs OmpA and OmpW was observed under the same conditions. These results suggest that together such mechanisms contribute to imipenem resistance in A. baumannii.
In the presence of efficient alternatives, such as CHDL production and/or porin loss, the selective pressure of β-lactams against PBPs in A. baumannii may be insufficient to select for structural changes that are directly involved in β-lactam resistance. On the other hand, it is possible that other complex mechanisms associated with the regulation of PBP genes, as well as posttranscriptional events, could cause β-lactam resistance in this pathogen. Our findings may represent a first step toward understanding of the actual role of the PBPs in the complex resistance of A. baumannii to β-lactams.
Supplementary Material
ACKNOWLEDGMENTS
This work was partially supported by the Instituto de Salud Carlos III, Spanish Network for Research in Infectious Diseases (REIPI, RD06/0008), and FIS (PI080209). We are grateful to the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), which gave a PDEE grant to R.C. (protocol 4149/08-4). We acknowledge the funding of MICINN (BFU2009-09200) to J.A.A.
L. Dijkshoorn is thanked for providing A. baumannii strains RUH-134 and RUH-875.
We have no conflicts of interest to report.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 26 September 2011.
REFERENCES
- 1. Baumann P. 1968. Isolation of Acinetobacter from soil and water. J. Bacteriol. 96:39–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bertsche U., Breukink E., Kast T., Vollmer W. 2005. In vitro murein peptidoglycan synthesis by dimers of the bifunctional transglycosylase-transpeptidase PBP1B from Escherichia coli. J. Biol. Chem. 280:38096–38101 [DOI] [PubMed] [Google Scholar]
- 3. Born P., Breukink E., Vollmer W. 2006. In vitro synthesis of cross-linked murein and its attachment to sacculi by PBP1A from Escherichia coli. J. Biol. Chem. 281:26985–26993 [DOI] [PubMed] [Google Scholar]
- 4. Cabeen M. T., Jacobs-Wagner C. 2005. Bacterial cell shape. Nat. Rev. Microbiol. 3:601–610 [DOI] [PubMed] [Google Scholar]
- 5. Carvalho K. R., et al. 2009. Dissemination of multidrug-resistant Acinetobacter baumannii genotypes carrying blaOXA-23 collected from hospitals in Rio de Janeiro, Brazil. Int. J. Antimicrob. Agents 34:25–28 [DOI] [PubMed] [Google Scholar]
- 6. Catalano M., Quelle L. S., Jeric P. E., Di Martino A., Maimonet S. M. 1999. Survival of Acinetobacter baumannii on bed rails during an outbreak and during sporadic cases. J. Hosp. Infect. 42:27–35 [DOI] [PubMed] [Google Scholar]
- 7. Chen T. L., et al. 2010. Emergence and distribution of plasmids bearing the blaOXA-51-like gene with an upstream ISAba1 in carbapenem-resistant Acinetobacter baumannii in Taiwan. Antimicrob. Agents Chemother. 54:4575–4581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clinical and Laboratory Standards Institute 2009. Methods for dilution antimicrobial susceptibility test for bacteria that grow aerobically. Eighth edition: approved standard M7-A8. CLSI, Wayne, PA [Google Scholar]
- 9. Clinical and Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing. Twenty-first edition. Informational supplement M100-S21. CLSI, Wayne, PA [Google Scholar]
- 10. Den Blaauwen T., Aarsman M. E., Vischer N. O., Nanninga N. 2003. Penicillin-binding protein PBP2 of Escherichia coli localizes preferentially in the lateral wall and at mid-cell in comparison with the old cell pole. Mol. Microbiol. 47:539–547 [DOI] [PubMed] [Google Scholar]
- 11. Dijkshoorn L., et al. 1996. Comparison of outbreak and nonoutbreak Acinetobacter baumannii strains by genotypic and phenotypic methods. J. Clin. Microbiol. 34:1519–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fernández-Cuenca F., et al. 2003. Relationship between beta-lactamase production, outer membrane protein and penicillin-binding protein profiles on the activity of carbapenems against clinical isolates of Acinetobacter baumannii. J. Antimicrob. Chemother. 51:565–574 [DOI] [PubMed] [Google Scholar]
- 13. Gehrlein M., Leying H., Cullmann W., Wendt S., Opferkuch W. 1991. Imipenem resistance in Acinetobacter baumannii is due to altered penicillin-binding proteins. Chemotherapy 37:405–412 [DOI] [PubMed] [Google Scholar]
- 14. Ghosh A. S., Chowdhury C., Nelson D. E. 2008. Physiological functions of d-alanine carboxypeptidases in Escherichia coli. Trends Microbiol. 16:309–317 [DOI] [PubMed] [Google Scholar]
- 15. Giamarellou H., Antoniadou A., Kanellakopoulou K. 2008. Acinetobacter baumannii: a universal threat to public health? Int. J. Antimicrob. Agents 32:106–119 [DOI] [PubMed] [Google Scholar]
- 16. Giannouli M., et al. 2010. Molecular epidemiology of multidrug-resistant Acinetobacter baumannii in a tertiary care hospital in Naples, Italy, shows the emergence of a novel epidemic clone. J. Clin. Microbiol. 48:1223–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goffin C., Ghuysen J. M. 1998. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 62:1079–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Héritier C., et al. 2005. Characterization of the naturally occurring oxacillinase of Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:4174–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Higgins P. G., Dammhayn C., Hackel M., Seifert H. 2010. Global spread of carbapenem-resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 65:233–238 [DOI] [PubMed] [Google Scholar]
