Abstract
Toxoplasma gondii is an obligate intracellular parasite that can cause disease in the developing fetus and in immunocompromised humans. Infections can last for the life of the individual, and to date there are no drugs that eliminate the chronic cyst stages that are characteristic of this parasite. In an effort to identify new chemical scaffolds that could form the basis for new therapeutics, we carried out a chemoinformatic screen for compounds that had the potential to interact with members of a superfamily of parasite-secreted kinases and assayed them for growth inhibition in vitro. Of 17 candidate compounds, we identified one with potent antiparasitic activity. The compound has a 50% inhibitory concentration (IC50) of ∼2 nM, and structure-function analyses implicate the benzodioxole moiety in its action. The compound does not appear to be cytotoxic to host cells. Using microarray analyses of both parasites and host cells treated with the compound, we found that the levels of very few host cell transcripts are altered by the compound, while a large number of parasite transcripts have a different abundance after compound treatment. Gene ontology analyses of parasite transcripts with a different abundance revealed an enrichment of cell cycle-related genes, suggesting that the compound alters progression of the parasite through the cell cycle. Assaying the nuclear content of treated parasites demonstrated that compound treatment significantly increased the percentage of parasites in the S/M phase of the cell cycle compared to controls. This compound and its analogs represent a novel scaffold with antiparasitic activity.
INTRODUCTION
Toxoplasma gondii is an obligate intracellular protozoan parasite that infects over a billion humans worldwide. It causes severe disease in individuals with compromised immune systems, such as HIV/AIDS patients and immunosuppressed organ transplant patients (19). In addition, primary Toxoplasma infections during pregnancy can result in severe disease in the developing fetus (2, 21). While in most cases healthy individuals do not become severely ill from Toxoplasma infection, in recent years strains that have caused severe disease in healthy, immunocompetent adults have been identified (17, 18). The accepted treatment regimen for toxoplasmosis is pyrimethamine and sulfadiazine (33). Pyrimethamine treatment requires supplementation with folinic acid to prevent bone marrow suppression, and sulfadiazine can cause severe kidney disease in children (15) and HIV/AIDS patients (7) and can cause rashes after long-term treatment (49). These data highlight the need for the development of new anti-Toxoplasma therapies that are more effective and have fewer adverse side effects. Moreover, there is still no cure for Toxoplasma infection, in that tissue cysts of this parasite can persist for the life of the individual even after treatment, and these cysts can reactivate to cause severe disease (9).
Toxoplasma protein kinases have been shown by us and others (44, 47) to be key virulence genes in Toxoplasma infections. Secreted protein kinases, such as ROP18, are highly polymorphic proteins that are major determinants of strain-specific disease outcome in the mouse model. For example, expression of virulent alleles of ROP18 in an avirulent genetic background can increase lethality in the mouse by over 4 log units (44, 47). ROP18 belongs to a superfamily of secreted kinases from Toxoplasma (the ROP2 superfamily) (24) with an estimated size of over 50 gene products (10). This superfamily includes both catalytically active kinases (such as ROP18) and a large number of kinases that are predicted to be catalytically inactive (so-called “pseudokinases”) (10, 34). In an effort to begin to identify molecules that might affect the activity of this superfamily of proteins, we took a focused chemoinformatics approach to identify small-molecule inhibitors of rhoptry kinases. Crystal structures are available for two members of the ROP2 superfamily (ROP2 and ROP8; Protein Data Bank identification numbers [PDBID] 3byv and 3dzo, respectively) (41), and we used these to construct a homology model of the type II allele ROP18 (ROP18II). We then carried out in silico similarity searches on 45,384 commercially available compounds to identify those that with structural similarity to ATP and then used virtual docking to select those compounds with the most favorable binding energy within the predicted ATP binding pocket of our ROP18II homology model. Of these, we selected 17 compounds and screened them for the ability to alter parasite growth in vitro. From this small number of compounds, we identified one with potent inhibitory effects against Toxoplasma in vitro and assessed the effects of the compound on parasites as well as host cells using microscopy and microarray analyses. While the active compound does not appear to target ROP18II directly, it is highly inhibitory to the growth of Toxoplasma in vitro. Its effect appears to be irreversible after 24 h of treatment, and it has no observed cytotoxic effects on host cells at its most effective antiparasitic concentration. Moreover, the compound may act via disruption of the cell cycle based on microarray and DNA content analyses.
MATERIALS AND METHODS
Structure prediction and evaluation.
The modeling of the three-dimensional (3D) structure of type II ROP18 was performed by the automated homology modeling program Swiss-Model (11, 32, 40). The ROP18 protein sequence from strain ME49 (www.toxodb.org; TGME49_005250) was used to search for appropriate templates using the Swiss-Model server. As expected, the best structural templates were the kinase-like domains of two other rhoptry kinases, ROP2 (PDBID 3dzo) (41) and ROP8 (PDBID 3byv) (41), whose catalytic domains are predicted to be inactive (34). Given that there was low sequence identity with the best templates (29% with 3byv and 28% with 3dzo), the alignment process was done manually using the program DeepView (45). In brief, the ROP18II sequence and templates were loaded into DeepView. 3dzo was then superimposed iteratively onto 3byv, and the ROP18II sequence was aligned onto the superimposed structure. Both templates were then chosen as modeling scaffolds and were used by the Swiss-Model server to generate the ROP18II homology model. Validation of the homology model was performed using PROCHECK (35), and energy minimization was performed by Verify3D (13). The overall stereochemical quality of the protein was assessed by Ramachandran plot analysis (42), and the structures were visualized using Swiss PdbViewer-4.0. To further validate the ROP18II model, it was visually compared to a published homology model for the type I allele of ROP18 (ROP18I) (41) that was based on the ROP8 structure (3byv) (41), T. gondii calcium-dependent protein kinase 1 bound to a “bumped” kinase inhibitor NA-PP2 (TgCDPK1; PDBID 3I7C) (39), and mouse protein kinase A bound to ATP (PDBID 1ATP) (55).
ATP similarity searches.
Two small-molecule libraries of compounds were screened in silico in this study. One is the Kinase Targeted Library from Life Chemicals (Orange, CT), composed of 26,573 compounds; the second one is the drug-like subset of the ZINC 8.0 database (approximately 3.5 million compounds) (29). The Kinase Targeted Library from Life Chemicals is a collection of synthetically tractable and commercially available small molecules that were generated through ligand- and receptor-based approaches and further filtered using Lipinski's rule of five (37). Both compound libraries were obtained in SDF format (2D) and converted to MOL2 format (3D) using the program OPEN BABELv2.2.0 (27). Morphological similarity to ATP was determined using Sybyl8.0 (Tripos, St. Louis, MO) (30), and all molecules with similarity scores of greater than 0.7 were chosen for further analysis.
