Abstract
The accessory gene regulator (agr) locus has been shown to be important for virulence in several animal models of Staphylococcus aureus infection. However, the role of agr in human infections, and specifically in antibiotic treatment, is controversial. Interestingly, agr dysfunction has been associated with reduced vancomycin responses. To systematically investigate the role of agr in virulence and treatment outcome in the context of endovascular infection, 10 well-characterized vancomycin-susceptible methicillin-resistant S. aureus (MRSA) bloodstream isolates (5 agr-I [clonal complex 45, or CC45] and 5 agr-II [CC5]) were studied for (i) agr function, (ii) RNAIII transcriptional profiles, (iii) agr locus sequences, (iv) intrinsic virulence and responses to vancomycin therapy in an experimental infective endocarditis (IE) model, and (v) in vivo RNAIII expression. Significant differences in agr function (determined by delta-hemolysin activity) correlated with the time point of RNAIII transcription (earlier RNAIII onset equals increased agr function). Unexpectedly, four MRSA strains with strong delta-hemolysin activities exhibited significant resistance to vancomycin treatment in experimental IE. In contrast, five of six MRSA strains with weak or no delta-hemolysin activity were highly susceptible to vancomycin therapy in the IE model. agr sequence analyses showed no common single-nucleotide polymorphism predictive of agr functionality. In vivo RNAIII expression in cardiac vegetations did not correlate with virulence or vancomycin treatment outcomes in the IE model. Inactivation of agr in two strains with strong delta-hemolysin activity did not affect virulence or the in vivo efficacy of vancomycin. Our findings suggest that agr dysfunction does not correlate with vancomycin treatment failures in this experimental IE model in two distinct MRSA genetic backgrounds.
INTRODUCTION
Staphylococcus aureus continues to be a predominant cause of community-acquired and health care-associated infections (14). It is the most prevalent cause of endovascular infections, including catheter sepsis and infective endocarditis (IE) (8, 46), and the second most common cause of bacteremia (8, 16). The increase in infections due to methicillin-resistant S. aureus (MRSA), the high rates of vancomycin clinical treatment failures, and the growing problems of linezolid and daptomycin resistance have all further complicated the management of patients with MRSA infections and have led to high health care costs (2, 9, 15, 30). In addition, MRSA bacteremia and IE are associated with high rates of morbidity and mortality (15 to 40%), even with seemingly appropriate antimicrobial therapy (9, 20).
There are a number of factors that appear to play key roles in the virulence of S. aureus infections. The accessory gene regulator (agr) locus, a well-characterized quorum-sensing two-component regulatory system, is a principal global regulator within the overall staphylococcal virulon. in vitro, agr upregulates secreted proteins and downregulates many pivotal surface adhesins during postexponential growth. The agr locus is complex, consisting of two divergent transcription units, driven by promoters P2 and P3 (reviewed in reference 34). The P2 operon encodes a two-component signaling module, of which AgrC is the receptor and AgrA is the response regulator. It also encodes two proteins, AgrB and -D, which combine to produce and secrete an autoinducing peptide (AIP) that is the ligand for AgrC. AgrA activates the agr P3 promoter, which drives the synthesis of RNAIII, the effector of target gene regulation. RNAIII also encodes delta-hemolysin, which makes semiquantitative scoring of delta-hemolysin elaboration a commonly used indicator of RNAIII expression and, thus, agr function (42, 43). There are four types of agr quorum-sensing systems (18, 19). Each of these agr systems, referred to as agr-I through agr-IV, recognizes a unique AIP structure (AIP-I through AIP-IV). Several distinct animal models have demonstrated that the agr system plays an important role in the infectious process, including arthritis (1), subcutaneous abscesses, mastitis, IE (3), and osteomyelitis (12); agr mutants have been shown to be attenuated in virulence in all these models.
There have been numerous reports suggesting that treatment of MRSA infections with vancomycin can be ineffective, despite in vitro susceptibility of the organism to this agent (11, 28, 29, 31). There are several potential mechanisms that have been coassociated with reduced vancomycin treatment success in both human and experimental staphylococcal infections, such as reduced susceptibility to host defense cationic peptides (9, 40) and increased biofilm formation (40). Interestingly, attenuated agr function, especially in association with agr-II strains, and development of glycopeptide intermediate resistance S. aureus (GISA) in association with the agr-II genotype have both been associated with reduced vancomycin responses (9, 27, 37, 38). To more systematically evaluate the role of the agr locus in virulence and vancomycin treatment outcome, we studied the composite correlations of agr activation, functionality, locus sequence, intrinsic virulence, and vancomycin treatment outcomes in a relevant animal model of endovascular infection by using 10 recent clinical MRSA bloodstream isolates and two agr mutants.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Ten MRSA bloodstream isolates were selected from a recent strain collection of 36 MRSA strains (Table 1) (7, 40). Because agr-I comprises a majority of current S. aureus clinical scenarios (44) and agr-II MRSA strains are putatively associated with vancomycin therapy failure in the clinical setting (31), we focused on isolates with these two agr types (Table 1). Within the agr-I group, we selected five isolates genotyped as clonal complex 45 (CC45), because this genetic background has been associated with reduced vancomycin responses clinically (11). Among the agr-II isolates in the overall strain collection, all were genotyped as CC5, the most common CC type in MRSA bacteremia and which has been associated with complicated MRSA infections (10, 24). Five isolates were randomly selected from the latter CC group. The overall clinical and demographic characteristics of the patient sources of these 36 strains in this collection have been previously detailed (7).
Table 1.