- 20. Huang L., Sun L., Xu G., Xia T. 2008. Differential susceptibility to carbapenems due to the AdeABC efflux pump among nosocomial outbreak isolates of Acinetobacter baumannii in a Chinese hospital. Diagn. Microbiol. Infect. Dis. 62:326–332 [DOI] [PubMed] [Google Scholar]
- 21. Jawad A., Seifert H., Snelling A. M., Heritage J., Hawkey P. M. 1998. Survival of Acinetobacter baumannii on dry surfaces: comparison of outbreak and sporadic isolates. J. Clin. Microbiol. 36:1938–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Michalopoulos A., Falagas M. E. 2010. Treatment of Acinetobacter infections. Expert Opin. Pharmacother. 11:779–788 [DOI] [PubMed] [Google Scholar]
- 23. Monterrubio-Villar J., et al. 2009. Outbreak of multiresistant Acinetobacter baumannii in a polyvalent intensive care unit: clinical, epidemiological analysis and PFGE-printing evolution. Eur. J. Clin. Microbiol. Infect. Dis. 28:1281–1284 [DOI] [PubMed] [Google Scholar]
- 24. Moya B., et al. 2009. Beta-lactam resistance response triggered by inactivation of a nonessential penicillin-binding protein. PLoS Pathog. 5:e1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mussi M. A., Limansky A. S., Viale A. M. 2005. Acquisition of resistance to carbapenems in multidrug-resistant clinical strains of Acinetobacter baumannii: natural insertional inactivation of a gene encoding a member of a novel family of β-barrel outer membrane proteins. Antimicrob. Agents Chemother. 49:1432–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Obara M., Nakae T. 1991. Mechanisms of resistance to β-lactam antibiotics in Acinetobacter calcoaceticus. J. Antimicrob. Chemother. 28:791–800 [DOI] [PubMed] [Google Scholar]
- 27. Orsi G. B., et al. 2008. Multidrug-resistant Acinetobacter baumannii outbreak in an intensive care unit. J. Chemother. 20:219–224 [DOI] [PubMed] [Google Scholar]
- 28. Pachón J., Vila J. 2009. Treatment of multiresistant Acinetobacter baumannii infections. Curr. Opin. Investig. Drugs 10:150–156 [PubMed] [Google Scholar]
- 29. Piette A., et al. 2004. Structural determinants required to target penicillin-binding protein 3 to the septum of Escherichia coli. J. Bacteriol. 186:6110–6117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Poirel L., Nordmann P. 2006. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 12:826–836 [DOI] [PubMed] [Google Scholar]
- 31. Poirel L., Naas T., Nordmann P. 2010. Diversity, epidemiology, and genetics of class D beta-lactamases. Antimicrob. Agents Chemother. 54:24–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Robledo I. E., et al. 2010. Detection of KPC in Acinetobacter spp. in Puerto Rico. Antimicrob. Agents Chemother. 54:1354–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sauvage E., Kerff F., Terrak M., Ayala J. A., Charlier P. 2008. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32:234–258 [DOI] [PubMed] [Google Scholar]
- 34. Seifert H., et al. 2005. Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J. Clin. Microbiol. 43:4328–4335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Spratt B. G., Zhou J., Taylor M., Merrick M. J. 1996. Monofunctional biosynthetic peptidoglycan transglycosylases. Mol. Microbiol. 19:639–640 [DOI] [PubMed] [Google Scholar]
- 36. Tayler A. E., et al. 2010. Induction of beta-lactamase production in Aeromonas hydrophila is responsive to beta-lactam-mediated changes in peptidoglycan composition. Microbiology 156:2327–2335 [DOI] [PubMed] [Google Scholar]
- 37. Tenover F. C., et al. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Urban C., Go E., Mariano N., Rahal J. J. 1995. Interaction of sulbactam, clavulanic acid and tazobactam with penicillin-binding proteins of imipenem-resistant and -susceptible Acinetobacter baumannii. FEMS Microbiol. Lett. 125:193–197 [Google Scholar]
- 39. Vaneechoutte M., et al. 1995. Identification of Acinetobacter genomic species by amplified ribosomal DNA restriction analysis. J. Clin. Microbiol. 33:11–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang H., et al. 2007. Molecular epidemiology of clinical isolates of carbapenem-resistant Acinetobacter spp. from Chinese hospitals. Antimicrob. Agents Chemother. 51:4022–4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wendt C., Dietze B., Dietz E., Rüden H. 1997. Survival of Acinetobacter baumannii on dry surfaces. J. Clin. Microbiol. 35:1394–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wilke M. S., Lovering A. L., Strynadka N. C. 2005. Beta-lactam antibiotic resistance: a current structural perspective. Curr. Opin. Microbiol. 8:525–533 [DOI] [PubMed] [Google Scholar]
- 43. Yun S. H., et al. 2011. Quantitative proteomic analysis of cell wall and plasma membrane fractions from multidrug-resistant Acinetobacter baumannii. J. Proteome Res. 10:459–469 [DOI] [PubMed] [Google Scholar]
- 44. Zamorano L., Moyá B., Juan C., Oliver A. 2010. Differential beta-lactam resistance response driven by ampD or dacB (PBP4) inactivation in genetically diverse Pseudomonas aeruginosa strains. J. Antimicrob. Chemother. 65:1540–1542 [DOI] [PubMed] [Google Scholar]
- 45. Zapun A., Contreras-Martel C., Vernet T. 2008. Penicillin-binding proteins and β-lactam resistance. FEMS Microbiol. Rev. 32:361–385 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.