ADME/toxicity predictions.
Computational modeling tools were used to estimate the bioavailability, aqueous solubility, blood brain barrier potential, human intestinal absorption, cytochrome P450 (i.e., CYP2D6) enzyme inhibition potential, mutagenicity, and hERG inhibition for the hits obtained from the database screening using the ACD/ADME Boxes software (ACD Labs, Toronto, Canada) (31). Mutagenicity and hERG and CYP2D6 inhibition were estimated with ACD/TOX screening (ACD Labs, Toronto, Canada). Compounds with the best ADME/TOX profiles were chosen for further analysis.
Virtual docking.
Docking studies were performed to see if the hits identified through similarity searches were able to interact with the type II ROP18 homology model using Molegro Virtual Docker (MVD) (48). This program has the ability to carry out flexible docking studies based on a hybrid search algorithm called guided differential evolution (48). Initially all the molecules imported into MVD's workspace were automatically prepared using MVD's procedure (48). The preparation procedure fixes and/or assigns any missing bonds, bond order, charges, and protonation states in order to make proper predictions during docking. Prior to docking, an ATP binding domain pocket with a volume of 338 Å3 was identified in our ROP18II homology model using MVD's built-in cavity detection algorithm (48). For the docking simulation, we employed the MolDock scoring function in combination with the MolDock SE search algorithm and Tabu clustering (11). A search space volume of radius 15 Å encompassing the ATP binding domain was chosen for docking, and both the ligands and catalytic pocket residues were allowed to be flexible during the simulation. Each ligand was docked iteratively into the chosen cavity in 10 independent runs, each of which consisted of 1,500 steps. Poses generated from each run were subjected to Tabu clustering whereby the lowest energy pose below an energy score (MolDock E score) (46) of 100 was generated as output. Thus, there were 10 poses per ligand ranked by energy. The lowest-energy pose of the 10 poses per ligand was selected for visual inspection by comparing the orientation of the molecule to a reference ATP pose docked in a similar fashion. The reference ATP pose (Fig. 1) had the lowest free energy and an orientation similar to that of bound ATP in mouse protein kinase A (PKA) (PDBID 1ATP) (55). Ligands that had docking orientations similar to that of the reference ATP pose were chosen for in vitro growth screens.
Fig. 1.
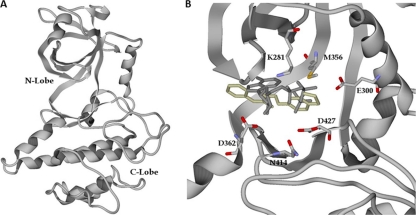
ClustalW alignment of ROP18II, ROP2I, and ROP8I, showing the sequence included in the homology model. For ROP18II, residue numbers are based on the first methionine of ToxoDB gene model TGME49_005250. Key residues are indicated by squares: M356 is the putative “gatekeeper” residue in the ATP binding pocket, D362 is the predicted catalytic aspartic acid, D427 and N414 coordinate interactions with Mg2+, K281 is predicted to bind the alpha and beta phosphates of ATP, and E300 is predicted to coordinate ATP positioning.
Passage of human fibroblast cell line.
Human foreskin fibroblasts (HFFs) were passed and maintained at 37°C and 5% CO2 in Dulbecco's modified Eagle medium with 10% fetal calf serum, 2 mM glutamine, and 50 μg/ml each of penicillin and streptomycin (cDMEM).
Parasite strains.
In most of the in vitro assays we used strain 5A10, which was constructed as follows. Strain CEPΔHXGPRT (a type III strain) was transfected with a construct containing both green fluorescent protein (GFP) and click beetle luciferase (under the control of the GRA2 and dihydrofolate reductase [DHFR] promoters, respectively) and sorted using flow cytometry, and GFP-positive clones were isolated using limiting dilution. To complement the hypoxanthine-xanthine-guanine phosphoribosyl transferase (HXGPRT) defect of this strain, it was then transfected with the vector pGRA-HA-HXGPRT (44), selected for the presence of the plasmid using selection with mycophenolic acid and xanthine (MPA-X) as described previously (22), and cloned by limiting dilution. Strain PB3-10 was derived from strain ME49B7 by transfection with the same click beetle luciferase-GFP plasmid as for 5A10, sorted using flow cytometry, and cloned by limiting dilution. To test the effects of ROP18II expression on compound sensitivity, strain CEPΔHXGPRT was transfected with a hemagglutinin (HA)-tagged version of the type II allele of ROP18 (including 538 bp of upstream promoter sequence) (44) and cloned by limiting dilution after MPA-X selection. When this strain was used to infect mice, it showed the same increase in virulence associated with expression of type II ROP18 in a type III strain as described previously (i.e., injection with 1,000 parasites was uniformly lethal) (44).
Parasite preparation for cell-based assays.
For each cell-based assay, infected host cell monolayers were scraped, parasites were isolated from host cells by passage through 25- and 27-gauge needles and quantified using a hemacytometer, and wells of 24- or 96-well plates were seeded with either 10,000 or 20,000 parasites in cDMEM. After 4 to 24 h of incubation at 37°C and 5% CO2 to allow for invasion, medium was removed and test compounds were added. All test compounds were diluted in cDMEM to a final dimethyl sulfoxide (DMSO) concentration of 1%. cDMEM-1% DMSO was used as vehicle control and is referred to as vehicle throughout.
Luciferase assays.
Samples were harvested for luciferase assays by aspirating medium and adding either 300 μl (for 24-well plate assays) or 50 μl (for 96-well plate assays) of 1× cell culture lysis reagent (Promega, Madison, WI) to each well. Twenty microliters of each sample was transferred to a white-bottom 96-well plate (Nunc, Denmark) and mixed with 50 μl of luciferase assay reagent (LAR) (Promega, Madison, WI). Parasite-derived luciferase activity was counted using a Centro XS3 LB960 luminometer (Berthold Technologies, Oak Ridge, TN).
Extracellular and intracellular parasite treatments.
To test the effects of compound treatment on extracellular parasites and HFF monolayers, freshly lysed parasites and confluent monolayers were treated separately for 16 h with either vehicle or 2 μM F1792-0016 and then washed three times with 200 μl cDMEM. Treated parasites were then used to infect untreated monolayers, while freshly lysed parasites were used to infect treated monolayers, for 3 days prior to harvest and luminescence assays. To test the effects of compound exposure on intracellular parasites, HFFs in 96-well plates were infected for 4 h and then treated for 24 h with either 2 μM F1792-0016 or vehicle. After washing with cDMEM, wells were then treated with vehicle or compound. In this assay we tested 3 conditions: (i) 24 h of vehicle followed by 48 h of vehicle (control), (ii) 24 h of compound treatment followed by 48 h of vehicle, and (iii) 24 h of compound followed by 48 h of compound. Plates were harvested and analyzed for luminescence as described above.