Strains used in this study
| Source and strain | Relevant characteristicsa | VAN MIC (μg/ml)b | Reference(s) |
|---|---|---|---|
| Laboratory strains | |||
| RN4220 | NCTC8325-4, α-hemolysin negative, β-hemolysin positive | 33 | |
| RN6911 | NCTC8325-4 agr::tet(M), Tcr | 33 | |
| SH1000 | rsbU-positive derivative of NCTC8325-4, agr-I | 17 | |
| SH1001 | SH1000 agr::tet(M), Tcr | 17 | |
| Clinical MRSA bacteremia isolates | |||
| 300-087 | agr-I, SCCmec IV, CC45c | 0.5 | 7, 37 |
| 324-136 | agr-I, SCCmec IV, CC45c | 0.5 | 7, 37 |
| 300-169 | agr-I, SCCmec IV, CC45c | 0.5 | 7, 37 |
| 300-103 | agr-I, SCCmec IV, CC45c | 0.5 | 7, 37 |
| 301-188 | agr-I, SCCmec IV, CC45c | 0.5 | 7, 37 |
| 300-246 | agr-II, SCCmec I, CC5c | 0.5 | 7, 37 |
| 010-016 | agr-II, SCCmec II, CC5c | 1 | 7, 37 |
| 077-107 | agr-II, SCCmec II, CC5c | 1 | 7, 37 |
| 088-180 | agr-II, SCCmec II, CC5c | 0.5 | 7, 37 |
| 088-237 | agr-II, SCCmec II, CC5c | 0.5 | 7, 37 |
| Mutants | |||
| 300-169Δagr | 300-169 agr::tet(M), Tcr | 0.5 | This study |
| 324-136Δagr | 324-136 agr::tet(M), Tcr | 0.5 | This study |
Tcr, tetracycline resistant.
Vancomycin MICs were determined according to CLSI guidelines.
spa types were used to predict clonal complexes, according to the methods of McCalla et al. (24).
Phage φ11 was used to transduce the agr::tet(M) mutation from S. aureus strain RN6911 into two agr-I (CC45) strains (300-169 and 324-136) (33). The generated strains, 300-169Δagr and 324-136Δagr, had a complete deletion of the agr locus. The agr locus deletions were confirmed by Southern blot analysis according to standard protocols using RNAIII-specific digoxigenin (DIG)-labeled probes produced by using the PCR DIG probe synthesis kit (Roche, Basel, Switzerland) and RNAIII-specific primers (Table 2). S. aureus was routinely grown in tryptic soy broth (TSB) or on tryptic soy agar (TSA) plates with 5 μg/ml tetracycline for mutant selection.
Table 2.
Primers used in this study
| Primer | Sequence (5′–3′) | Purpose | Reference |
|---|---|---|---|
| RNAIII+ | AGATCACAGAGATGTGATGG | Northern/Southern blotting | 41 |
| RNAIII− | TCGATGTTGTTTACGATAGC | Northern/Southern blotting | 41 |
| spa-F | TGAATTCGTAAACTAGGTGTAGG | Northern blotting | 41 |
| spa-R | CGGTACCAGGCTTGTTATTGTCTTCC | Northern blotting | 41 |
| hla+ | AAGGTACAGTTGCAACTACC | Northern blotting | 41 |
| hla− | ATAACTGTAGCGAAGTCTGG | Northern blotting | 41 |
| gyrB.MB-F2 | CGCAGGCGATTTTACCATTA | RT-PCR | This study |
| gyrB.MB-R2 | GCTTTCGCTAGATCAAAGTCG | RT-PCR | This study |
| RNA3.MB-F | GCCATCCCAACTTAATAACCA | RT-PCR | This study |
| RNA3.MB-R | TGTTGTTTACGATAGCTTACATGC | RT-PCR | This study |
Determination of MICs, kill curve experiments, and population analyses.
Determination of vancomycin MICs were conducted by broth microdilution as recommended by the CLSI (4). in vitro vancomycin kill curves and vancomycin population analyses were carried out as previously described (6, 48). All experiments were performed at least twice for each strain on different days.
Assessment of delta-hemolysin activity.
Delta-hemolytic activities were determined by cross-streaking test strains perpendicularly to RN4220, a strain which is a hyperproducer of beta-hemolysin but does not produce alpha-hemolysin (42, 43). Strains SH1000 and SH1001 (agr mutant of SH1000) were used as positive and negative controls, respectively (Table 1). Delta-hemolytic activity was denoted by an enhanced area of hemolysis at the intersection of RN4220 and test strain streaks. All experiments were conducted at least three times on separate days.
Northern blot analyses.
Overnight cultures of S. aureus were diluted to ∼107 CFU/ml (optical density at 600 nm [OD600], 0.05; dilution factor, >1:200) in fresh TSB medium and grown at 37°C in a shaking incubator. Samples were removed at 2, 4, 6, and 8 h of growth. RNA extraction and Northern blotting were performed as previously described (41). Digoxigenin-labeled DNA probes were used for the detection of RNAIII, spa, and hla transcripts by Northern hybridization. Primers used for probe amplification are listed in Table 2. Northern blot analyses were performed with samples from three different days.
Quantitative RT-PCR.
To confirm RNAIII expression levels at the early time point, RNAIII transcripts from 2-h samples (OD600, 0.6 ± 0.05 [mean ± standard deviation, SD] for all tested strains) were quantified by quantitative reverse transcription-PCR (RT-PCR). In addition, RNA was isolated from in vitro cultures that were grown for 24 h in order to compare in vivo cardiac vegetation samples collected 24 h postinfection (see below). RNA samples were treated with Turbo DNA free (Ambion) for 30 min at 37°C. DNase treatment was stopped using DNase inactivation reagent (Ambion). Two micrograms of RNA was transcribed into cDNA by using a RETROscript kit and 5 μM random decamer primers (Ambion). Quantitative real-time PCR was carried out using an ABI Prism 7000 instrument (Applied Biosystems) and the SYBR green PCR master mix (Applied Biosystems). Reaction mixtures were prepared using 100 nM primers listed in Table 2. gyrB was used to normalize for transcript quantification, because it has been shown to be not affected by cell density and/or agr (13). Relative RNAIII expression was calculated as the differences in cycle thresholds (ΔCT) (gyrBCT − RNAIII CT) for all samples as previously described (25, 47, 53). PCR experiments were performed using two biological replicates, each tested in triplicate.
DNA isolation and agr locus sequencing.
Single colonies of S. aureus were isolated and restreaked onto TSA plates and grown overnight at 37°C. Chromosomal DNA was isolated using a Wizard genomic DNA purification kit (Promega, Madison, WI), with 20 μg/ml lysostaphin (Sigma-Aldrich, St. Louis, MO) for 30 min at 37°C. The agr loci were PCR amplified using a method published previously (42). Both strands of the PCR product were commercially sequenced by the standard Sanger dideoxynucleotide method. DNA sequences were assembled and analyzed using the DNAStar (Madison, WI) sequence analysis suite. Each agr sequence was compared to a prototypical strain with each specific agr genotype (23, 32).
Experimental rabbit IE model.
Rabbits were maintained in accordance with the American Association for Accreditation of Laboratory Animal Care criteria. The Animal Research Committee (IACUC) of the Los Angeles Biomedical Research Institute at Harbor—UCLA Medical Center approved all animal studies. A model of left-sided catheter-induced IE in rabbits was used to study composite virulence and responsiveness to vancomycin therapy among the study MRSA strains (51). Briefly, female New Zealand White rabbits (Harlan Laboratories; 2.0 to 2.5 kg body weight) underwent transcarotid-transaortic valve catheterization, and IE was induced by intravenous (i.v.) injection of ∼105 CFU of each MRSA strain at 24 h after catheterization. At 24 h after infection, animals were euthanized by a rapid intravenous injection of sodium pentobarbital (100 mg/kg; Abbott Laboratories), and their cardiac vegetations, kidneys, and spleen were removed and quantitatively cultured (51). Tissue MRSA counts are given as the mean log10 CFU/g of tissue.
To assess responses to vancomycin therapy, at 24 h postinfection, animals with IE were randomized to receive vancomycin therapy (15 mg/kg i.v., twice per day for 3 days). This is a standard effective dose of vancomycin in the experimental IE model caused by vancomycin-susceptible strains (5, 51). At 24 h after the last vancomycin dose, animals were sacrificed, and target tissues were removed and quantitatively cultured as above.
The following definitions were used for grading the vancomycin treatment response: responders were defined as strains in which treatment led to a ≥5-log10 reduction in the number of CFU/g of vegetations and ≥3-log10 reduction in CFU/g of kidney and spleen. Nonresponders were defined as strains with <1.5-log10 reductions in CFU/g in all three target tissues (vegetations, kidneys, and spleen). These definitions were based on extensive pilot studies related to vancomycin responsiveness of MRSA IE in this model.
In vivo RNAIII expression in cardiac vegetations by quantitative RT-PCR.
At 24 h postinfection, catheterized animals were sacrificed, and cardiac vegetations were quick-frozen in liquid nitrogen. Total RNA was isolated using Tri reagent (Ambion) according to the manufacturer's instructions, including a -in. ceramic sphere in the cell disruption step. Samples were treated with DNase, and 2 μg of total in vivo RNA was transcribed into cDNA as described above. Quantitative RT-PCR analysis for RNAIII expression was performed on at least two different animals for each strain.
Statistical analysis.
Means and SDs were calculated for MRSA tissue counts and relative RNAIII expression levels. To compare tissue MRSA counts of treated versus untreated animals and for the analysis of the relationship between RNAIII expression levels and vancomycin responses (responders versus nonresponders), univariate analyses were performed using Student's t test. To assess the relationship between RNAIII expression levels and virulence in terms of bacterial density in target tissues, univariate analyses were performed with simple linear regression. P values of <0.05 were considered statistically significant.
RESULTS
in vitro susceptibility to vancomcyin.
All 10 MRSA isolates were susceptible to vancomycin, with MICs of 0.5 or 1.0 μg/ml (Table 1). No strain exhibited vancomycin tolerance based on in vitro kill curve kinetics, and population analyses revealed no evidence of vancomycin-heteroresistant subpopulations (data not shown).
Delta-hemolysin activities and RNAIII transcription profiles.
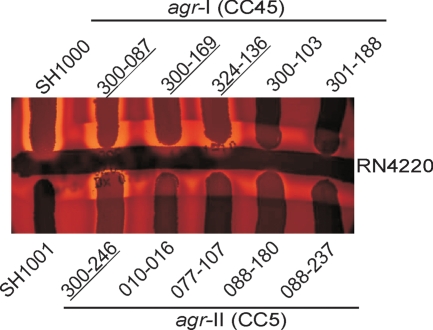
As a first screen for agr function, we tested the 10 clinical MRSA isolates for delta-hemolysin production. Three agr-I (CC45) strains and one agr-II (CC5) strain produced strong delta-hemolysin zones (Fig. 1). In contrast, two agr-I (CC45) strains and three agr-II (CC5) strains exhibited weak delta-hemolysin production. Strain 088-237 produced no delta-hemolysin (Fig. 1).
Fig. 1.
Delta-hemolysin activities of 10 clinical MRSA isolates. Strains were tested against RN4220, and strains SH1000 and SH1001 (SH1000Δagr) served as positive and negative controls, respectively. MRSA isolates with strong delta-hemolysin activities are underlined.