Cell viability and compound toxicity.
Host cell viability in the presence of compounds was determined using both microscopic observation and the Cell Titer 96 Aqueous One solution assay (Promega, Madison, WI), which measures the number of living cells based on mitochondrial dehydrogenase conversion of MTS [3-(4,5-dimethylthiazol-2yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] to formazan, which absorbs at 490 nm. HFFs were seeded into 96-well plates (BD Falcon, NJ) and incubated for 72 h. Medium was removed, and the cells were treated with each compound at a final concentration 10 μM in phenol-red free cDMEM for 72 h. Staurosporine (10 μM) and vehicle were included as positive and negative controls, respectively. Three independent experiments were done for each compound in replicates of 10 per experiment. Absorbance at 490 nm was recorded using an iMark microplate absorbance reader (Bio-Rad, Hercules, CA).
Fluorescence microscopy and electron microscopy.
Confluent HFF monolayers on coverslips were infected with 20,000 parasites/well for 4 h, washed, and then treated with 2 μM and 0.01 μM F1792-0016 or vehicle. Cells were fixed with 4% formaldehyde and processed for fluorescence microscopy on days 1, 2, 3, and 4 posttreatment. Slides were examined using a Zeiss Axiophot upright microscope equipped with a 100× oil immersion objective. The F1656-0035 phenotyping was performed using confluent cells in T25 flasks infected with 100,000 parasites per flask and treated with 10 μM F1656-0035. The cells were visualized using a Zeiss Axiovert microscope equipped with a 40× objective on each day.
For electron microscopy, confluent HFF monolayers in 6-well plates were infected with parasites at a multiplicity of infection (MOI) of 1 for 4 h, washed, and then treated with 2 μM compound or vehicle. After 56 h of exposure, the monolayers were washed briefly in phosphate-buffered saline (PBS) (pH 7.4) and fixed with 2.5% glutaraldehyde in PBS for 12 h at room temperature. The monolayers were then washed 3 times in PBS and postfixed in 1% osmium tetroxide in 1% potassium ferricyanide for 1 h at 4°C. Monolayers were washed 3 times in PBS and then dehydrated in a graded ethanol series (30, 50, 70, and 90%). The final dehydration step was done 3 times in 100% ethanol with 15-min incubations between changes. Upon dehydration, the monolayers were then incubated for 3 h in Epon, which was changed every hour. After the final Epon incubation, Beem capsules were inserted into the monolayer and allowed to polymerize overnight at 37°C and then for 48 h at 60°C. Sections were visualized using a JEM-1011 transmission electron microscope (JEOL Ltd., Tokyo, Japan).
Microarray analyses.
Parasites growing in confluent HFF monolayers were syringe released and quantified as described above. Resuspended parasites were then centrifuged at 800 × g and washed twice with 10 ml cDMEM. Washed parasites were resuspended in enough fresh medium to infect HFFs in T25 flasks at an MOI of 1.5. The parasites were allowed to grow overnight and divided into three different treatment groups: group 1 had medium replaced with 2 μM F1792-0016, group 2 with 10 μM F1792-0007, and group 3 with vehicle. In parallel, uninfected HFFs were divided into three sets of three and treated similarly. Both infected and uninfected flasks were incubated for 24 and 48 h, and RNA extraction and labeling were performed per the manufacturer's recommendations using TRIzol reagent (Invitrogen, Carlsbad, CA) and an Illumina TotalPrep RNA amplification kit (Applied Biosystems, Carlsbad, CA). For F1792-0016, F1792-0007, and vehicle, we performed three biological replicates for the 48-h time point for both infected and uninfected cells. For F1792-0016 only, we also performed one biological replicate for the 24-h time point for infected cells only. cRNA samples containing parasite and host cell RNA were hybridized onto Toxoplasma 169 chips (available through the University of Pennsylvania Genomics Core Facility) (3), while those containing only host cells were hybridized to human WG-6 v3 expression bead chips (Illumina, San Diego, CA).
For the human WG-6 Illumina bead chips, data were normalized using the cubic spline method in BeadStudio v3 (23), while the data from the Toxoplasma Affymetrix chip were normalized using the Affy package implemented in R using the quantile normalization method and log2 transformed using median polishing (26). To identify genes with significantly different abundances between treatment groups for both sets of microarrays, we used the Linear Models for Microarray Analysis package as implemented in R (R/Limma) (46). After linear model fitting, raw P values were calculated for each gene for contrasts of vehicle versus F1792-0016 treatment and vehicle versus F1792-0007 treatment for the 48-h time point. To control for the testing of multiple hypotheses, the false discovery rate method was used to adjust P values. Genes were deemed to be of significant differential abundance if the adjusted P value was less than 0.05 and the absolute value of the fold difference was ≥2.
Gene ontology (GO) analyses of Toxoplasma transcripts were carried out using GeneMerge (14) and utilized the most recently released version of the annotated Toxoplasma gondii genome, for which there is at least one GO annotation for 3,202 genes (www.toxodb.org). This software uses the hypergeometric distribution to determine if particular GO categories are enriched in groups of differentially expressed genes compared to all of the genes on the microarray (14). In addition, it uses a highly stringent Bonferroni P-value correction (based on the total number of GO categories represented on the microarray) to adjust P values and control the experiment-wise error rate (14). In separate analyses, the GO annotations for all genes on the array that were of either significantly higher or lower abundance after 48 h of treatment with F1792-0016 (identified as described in the preceding paragraph) were loaded as the “study” group, and all of the GO annotations were loaded as the “population group.” GO categories were deemed significantly enriched in the higher- or lower-abundance gene sets if they had adjusted P values of <0.05.
Statistical analyses of compound treatment data.
Raw luminescence data for all runs were log2 transformed, and significant differences between vehicle and compound treatments were identified by one-way analysis of variance (ANOVA) followed by a Bonferroni-Holm post hoc test. Bonferroni corrections were always based on the number of predetermined comparisons (in most cases, vehicle versus all compound treatments). Differences in treatment results were considered significant at a P value of <0.05.
Flow cytometric analyses of cell cycle.
Parasites were used to infect monolayers for 24 h, then treated as described above with either vehicle or 2 μM F1792-0016 for 24 and 48 h, and then harvested for cell cycle analyses as described previously (36). Briefly, intracellular parasites were isolated by passage of host cells through a 30-gauge needle followed by filtration through a 3.0-μm-pore-size membrane (Whatman). Parasites were then fixed in 70% ethanol and their DNA stained using 1 μM Sytox Green (Invitrogen, Carlsbad, CA) plus 50 units RNase A and 200 units RNase T1 (Ambion, Foster City, CA) per ml in 50 mM Tris, pH 7.5. Samples were analyzed on a FACSAria flow cytometer (BD Biosciences, Franklin Lakes, NJ). Results were analyzed using FlowJo software (Tree Star), and the percentage of parasites in each phase of the cell cycle (G1 or S/M) was determined by gating. All assays were performed in triplicate, and the percentages of parasites in the S/M phase of the cell cycle were compared for statistical significance between control and test groups by use of a two-tailed t test (P < 0.05) using JMP software (SAS Institute, Cary, NC). At least 10,000 events were collected per sample.