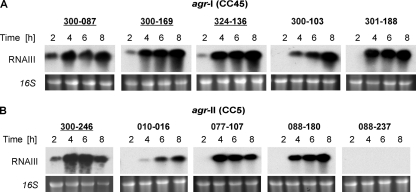
To further evaluate agr function, we assessed agr RNAIII expression during growth by Northern blot analyses. The four strains with strong delta-hemolysin activity exhibited a relatively early onset of RNAIII expression, beginning at 2 h of growth (Fig. 2). The other six strains had no RNAIII transcripts detectable before 4 h. Strain 088-237 exhibited no RNAIII transcripts at any of the time points tested, compatible with the delta-hemolysin-negative phenotype of this strain. These data indicated that early onset of RNAIII transcription correlated with strong delta-hemolysin production, independent of the agr genotype. Growth rates for all 10 strains were similar, suggesting that the observed differences in RNAIII expression onset were not due to differences in growth dynamics (data not shown). To exclude any differences in processing (e.g., blotting time, film exposure time) that might lead to differences in transcript detection, quantitative RT-PCR of the 2-h samples was carried out (Fig. 3A). Within the agr-I (CC45) group, the strains with strong delta-hemolysin production (300-087, 300-169, and 324-136) exhibited significantly higher RNAIII transcript levels than the other strains at the 2-h time point. Accordingly, within the agr-II (CC5) group, strain 300-246, which exhibited strong delta-hemolysin activity, showed the strongest RNAIII transcription at this time point. Thus, quantitative RT-PCR results were in accordance with the Northern blotting results, as shown above (Fig. 2).
Fig. 2.
Northern blot analyses of RNAIII, transcripts of five clinical MRSA isolates with an agr-I (CC45) background (A) and five clinical MRSA isolates with an agr-II (CC5) background (B). Transcripts and time points are indicated. Ethidium bromide-stained 16S rRNA patterns are shown as an indication of RNA loading. Isolates with strong delta-hemolysin activities are underlined.
Fig. 3.
Quantitative RT-PCR of RNAIII transcription at 2 h (A) and 24 h (B) in vitro and in infected cardiac vegetations in the IE model 24 h postinfection (C). Relative transcript levels of RNAIII were determined by RT-PCR. Results are presented as the ΔCT (gyrBCT − RNAIII CT) and represent the mean (+SD) of two biological replicates in vitro and of at least two animals in vivo. Isolates with strong delta-hemolysin activities are underlined.
spa and hla expression profiles.
To further characterize agr function of the strains, we examined the expression of spa (encoding protein A), generally repressed by agr, and of hla (encoding alpha-toxin), generally activated by agr (35). In all strains with strong delta-hemolysin activity, spa transcripts were detectable at only 2 h but not at later time points (data not shown). Most strains with weak or no delta-hemolysin production exhibited spa transcripts throughout the 2- to 8-h time points. Even though strain 300-103 exhibited weak delta-hemolysin activity, it exhibited spa transcripts at 2 h, with only smears detectable at later time points. All strains, except strains 010-016 and 088-237, expressed hla at 4 h and later time points. Strain 010-016 exhibited hla transcripts at 6 and 8 h only. In strain 088-237, only weak hla transcripts were detectable at 8 h, which is in accordance with the absence of RNAIII transcripts in this strain (data not shown).
agr locus sequence analyses.
We sequenced the entire agr locus of the 10 strains (including RNAII, RNAIII, and promoter regions). The agr sequences of all five agr-I (CC45) strains studied were similar to the previously published agr sequence of strain Germ825/96, a strain with a similar genetic background (36). Strains 300-087, 300-169, 324-136, and 301-188 had identical agr locus sequences, suggesting that the weak delta-hemolysin activity of 301-188 was not mediated by agr mutational events. The agr sequence of 300-103, the other agr-I (CC45) strain with weak delta-hemolysin activity, differed by 1 amino acid within agrA: V232L.
The agr sequences of agr-II (CC5) strains 010-016, 077-107, and 088-180 (all having weak delta-hemolysin activities) were identical to the agr sequence of strain JH1 (32). The agr sequence of 088-237 differed by 1 nucleotide from these three strains in agrA (L96S), which is a potential explanation for the lack of delta-hemolysin activity in this strain. The agr sequence of strain 300-246, which exhibited strong delta-hemolysin activity, differed from the other four agr-II (CC5) strains by 1 nucleotide within the intergenic region between RNAII and RNAIII: A→G. This mutation was located 34 bp downstream of the P2 region, and it is unclear if this mutation was the reason for the strong delta-hemolysin activity observed in this strain.
IE model.
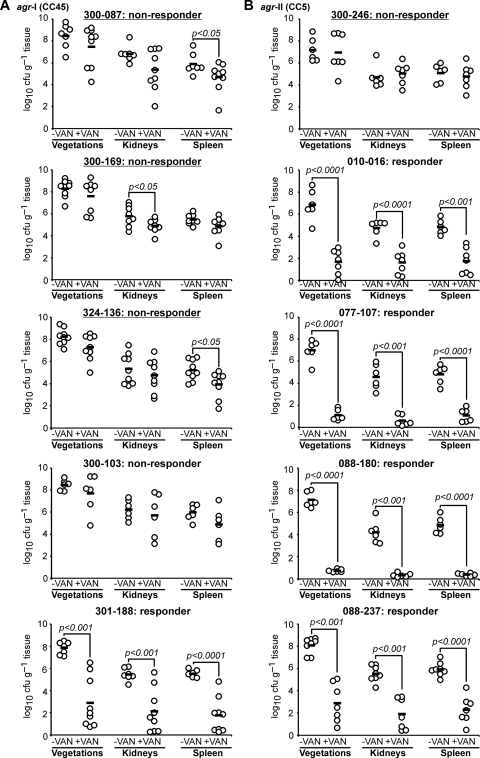
To define whether the above in vitro agr RNAIII transcription, agr functional, and whether sequence profile differences translated to infection outcomes in vivo, the 10 strains were studied in experimental IE for their intrinsic virulence and vancomycin responsiveness (Fig. 4). The intrinsic virulence of the study strains in the IE model (based on achievable target tissue MRSA counts) was similar among the isolates within the agr-I (CC45) group (Fig. 4A). Interestingly, within the agr-II (CC5) group, strain 088-237, a natural agr mutant, had significantly higher bacterial densities in vegetations and kidneys than strains 077-107 and 088-180 (Fig. 4B) (P < 0.05). Animals infected with agr-I (CC45) strains had significantly higher MRSA densities in all three target tissues 24 h after infection than animals infected with strains from the agr-II (CC5) group (P < 0.05). Overall, there was no obvious relationship between agr functionality and intrinsic fitness of the 10 strains.
Fig. 4.
Densities of MRSA in target tissues in the IE model due to 105-CFU challenges with and without vancomycin therapy (15 mg/kg, i.v., two times per day for 3 days, starting 24 h postinfection). (A) agr-I isolates; (B) agr-II isolates. Each dot represents one rabbit. Horizontal black bars indicate means of observations. Isolates with strong delta-hemolysin activities are underlined. A responder showed a ≥5-log10 reduction in the number of CFU per g of vegetation and ≥3-log10 reductions in CFU per g kidney and spleen due to vancomycin treatment. A nonresponder showed <-1.5-log10 reductions in CFU per g of the three target tissues (vegetations, kidneys, spleen). Isolates with strong delta-hemolysin activities are underlined.
Strains with strong delta-hemolysin activity and early RNAIII onset (e.g., 300-087, 300-169, 324-136, and 300-246) did not respond to vancomycin treatment; residual target tissue MRSA densities in vancomycin-treated animals infected with these strains were similar to those in their respective untreated control groups (Fig. 4A and B). In contrast, vancomcyin treatment of rabbits infected with five strains that exhibited weak or no delta-hemolysin activity in vitro resulted in uniform and highly significant reductions of MRSA counts in all target tissues compared with their respective control groups (P < 0.001 for all strains and all three target tissues). The only outlier was strain 300-103, which exhibited weak delta-hemolysin activity but which was not responsive to vancomycin treatment in the IE model.
RNAIII expression in cardiac vegetations.
To assess whether there was a correlation between in vivo and in vitro agr activities and treatment outcomes, we quantified RNAIII expression in cardiac vegetations at 24 h postinfection. Figure 3B and C show RNAIII expression levels of the 10 strains grown in vitro for 24 h and in cardiac vegetations in the IE model 24 h postinfection, respectively. All strains except strain 088-237 exhibited high RNAIII expression levels at 24 h in vitro. RNAIII expression levels in cardiac vegetations in the IE model at 24 h postinfection were significantly lower than the 24-h in vitro samples (P < 0.01 for all strains except 088-237, which is a natural agr mutant strain and exhibited low ΔCT values under all conditions tested). There was no correlation between RNAIII expression levels in vivo and in vitro (2 h and 24 h; P > 0.05). Also, there was no correlation between in vivo RNAIII expression levels and virulence in terms of bacterial counts in infected target tissues and/or treatment outcomes in the IE model (P > 0.05 for all comparisons).
Effect of agr deletion on virulence and vancomycin response in the IE model.
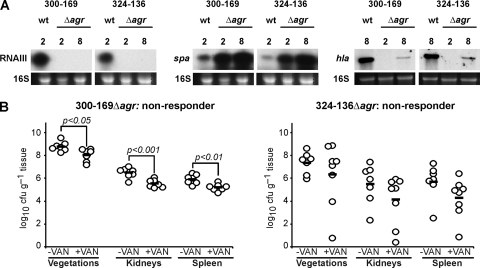
To further evaluate the role of agr function in virulence and vancomycin responses in the IE model, the entire agr locus of two agr-I (CC45) strains that were vancomycin nonresponders in the IE model (300-169 and 324-136) was deleted. As expected, and in contrast to their parental strains, both agr deletion mutants exhibited strong spa and weak hla transcripts in the later growth phases (Fig. 5A). We were not successful in deleting the agr locus of strain 300-246, the only agr-II (CC5) strain that exhibited strong delta-hemolysin activity and did not respond to vancomycin treatment.
Fig. 5.
(A) Northern blot analyses of RNAIII, spa, and hla transcripts of strains 300-169 and 324-136 and their agr mutants. Transcripts and time points are indicated, and ethidium bromide-stained 16S rRNA patterns are shown as an indication of RNA loading. (B) Densities of MRSA in target tissues in the IE model following inoculation of 105 CFU of strain 300-169Δagr or 324-136Δagr, with and without vancomycin therapy (15 mg/kg, i.v., twice a day for 3 days, starting 24 h postinfection). Each dot represents one rabbit. Horizontal black bars indicate means of observations. A nonresponder showed <1.5-log10 reductions in CFU load per g of the three target tissues (vegetations, kidneys, and spleen).
No significant changes were observed in the intrinsic virulence in these agr mutants (Fig. 5B) compared to their respective parental strains (Fig. 5A) in experimental IE in terms of target tissue MRSA counts (P > 0.1 for all comparisons). Even though statistically relevant, only relatively small reductions in target tissue counts were seen following vancomycin treatment in animals infected with 300-169Δagr versus the corresponding untreated controls (<1-log10 CFU reduction/g of target tissue) (Fig. 5B). In animals infected with mutant strain 324-136Δagr, vancomycin treatment did not significantly reduce bacterial counts in any target tissue compared with untreated controls (Fig. 5B), or with its parent strain (Fig. 5A). Thus, both agr mutations in initially vancomycin-nonresponding strains did not alter their response profiles in the experimental IE model.