RESULTS
Sequence alignment and model building.
Since the percent identity of ROP18II to the two best available templates (ROP2 and ROP8) falls below 30%, we manually aligned the target sequence with the templates as described in Materials and Methods. Ramachandran plot analysis showed that 90% of the residues were in the “most favored” region, 9.4% in the “allowed” region, and 0.7% in the “disallowed” region, and the root mean square deviations (RMSD) between the templates and the model were 0.66 Å and 0.37 Å with ROP2 and ROP8, respectively. The overall topology of the modeled type II ROP18 consists of 13 beta strands and 15 alpha helices, encompassing residues 224 to 536 (Fig. 1 and 2). To validate our model, we compared our modeled ATP binding site to a recently published homology model of another ROP18 allele (that from the type I strain RH) that was also based on the ROP2 and ROP8 structures (3dzo and 3byv, respectively) (41), TgCDPK1 (PDBID 3I7C) (39), and mouse protein kinase A bound to ATP (PDBID 1ATP) (55). The type I and type II alleles of ROP18 share 96% identity, and there is a significant amount of concordance between our ROP18II model and that for ROP18I (41). In the published homology model for ROP18I, lysine 266 and glutamine 285 were predicted to coordinate interactions with the alpha and beta phosphates on the ATP molecule (41), and these are found within the same locations in our model (K281 and E300 in the ROP18II sequence) (Fig. 2B). Moreover, these residues are in close proximity to the alpha and beta phosphates of ATP derived from the structure overlay with PKA. Other general features are very similar in the two homology models, including the position of the activation loop and the novel N-terminal extension characteristic of the ROP2 superfamily of secreted pseudokinases and kinases (24, 34). Compared to PKA, the ROP18II Lys 281 and Glu 300 are in the same locations as Lys 72 and Glu 91 of PKA, although Glu 300 is slightly further away from the alpha phosphate of ATP than the corresponding glutamine in PKA (see Fig. S1 in the supplemental material). Asp 427 and Asn 414 of ROP18II are in the same locations and orientations as Asp 184 and Asn 171 of PKA, which coordinate interactions with 2 manganese ions and the alpha and beta phosphates of ATP (see Fig. S1 in the supplemental material). TgCDPK1 bound to the bumped kinase inhibitor NA-PP2 also shows high concordance with our ROP18II model. In particular, while TgCDPK1 has a glycine residue at the “gatekeeper” position (Gly 128) which allows for binding of NA-PP2, ROP18II has a methionine at this position (Met 356), and this bulky residue overlaps with and would be predicted to sterically hinder NA-PP2 binding (data not shown), as expected (39). The concordance of our model with TgCDPK1 and PKA further supports the model for kinase activity and ability to coordinate cofactors in the pocket during docking.
Fig. 2.
(A) Homology model of ROP18II based on residues 224 to 536. The predicted N lobe and C lobe are indicated. (B) ATP binding pocket of the ROP18II homology model, showing the lowest-energy poses for ATP (gray) and F1792-0016 (beige). The residues discussed in the legend to Fig. 1 are also indicated.
Similarity searches, toxicity predictions, and virtual docking.
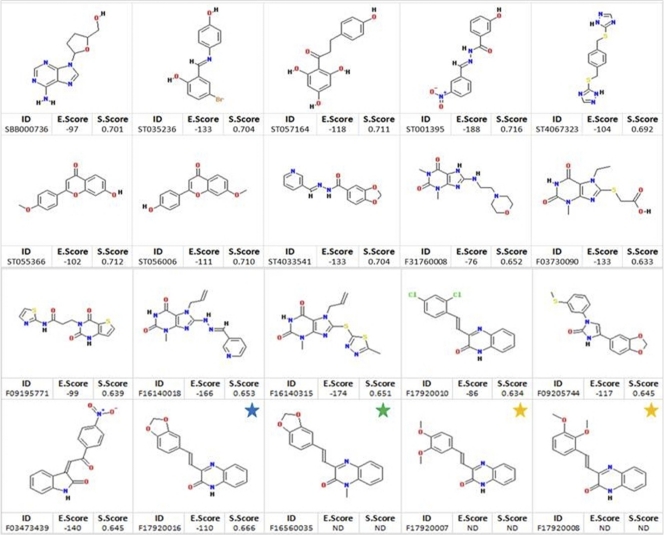
Recognition of small molecules by proteins is mediated largely by molecular surface complementarities. Structure-based drug design approaches use this as the fundamental guiding principle; that is, closely related molecules will elicit similar activities in a biological assay (38, 54). We searched our selected libraries for compounds with the highest similarity to ATP. Three hundred five compounds from the Life Chemical Library and 336 compounds from the ZINC 8.0 drug-like data set showed a similarity to ATP of higher than 70% and were subjected to further analyses. To these molecules we applied in silico assessment of ADME/TOX properties using ACD ADME/TOX software (1). Based on this screen, we selected 199 compounds from the Life Chemicals library and 100 molecules from the ZINC8.0 drug-like data set with the best ADME/TOX properties. These 299 compounds were docked using Molegro Virtual Docker (MVD) software (48). A total of 17 molecules were selected based on both the MolDock energy score (48) and the way they fit into the ATP binding site (Fig. 3).
Fig. 3.
Structures of the 20 compounds assayed for antiparasitic activity in this study. Compounds were selected based on cavity fit into the homology model and docking energy score, except for the three analogs of F1792-0016 (blue star) that were obtained from PubChem similarity searches (green and yellow stars). S.Score, similarity score to ATP; E.Score, docking energy score; ND, not determined.
F1792-0016 inhibits Toxoplasma growth in vitro.
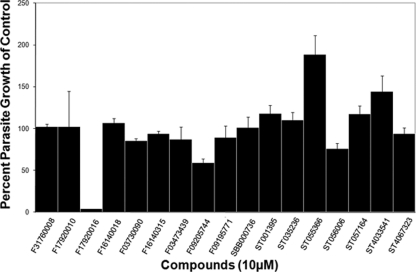
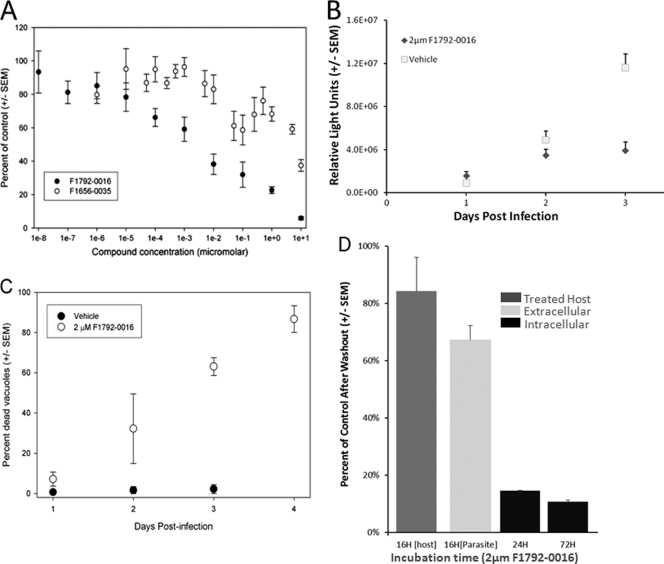
We hypothesized that small molecules designed to fit into the ATP binding pocket of ROP18 may make excellent candidate compounds with the capacity to interact with members of this superfamily. By testing only a small number of compounds (i.e., 17), we identified one compound that significantly affected the growth of strain 5A10 (a type III strain) in vitro (Fig. 4). This compound, F1792-0016 (structures are shown in Fig. 3), was the most potent, decreasing parasite growth by 96% ± 0.14% (P < 0.001 compared to vehicle control). Dose curves showed that the 50% inhibitory concentration (IC50) of F1792-0016 is 2.1 ± 1.8 nM (Fig. 5A), but there is also a reproducible plateau between 10 and 100 nM, suggesting that this compound may have multiple binding sites and/or targets with different inhibitory concentrations. In that 5A10 is a type III strain and is known to express very low levels of ROP18 (44), we also determined the IC50 of F1792-0016 against a type III strain engineered to express a type II allele of ROP18 and found that its sensitivity to the compound was similar to that of 5A10 (IC50 = 3.6 ± 2.3 nM). This suggests that while we chose this compound based on the potential to inhibit ROP18, it is unlikely that the mechanism of killing is due to direct interaction with this kinase.
Fig. 4.
Results of a primary screen with 17 compounds identified by ATP similarity searches and virtual docking. All compounds were screened at 10 μM, and parasite growth was measured after 3 days of treatment. One compound (F1792-0016) inhibited parasite growth by 96% ± 0.14% (P = 4.63 × 10−8). In addition, F0920-5744 mildly inhibited growth by 40.60% ± 4.73% (P > 0.05), but upon visual inspection it was found to be toxic to the host cells. ST055366 mildly enhanced parasite growth by 188.60% ± 22.82% (P > 0.05), while ST056006 was mildly inhibitory, at 23.93% ± 6.41% (P > 0.05). ST056006 and ST055366 are identical except for two switched functional groups (Fig. 2).
Fig. 5.
(A) Dose-response curves for F1792-0016 and F1656-0035. The observed EC50 for F1792-0016 is 2.1 ± 1.8 nM, and F1656-0035 is much less potent, with an EC50 of 5.03 ± 0.58 μM. (B) Growth time course for parasites treated with 2 μM F1792-0016 or vehicle alone. Parasites were harvested on days 1, 2, and 3 posttreatment. F1792-0016 does not show significant inhibitory effects until the third day of treatment. (C) Time course examining the percentage of destroyed vacuoles after treatment with 2 μM F1792-0016 or vehicle. The percentage of destroyed vacuoles increases linearly over time after F1792-0016 treatment from 7.2% ± 3.5% after 24 h to 86.7% ± 6.7% after 96 h of treatment. (D) Treatment of extracellular parasites and host cells with 2 μM F1792-0016 prior to invasion has only moderate effects on parasite growth over a 3-day period after treatment (62% ± 7.0% and 85% ± 17% compared to control, respectively). In contrast, treatment of intracellular parasites with F1792-0016 for 24 h appears to be irreversible: there is no significant difference between the effects of 1 day of treatment and 3 days of treatment with 2 μM F1792-0016 (P > 0.05).
While F1792-0016 was soluble in liquid media at all tested concentrations, it crystallized only after it was placed in the presence of host cells at 10 μM. F0920-5744 also inhibited parasite growth by 40.60% ± 4.73%, but the host cells were also rounded and distorted, suggesting that this compound may be toxic to host cells. Therefore, for this study we focused on further characterizing the effects of F1792-0016 on parasite growth.
Two other compounds showed moderate enhancing effects in our primary screen of our 17 compound set (ST055366 and ST4033541; 190 and 145% of control value, respectively) (Fig. 4), although these increases were not found to be statistically significant (P > 0.05). We conducted further experiments with ST055366 and found that it consistently enhanced parasite growth in a type III strain (clone 5A10; 220% of control) and equally well in a type III strain engineered to express high levels of ROP18II (223% of control), indicating that this enhancing effect is unlikely due to interactions with ROP18II. Compound ST4033541 did not show any significant enhancement of growth in subsequent assays (data not shown).
A screen of second-generation compounds shows a potential relationship between structure and function.
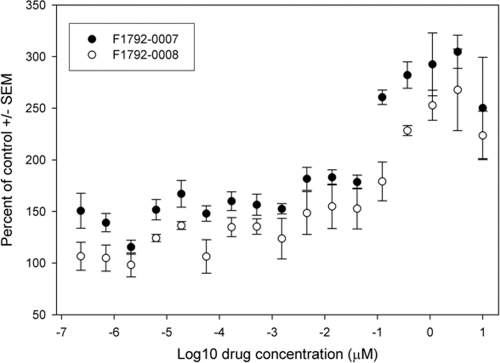
To identify analogs of F1792-0016, we used it as a scaffold in similarity searches using PubChem (pubchem.ncbi.nlm.nih.gov) and identified 3 other compounds with similarity to F1792-0016 of at least 0.9 (based on Tanimoto scores [28]) (Fig. 3). One compound, F1656-0035, showed mild inhibition of parasite growth (62.90% ± 9.44%; P < 0.01) and is structurally identical to F1792-0016 except for an additional methyl group. This modification increased the solubility of the compound, in that concentrations of as high at 30 μM were soluble in DMSO (compared to 2 μM for F1792-0016). In contrast, two compounds (F1792-0007 and F1792-0008) consistently increased parasite growth after 3 days of treatment (250% ± 49.0% and 223% ± 23.5%, respectively; P < 0.0001 for both) and are structurally similar to F1792-0016 except that the bond between the two oxygens on the benzodioxole moiety has been broken. F1792-0007 and F1792-0008 have 50% effective concentrations (EC50s) of 0.04 ± 0.02 μM and 0.1 ± 0.07 μM, respectively (Fig. 6).