DISCUSSION
Several recent studies have reported an increased risk for vancomycin treatment failure in bacteremia associated with vancomycin-susceptible MRSA strains with phenotypic agr dysfunction (9, 27, 37, 38). Such agr dysfunction may be multifactorial, caused by mutations in the agrA and/or agrC regions of this operon that impact RNAIII transcription, function, or stability, or which inactivate delta-hemolysin translation (39, 45). In our current study, most isolates with low or no delta-hemolysin production initiated RNAIII transcription at later time points compared to isolates that exhibited strong delta-hemolysin production. Previous investigations suggested that late onset of RNAIII expression (at 4 h and later) resulted in a failure to translate delta-hemolysin and alpha-hemolysin, and hence, a weak or nonhemolytic phenotype (42, 43). Therefore, even though the lack of delta-hemolysin production is a phenotypic marker for a nonfunctional agr locus (9, 27, 37), its absence does not necessarily correlate with the absence of RNAIII transcripts (49).
Several studies have demonstrated that S. aureus agr mutants are substantially less virulent in experimental S. aureus infection models (1, 12, 26), and administration of an inhibitory agr-encoded heterotypic AIP blocked experimental murine abscess formation (50). Interestingly, one of the current study strains (088-237) was a natural agr mutant that did not transcribe RNAIII at any of the time points tested and did not produce delta-hemolysin. This strain was highly virulent in experimental IE, suggesting that a functional agr locus is not required for in vivo fitness in the experimental IE model. This is in accordance with previous findings (3), in which inactivation of agr had only minor effects on virulence in the IE model. Interestingly, the latter agr dysfunctional strain was highly susceptible to vancomycin treatment in the IE model. It should be emphasized that most recent studies comparing agr function with vancomycin treatment outcomes have been based in clinical settings and thus might well be influenced by host factors varying from patient to patient.
In the present investigation, we attempted to evaluate the role of the agr locus in vancomycin treatment outcomes in a relevant model of endovascular infection in which host factors were similar from animal to animal. Of note, the current study also correlated such outcomes with composite phenotypic and genotypic metrics involving agr, including gene transcription, functionality, sequence profiles, and in vivo gene expression. A number of interesting observations emerged from these investigations. Our data clearly indicate that agr dysfunction does not correlate with suboptimal vancomycin treatment outcomes in experimental IE in two distinct genetic backgrounds (agr-I [CC45] and agr-II [CC5]). Five of six MRSA strains tested with relatively late or no agr activation and weak or no delta-hemolysin production were very responsive to vancomycin therapy in experimental IE. In contrast, all four strains with strong delta-hemolysin activities did not respond to vancomycin treatment. Importantly, all isolates in this study were susceptible and nontolerant to vancomycin in vitro and showed no evidence of hetero-vancomycin intermediate resistance S. aureus subpopulations. Therefore, the different vancomycin treatment outcomes observed in the IE model were not related to any overt differences in the in vitro vancomycin susceptibility properties of the study strains. Interestingly, the agr-I (CC45) group had a higher proportion of strains that did not respond to vancomycin treatment (80%) than the agr-II (CC5) group (20%) in the IE model (P < 0.05), indicating that the vancomycin response may be linked to specific genetic backgrounds. These findings differ from other recent clinical investigations (31), in which MRSA infections caused by agr-II genotype strains correlated with vancomycin treatment failure. In vivo and in vitro agr RNAIII expression profiles differed in the present investigation, with RNAIII transcription levels in vivo significantly lower than in vitro RNAIII expression at 24 h. This is in accordance with previous publications (13, 52), in which RNAIII expression was repressed in selected in vivo settings.
In order to determine whether perturbations in activation and function of agr might be due to acquisition of single-nucleotide polymorphisms (SNPs), we sequenced the entire agr loci of the 10 study strains. Our data indicated that there was no common SNP(s) linked to the temporal RNAIII expression profile within the agr-I (CC45) group, suggesting the possibility that loci outside agr contribute to the observed differences in agr RNAIII activation. The role of agr in the in vivo virulence of S. aureus is somewhat controversial. A number of recent investigations have reported that agr mutants reduce virulence in various animal models (1, 12, 22, 26). In contrast, other studies showed that inactivation of agr did not affect virulence in vivo various (21, 22). Therefore, to further assess the role of agr in vancomycin treatment responses in vivo, we selected two strains that did not respond to vancomycin treatment in vivo (300-169 and 324-136) and deleted their whole agr loci. Such agr inactivation neither reduced virulence nor significantly enhanced vancomycin responsiveness compared to the parental strains in the IE model. These data further suggest that agr itself does not affect virulence or vancomycin susceptibility in the IE model and that regulatory loci outside agr are in play in this group. The agr locus of the only strain with strong hemolytic activity (300-246) in the agr-II (CC5) group differed by 1 nucleotide from the agr loci of the other isolates within this group, which might explain the early agr activation of this strain. This strain was also the only strain in the agr-II (CC5) that did not respond to vancomycin treatment in the IE model. Unfortunately, we were not able to inactivate the agr locus of this strain. Therefore, it remains undefined as to how agr activity impacts treatment outcome within the agr-II (CC5) group. Although beyond the scope of the present study, future investigations could examine the in vivo impacts of agr knockouts in strains that respond to vancomycin treatment in the IE model.
In summary, we present here a systematic approach to assess the role of the agr locus in virulence and vancomycin treatment outcome in a relevant model of endovascular infection. We found that agr function, transcription, and sequence did not influence virulence and vancomycin treatment outcomes in the IE model among the agr-I (CC45) strains investigated. Studies are in progress in our laboratory to further examine the detailed roles of the agr locus in virulence and antibiotic treatment outcomes in the experimental IE model, using different MRSA genetic backgrounds (e.g., agr-I [CC22], agr-I [CC8], agr-III [CC30], and agr-IV [CC121]), and also more agr-II (CC5) isolates (including isogenic agr mutants).
ACKNOWLEDGMENTS
This work was supported by the Swiss National Science Foundation (grant PBZHP3-123284 to K.S.), the U.S. National Institutes of Health (grant AI-39108 to A.S.B.), and American Heart Association grants SDG 0630219N and AID 09GRNT2180065 to Y.Q.X.
We thank Vance G. Fowler, Jr., for providing the 10 clinical MRSA bacteremia isolates.
Footnotes
Published ahead of print on 3 October 2011.
REFERENCES
- 1. Abdelnour A., Arvidson S., Bremell T., Ryden C., Tarkowski A. 1993. The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infect. Immun. 61:3879–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chang F. Y., et al. 2003. A prospective multicenter study of Staphylococcus aureus bacteremia: incidence of endocarditis, risk factors for mortality, and clinical impact of methicillin resistance. Medicine (Baltimore) 82:322–332 [DOI] [PubMed] [Google Scholar]
- 3. Cheung A. L., et al. 1994. Diminished virulence of a sar−/agr− mutant of Staphylococcus aureus in the rabbit model of endocarditis. J. Clin. Invest. 94:1815–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. CLSI. 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 8th ed. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 5. Dhawan V. K., Yeaman M. R., Bayer A. S. 1999. Influence of in vitro susceptibility phenotype against thrombin-induced platelet microbicidal protein on treatment and prophylaxis outcomes of experimental Staphylococcus aureus endocarditis. J. Infect. Dis. 180:1561–1568 [DOI] [PubMed] [Google Scholar]
- 6. Drugeon H., Juvin M., Pirault J., Dellamonica P. 1988. Bactericidal activity of pefloxacin in combination with rifampicin, fosfomycin and vancomycin against Staphylococcus aureus: determination by a dynamic method. Rev. Infect. Dis. 10:80–81 [Google Scholar]
- 7. Fowler V. G., Jr., et al. 2006. Daptomycin versus standard therapy for bacteremia and endocarditis caused by Staphylococcus aureus. N. Engl. J. Med. 355:653–665 [DOI] [PubMed] [Google Scholar]
- 8. Fowler V. G., Jr., et al. 2005. Staphylococcus aureus endocarditis: a consequence of medical progress. JAMA 293:3012–3021 [DOI] [PubMed] [Google Scholar]
- 9. Fowler V. G., Jr., et al. 2004. Persistent bacteremia due to methicillin-resistant Staphylococcus aureus infection is associated with agr dysfunction and low-level in vitro resistance to thrombin-induced platelet microbicidal protein. J. Infect. Dis. 190:1140–1149 [DOI] [PubMed] [Google Scholar]
- 10. Fowler V. G., et al. 2007. Potential associations between hematogenous complications and bacterial genotype in Staphylococcus aureus infection. J. Infect. Dis. 196:738–747 [DOI] [PubMed] [Google Scholar]
- 11. Fusco D. N., et al. 2009. Clinical failure of vancomycin in a dialysis patient with methicillin-susceptible vancomycin-heteroresistant S. aureus. Diagn. Microbiol. Infect. Dis. 65:180–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gillaspy A. F., et al. 1995. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect. Immun. 63:3373–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goerke C., et al. 2000. Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infect. Immun. 68:1304–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grundmann H., Aires-de-Sousa M., Boyce J., Tiemersma E. 2006. Emergence and resurgence of methicillin-resistant Staphylococcus aureus as a public-health threat. Lancet 368:874–885 [DOI] [PubMed] [Google Scholar]
- 15. Hardy K. J., Hawkey P. M., Gao F., Oppenheim B. A. 2004. Methicillin resistant Staphylococcus aureus in the critically ill. Br. J. Anaesth. 92:121–130 [DOI] [PubMed] [Google Scholar]
- 16. Hoen B., et al. 2002. Changing profile of infective endocarditis: results of a 1-year survey in France. JAMA 288:75–81 [DOI] [PubMed] [Google Scholar]
- 17. Horsburgh M. J., et al. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jarraud S., et al. 2000. Exfoliatin-producing strains define a fourth agr specificity group in Staphylococcus aureus. J. Bacteriol. 182:6517–6522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ji G., Beavis R., Novick R. P. 1997. Bacterial interference caused by autoinducing peptide variants. Science 276:2027–2030 [DOI] [PubMed] [Google Scholar]
- 20. Khatib R., et al. 2006. Persistence in Staphylococcus aureus bacteremia: incidence, characteristics of patients and outcome. Scand. . J. Infect. Dis. 38:7–14 [DOI] [PubMed] [Google Scholar]
- 21. Kielian T., Cheung A., Hickey W. F. 2001. Diminished virulence of an alpha-toxin mutant of Staphylococcus aureus in experimental brain abscesses. Infect. Immun. 69:6902–6911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kong K. F., Vuong C., Otto M. 2006. Staphylococcus quorum sensing in biofilm formation and infection. Int. J. Med. Microbiol. 296:133–139 [DOI] [PubMed] [Google Scholar]
- 23. Kuroda M., et al. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225–1240 [DOI] [PubMed] [Google Scholar]
- 24. McCalla C., et al. 2008. Microbiological and genotypic analysis of methicillin-resistant Staphylococcus aureus bacteremia. Antimicrob. Agents Chemother. 52:3441–3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McElroy J., Nelson M., Caillier S., Oksenberg J. 2009. Copy number variation in African Americans. BMC Genet. 10:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McNamara P. J., Bayer A. S. 2005. A rot mutation restores parental virulence to an agr-null Staphylococcus aureus strain in a rabbit model of endocarditis. Infect. Immun. 73:3806–3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moise P. A., et al. 2010. Factors influencing time to vancomycin-induced clearance of nonendocarditis methicillin-resistant Staphylococcus aureus bacteremia: role of platelet microbicidal protein killing and agr genotypes. . J. Infect. Dis. 201:233–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moise P. A., Forrest A., Birmingham M., Schentag J. J. 2002. The efficacy and safety of linezolid as treatment for Staphylococcus aureus infections in compassionate use patients who are intolerant of, or who have feiled to respond to, vancomycin. J. Antimicrob. Chemother. 50:1017–1026 [DOI] [PubMed] [Google Scholar]