Fig. 6.
Dose-response curves for the growth-enhancing compounds F1792-0007 and F1792-0008. Both compounds are maximally effective at 3.33 μM, enhancing growth by 304.6% ± 16.0% and 267.8% ± 39.5%, respectively (P < 0.02 for both). Compound F1792-0007 has an EC50 of 0.04 ± 0.02 μM, while F1792-0008 has an EC50 of 0.1 ± 0.07 μM.
Active compounds do not show evidence for toxicity to human fibroblast host cells.
Using a tetrazolium reduction assay (MTS) (16), we compared MTS reduction in vehicle-treated host cells to that in cells treated for 72 h with each of the compounds. Based on this assay, cells treated with F1792-0016, F1792-0007, F1792-0035, and F1792-0008 were 95% ± 0.03%, 100% ± 0.02%, 103% ± 0.02%, and 101% ± 0.02% viable compared to those treated with vehicle alone. In contrast, positive-control cells treated with staurosporine had 0.4% ± 0.0005% viability compared to the negative-control, vehicle-treated cells. Thus, all four effective compounds were found to be noncytotoxic by this assay at concentrations that were effective at inhibiting parasite growth. In addition, visual inspection showed the monolayer to be intact for all the effective compounds, while those treated with staurosporine were detaching from the substratum at 4 h posttreatment.
The inhibitory effects of F1792-0016 on growth are delayed and irreversible.
To examine the timing of the effects of F1792-0016 treatment on parasite growth, we treated parasites with 2 μM F1792-0016 and quantified the luminescent signal at 24, 48, and 72 h posttreatment. The compound significantly reduced the luminescent signal compared to vehicle controls only at the 72-h time point (P < 0.001; 34% ± 10% of control) (Fig. 5B). To determine whether the inhibitory effects of the compound were reversible, we treated intracellular parasites for 24 h with either vehicle or 2 μM F1792-0016, thoroughly washed the monolayers to remove the compound, and allowed them to grow for another 48 h in the presence of either vehicle or 2 μM F1792-0016. Interestingly, growth compared to controls after 24 h of treatment and washing (14.6% ± 4.2% of control) was very similar to, and not significantly different (P = 0.48) from, that observed by growing the parasites in the presence of the compound for the full 3-day period (10.6% ± 2.2% of control) (Fig. 5D). Overall these data suggest that while the effects of the compound on total parasite numbers (as measured by luminescent signal from the parasites) are not observed until 3 days after drug exposure (Fig. 5B), the binding of the compound to its target(s) may be irreversible, or exposure to the compound for 24 h initiates a cascade that eventually leads to parasite growth inhibition and/or death. Despite the irreversible effect of the compound on intracellular parasites after only 24 of h exposure, the compound showed only mild effects that were not statistically significant (P = 0.56; 62% ± 7.0% of control) (Fig. 5D) on parasites pretreated for 16 h and washed prior to invasion and 3 days of growth (Fig. 5D). Similarly, only mild inhibitory effects (P = 0.60; 85% ± 17% of control) were observed when parasites were allowed to invade pretreated and washed host cells, showing that compound disruption of key host cellular processes is not responsible for reduced parasite growth.
F1792-0016 destroys parasite-containing vacuoles.
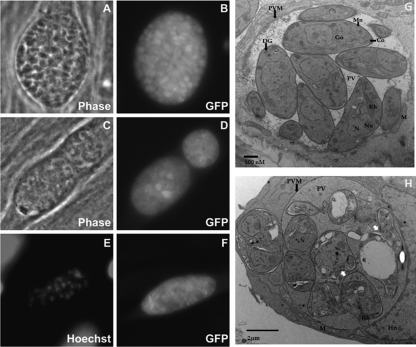
To visualize the effects of F1792-0016 on parasite morphology, we treated GFP-expressing parasites with either vehicle or 2 μM F1792-0016 and examined them after 24, 48, and 72 h of treatment using fluorescence microscopy and after 56 h of treatment using electron microscopy. We observed that a characteristic of F1792-0016 treatment is the appearance of vacuoles that fluoresce in the GFP channel but do not contain any individually defined parasites (Fig. 7). We quantified the percentage of vacuoles with this phenotype over time and found that their numbers increase linearly over the span of 3 days, from 7.2% ± 3.5% on day 1 to 63.0% ± 8.0% on day 3 (P < 0.001) (Fig. 5C). In contrast, on days 1, 2, and 3 a very low percentage (<1% for all time points) of these destroyed vacuoles are seen in the controls exposed to cDMEM-1% DMSO (Fig. 5C). This phenotype is less evident in parasites treated with 0.01 μM F1792-0016, in that after 2, 3, or 4 days of treatment with 0.01 μM F1792-0016, only 9, 18, and 21% of vacuoles were found to be lysed, respectively. A known lethal inhibitor of Toxoplasma gondii, 5-fluorodeoxyuridine (FUDR), also produces this same lysed phenotype (T. Meehan and J. Boyle, unpublished data), suggesting that 2 μM F1792-0016 kills T. gondii, which then results in destruction by the host cell. Since 2 μM F1792-0016 appears to kill the parasites, this explains why the effect is irreversible after 24 h (Fig. 5D) and suggests that F1792-0016 has an early effect on T. gondii that is not observed until 72 to 96 h posttreatment. In addition, while F1792-0016-treated vacuoles appear to be disrupted by both phase-contrast and GFP fluorescence, DNA staining with Hoechst dye reveals that the parasite nuclei are still capable of binding DNA dyes (Fig. 7). Treatment with 10 μM F1656-0035 (the F1792-0016 analog) also resulted in a gradual increase in the same phenotype as observed with F1792-0016 treatment (Meehan and Boyle, unpublished data).
Fig. 7.
Phenotypic effects of F1792-0016 on parasites in vitro. (A to F) Parasites were treated with vehicle (A and B) or 2 μM F1792-0016 (C to F), fixed, and observed using fluorescence microscopy and electron microscopy. (A and B) Parasites treated with vehicle for 48 h. (C and D) Parasites treated for 72 h with F1792-0016 exhibit significant vacuole disruption that is evident both in phase-contrast and in the GFP channel. (E and F) Nuclear staining and GFP fluorescence of a disrupted vacuole from parasites treated for 48 h with F1792-0016. (G and H) Parasite ultrastructure observed using transmission electron microscopy. (G) Vehicle-treated cells. (H) F1796-0016 causes disruption of the parasite plasma membrane (white arrows), and the PV is more electron dense than in the controls, possibly containing the contents of lysed parasites. Co, conoid; DG, dense granules; Go, Golgi complex; Hn, host nucleus; M, host mitochondria; Mn, micronemes; N, nucleus, Nu, nucleolus; PVM, parasitophorous vacuolar membrane; PV, parasitophorous vacuole; Rh, rhoptries. Magnifications, ×15,000 (G) and ×12,000 (H).