- 29. Moise P. A., Schentag J. J. 2000. Vancomycin treatment failures in Staphylococcus aureus lower respiratory tract infections. Int. J. Antimicrob. Agents 16:S31–S34 [DOI] [PubMed] [Google Scholar]
- 30. Moise P. A., et al. 2009. Genotypic and phenotypic relationships among methicillin-resistant Staphylococcus aureus from three multicentre bacteraemia studies. J. Antimicrob. Chemother. 63:873–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moise-Broder P. A., et al. 2004. Accessory gene regulator group II polymorphism in methicillin-resistant Staphylococcus aureus is predictive of failure of vancomycin therapy. Clin. Infect. Dis. 38:1700–1705 [DOI] [PubMed] [Google Scholar]
- 32. Mwangi M. M., et al. 2007. Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc. Natl. Acad. Sci. U. S. A. 104:9451–9456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Novick R., et al. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12:3967–3975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Novick R. P., Geisinger E. 2008. Quorum sensing in staphylococci. Annu. Rev. Genet. 42:541–564 [DOI] [PubMed] [Google Scholar]
- 35. Recsei P., et al. 1986. Regulation of exoprotein gene expression in Staphylococcus aureus by agr. Mol. Gen. Genet. 202:58–61 [DOI] [PubMed] [Google Scholar]
- 36. Robinson D. A., Monk A. B., Cooper J. E., Feil E. J., Enright M. C. 2005. Evolutionary genetics of the accessory gene regulator (agr) locus in Staphylococcus aureus. J. Bacteriol. 187:8312–8321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sakoulas G., et al. 2003. Staphylococcus aureus accessory gene regulator (agr) group II: is there a relationship to the development of intermediate-level glycopeptide resistance? . J. Infect. Dis. 187:929–938 [DOI] [PubMed] [Google Scholar]
- 38. Sakoulas G., et al. 2006. Effects of prolonged vancomycin administration on methicillin-resistant Staphylococcus aureus (MRSA) in a patient with recurrent bacteraemia. J. Antimicrob. Chemother. 57:699–704 [DOI] [PubMed] [Google Scholar]
- 39. Schwan W. R., Langhorne M. H., Ritchie H. D., Stover C. K. 2003. Loss of hemolysin expression in Staphylococcus aureus agr mutants correlates with selective survival during mixed infections in murine abscesses and wounds. FEMS Immunol. Med. Microbiol. 38:23–28 [DOI] [PubMed] [Google Scholar]
- 40. Seidl K., et al. 2011. Combinatorial phenotypic signatures distinguish persistent from resolving methicillin-resistant Staphylococcus aureus bacteremia isolates. Antimicrob. Agents Chemother. 55:575–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Seidl K., et al. 2006. Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob. Agents Chemother. 50:1183–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Traber K., Novick R. 2006. A slipped-mispairing mutation in AgrA of laboratory strains and clinical isolates results in delayed activation of agr and failure to translate δ- and α-haemolysins. Mol. Microbiol. 59:1519–1530 [DOI] [PubMed] [Google Scholar]
- 43. Traber K. E., et al. 2008. agr function in clinical Staphylococcus aureus isolates. Microbiology 154:2265–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. van Leeuwen W., van Nieuwenhuizen W., Gijzen C., Verbrugh H., van Belkum A. 2000. Population studies of methicillin-resistant and -sensitive Staphylococcus aureus strains reveal a lack of variability in the agrD gene, encoding a staphylococcal autoinducer peptide. J. Bacteriol. 182:5721–5729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Van Wamel W. J., van Rossum G., Verhoef J., Vandenbroucke-Grauls C. M., Fluit A. C. 1998. Cloning and characterization of an accessory gene regulator (agr)-like locus from Staphylococcus epidermidis. FEMS Microbiol. Lett. 163:1–9 [DOI] [PubMed] [Google Scholar]
- 46. Wisplinghoff H., et al. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309–317 [DOI] [PubMed] [Google Scholar]
- 47. Wong Y. W., Chew J., Yang H., Tan D. T. H., Beuerman R. 2006. Expression of insulin-like growth factor binding protein-3 in pterygium tissue. Br. J. Ophthalmol. 90:769–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wootton M., et al. 2001. A modified population analysis profile (PAP) method to detect hetero-resistance to vancomycin in Staphylococcus aureus in a UK hospital. J. Antimicrob. Chemother. 47:399–403 [DOI] [PubMed] [Google Scholar]
- 49. Wright J. S., et al. 2005. The agr radiation: an early event in the evolution of staphylococci. J. Bacteriol. 187:5585–5594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wright J. S., Jin R., Novick R. P. 2005. Transient interference with staphylococcal quorum sensing blocks abscess formation. Proc. Natl. Acad. Sci. U. S. A. 102:1691–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xiong Y. Q., et al. 2009. Phenotypic and genotypic characteristics of persistent methicillin-resistant Staphylococcus aureus bacteremia in vitro and in an experimental endocarditis model. . J. Infect. Dis. 199:201–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yarwood J. M., McCormick J. K., Paustian M. L., Kapur V., Schlievert P. M. 2002. Repression of the Staphylococcus aureus accessory gene regulator in serum and in vivo. J. Bacteriol. 184:1095–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zannoni A., et al. 2007. Prostaglandin F2-alpha receptor (FPr) expression on porcine corpus luteum microvascular endothelial cells (pCL-MVECs). Reprod. Biol. Endocrinol. 5:31. [DOI] [PMC free article] [PubMed] [Google Scholar]