Visualizing the parasite ultrastructure after 56 h of treatment with vehicle or 2 μM F1792-0016 using electron microscopy provides a clearer picture of parasite fate. Treated parasites have an intact parasitophorous vacuole membrane (PVM) and nuclei, but individual parasites are enlarged with a rounded feature that lacks a defined anterior or posterior position compared to vehicle-treated controls (Fig. 7G and H). Most of the parasites organelles are either missing or deformed, while some of the parasite's plasma membrane is broken, leading to organelle leakage into the parasitophorous vacuole (PV). The PV is also very dense compared to that of controls. The compound mode of killing seems to involve the destruction of parasite membrane integrity, which leads to the lysis of the parasites.
F1792-0016 does not stop replication or kill at a certain stage of replication or based on size.
As shown in Fig. 5C, F1792-0016 treatment results in destroyed vacuoles after 24 h, but parasite growth appears to continue in the survivors at least for the next 24, h since the average number of parasites per vacuole increases during this time period (see Fig. S2 in the supplemental material). To determine if this delayed effect was due to the completion of a certain number of replication cycles after compound treatment, we compared the size distributions of vacuoles containing parasites that were still defined based on GFP signal and those that exhibited the lysed phenotype (e.g., Fig. 7D and F) at 48 h after compound treatment. We found that the size distributions of live and dead vacuoles were very similar. Vacuoles destroyed by F1792-0016 treatment ranged in size from 12 to 557.6 μm2 with a mean of 106.34 ± 98.57 μm2, while live vacuoles ranged in size from 3.2 to 768 μm2 with a mean of 118.83 ± 158.26 μm2. This suggests that F1792-0016 treatment leads to the destruction of vacuoles regardless of size and does not kill based on a strict number of replication cycles after compound exposure.
Microarray analyses of host cells treated with F1792-0016 and F1656-0007.
While the data summarized above suggest that F1792-0016 and F1656-0007 act directly on parasites, it is possible that these compounds may affect parasite growth and/or viability by altering the host cell environment. As a first step to examine the effects of these compounds on host cells, uninfected fibroblast monolayers were treated for 48 h with compound or vehicle and RNA was harvested for microarray analyses. Transcript abundance was determined using Illumina WG-6 human gene chips. Out of the 37,805 host genes queried on the chip, only 3 were found to be of different abundances in F1792-0016-treated cells compared to controls (at least 2-fold different between treatments and an adjusted P value of <0.05; see Materials and Methods). The gene with the largest difference was that for cytochrome P450, which was 3.4-fold lower in F1792-0016-treated cells (adjusted P = 3 × 10−6). Interestingly, all 3 genes with different abundances in F1792-0016-treated cells were also significantly different in F1656-0007-treated cells, suggesting that some of these genes may be characteristic responses of human cells for processing chemical insults. Again, the cytochrome P450 gene was the gene with the greatest abundance difference in the presence of F1656-0007 (4.0-fold-lower abundance; adjusted P = 1 × 10−6). The second most altered transcript in both compound treatments was that coding for 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD)-inducible poly(ADP-ribose) polymerase (TIPARP), which was 2.1-fold lower in treated cells (P < 4 × 10−5 for both compounds). TIPARP is involved in cellular responses to exposure to dioxin (12), although it is typically upregulated by toxin exposure rather than being downregulated as we saw in our experiments (21). Overall, the effects of these compounds on uninfected human fibroblasts in vitro appears to be subtle, and the changes that do occur are in a small number of genes involved in dealing with chemical-induced stress.
Microarray analyses of parasites treated with F1792-0016 and F1656-0007.
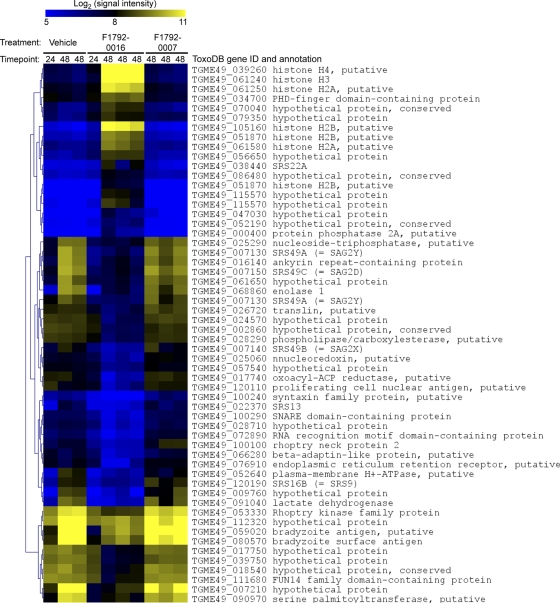
In contrast to the paucity of changes in host cell transcript levels due to compound exposure, parasite transcripts were dramatically altered due to treatment with F1792-0016. In all, 629 Toxoplasma genes (out of 8,059 queried on the chip) were found to be of different abundance after 48 h of F1792-0016 treatment. In a separate experiment, we also examined transcript abundance at 24 h after compound exposure and compared it to that observed at the 48-h time point. Of the 629 T. gondii genes that were found to be of different abundance at the 48-h time point, 142 (∼22%) were at least 2-fold different compared to after the 24-h vehicle control treatment (Fig. 8). While statistical analysis was not possible for the 24-h data since we only had one biological replicate, they suggest that some of the effects on parasite transcription are evident as early as 24 h after compound exposure. However, clearly the most potent effects on transcription occur at the 48-h time point (Fig. 8).
Fig. 8.
Heat map showing the effects of compound treatment on Toxoplasma gene expression. Parasites were treated with either vehicle, 2 μM F1792-0016, or 10 μM F1792-0007 for either 24 or 48 h, and RNA was hybridized to T. gondii Affymetrix microarrays. Data were normalized and analyzed as described in Materials and Methods, and all T. gondii genes with a fold difference of at least 3.0 and a adjusted P value of <0.05 are shown. The most significant changes in gene expression occur in F1792-0016-treated parasites after 48 h of exposure, while very few genes changed significantly in abundance after F1792-0007 exposure.
This is not surprising, given the extreme phenotypic effects observed and that at this time point ∼30% of the vacuoles have been destroyed (Fig. 5 and 7). Gene ontology analyses of those genes of higher and lower abundances in the presence of F1792-0016 for 48 h revealed a number of GO categories that were significantly enriched in each of these gene populations (Table 1). Specifically, genes that were of higher abundance in F1792-0016-treated parasites were enriched for the GO annotations for nucleosome and nucleosome assembly (P = 2 × 10−5 and 1.3 × 10−4, respectively), including histone H2A and its paralogs. Genes that were of lower abundance in the presence of F1792-0016 were significantly enriched for a number of GO categories, including the proteasome (prosome; P = 4.3 × 10−6) and other genes involved in intracellular protein transport and degradation (Table 1 and Fig. 8). In addition, of the 27 genes of at least 3-fold lower abundance in F1792-0016-treated parasites, 11 are genes that are either annotated as specific to cyst stages of the parasite (bradyzoites) or have a clear pattern of upregulation in bradyzoites based on publicly available data (www.toxodb.org). In the present study, many of these bradyzoite-specific genes were found to be of relatively high abundance in parasites treated with vehicle alone, and based on unpublished data from our laboratory as well as others (www.toxodb.org) with the same strain (ME49), these levels are much higher than is typical for tachyzoites in culture. This could be due to stresses that are placed on the parasite by the presence of 1% DMSO (although we have found that this treatment does not significantly alter the growth of parasites in vitro), and regardless of the reasons for atypically high levels of these bradyzoite-specific genes, it is clear from our data that parasites treated with F1792-0016 have significantly lower levels of these transcripts than controls. This suggests that the compound may downregulate the expression of key stress-related genes in the parasite and/or the parasite may be locked in a phase of the cell cycle during which these genes tend to be expressed at lower levels (8).
Table 1.
Enriched GO terms in Toxoplasma transcripts with either higher or lower abundance after 48 h of treatment with 2 μM F1792-0016
| Gene ontology term | Population frequency | Study frequency | P valuea | Description | No. of differentially expressed genes | F1792-0016 effect on transcript levels |
|---|---|---|---|---|---|---|
| 0006886 | 0.0329 | 0.1069 | 2.0E−06 | Intracellular protein transport | 28 | Lower |
| 0016192 | 0.0233 | 0.0878 | 2.1E−06 | Nonselective vesicle transport | 23 | Lower |
| 0004298 | 0.0093 | 0.0534 | 4.3E−06 | Prosome | 14 | Lower |
| 0005839 | 0.0093 | 0.0534 | 4.3E−06 | Macropain | 14 | Lower |
| 0008565 | 0.0115 | 0.0534 | 1.1E−04 | Protein transporter activity | 14 | Lower |
| 0030117 | 0.0103 | 0.0458 | 1.2E−03 | Membrane coat | 12 | Lower |
| 0008094 | 0.0034 | 0.0267 | 1.3E−03 | DNA-dependent ATPase activity | 7 | Lower |
| 0016021 | 0.0547 | 0.1221 | 1.8E−03 | Integral to membrane | 32 | Lower |
| 0005741 | 0.0012 | 0.0153 | 1.0E−02 | Outer mitochondrial membrane | 4 | Lower |
| 0004748 | 0.0012 | 0.0153 | 1.0E−02 | Ribonucleoside diphosphate reductase activity | 4 | Lower |
| 0008308 | 0.0012 | 0.0153 | 1.0E−02 | Voltage-gated anion channel activity | 4 | Lower |
| 0004175 | 0.0031 | 0.0229 | 1.1E−02 | Proteinase | 6 | Lower |
| 0006511 | 0.0227 | 0.0649 | 1.2E−02 | Ubiquitin-dependent protein catabolic process | 17 | Lower |
| 0015031 | 0.0149 | 0.0496 | 1.8E−02 | Protein transport | 13 | Lower |
| 0051603 | 0.0047 | 0.0267 | 1.9E−02 | Proteolysis during cellular protein catabolic process | 7 | Lower |
| 0016020 | 0.1603 | 0.2443 | 3.9E−02 | Membrane | 64 | Lower |
| 0006820 | 0.0016 | 0.0153 | 4.9E−02 | Anion transport | 4 | Lower |
| 0000786 | 0.0043 | 0.0574 | 2.5E−05 | Nucleosome | 7 | Higher |
| 0006334 | 0.0053 | 0.0574 | 1.3E−04 | Nucleosome assembly | 7 | Higher |
| 0004672 | 0.0957 | 0.2295 | 6.0E−04 | Protamine kinase activity | 28 | Higher |
| 0006468 | 0.0966 | 0.2295 | 7.3E−04 | Protein amino acid phosphorylation | 28 | Higher |
| 0003723 | 0.0503 | 0.1311 | 3.0E−02 | RNA binding | 16 | Higher |
Bonferroni-corrected P value based on the number of GO categories in the study population.
An advantage of the Toxoplasma microarrays used in this study (available through www.upenn.edu) (3) is that they also have probes for 297 human genes that are detectable when hybridizing labeled RNA from Toxoplasma-infected host cells prepared as we have for this study. Interestingly, while as described above, very few genes (11 in all) were of different abundance in host cells treated with F1792-0016 in the absence of Toxoplasma, 182 of the 297 human host genes on the Toxoplasma expression chip were of different abundance in infected cells treated with F1792-0016 (see Fig. S1 in the supplemental material). Of these, the vast majority (179/182) were of higher abundance in the presence of F1792-0016. The most dramatically upregulated genes were those for stromal cell-derived factor 1 (SDF1) (12.0-fold higher; P = 2.3 × 10−4), LIM domain binding protein 2 (LDB2) (11.0-fold higher; P = 2.5 × 10−4), and small inducible-cytokine subfamily B, member 10 (SCYB10) (9.0-fold higher; P = 0.002). Importantly, none of these genes were altered by F1792-0016 treatment in cells that were not infected (all data are available through the Geo Repository). Other host cell transcripts of higher abundance in F1792-0016-treated, Toxoplasma-infected cells include Toll-like receptor 4 (TLR4), interleukin-10 (IL-10) receptor beta, and STAT2. A subset of these data can be found in Fig. S1 in the supplemental material.
Overall, our microarray analyses found that (i) host cells treated with these active compounds alone appear to be only moderately affected in terms of their transcriptional profile and (ii) treatment of Toxoplasma with F1792-0016 leads to rampant changes in gene transcription in both Toxoplasma genes and a subset of host cell genes, and this may reflect alterations or blocks in the cell cycle.
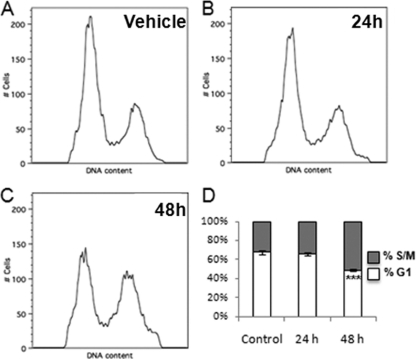
F1792-0016 treatment alters the cell cycle of T. gondii.
Given the significant enrichment for nucleosome-related genes in F1792-0016-treated parasites, we wanted to determine if progression through the cell cycle was blocked or altered by compound treatment. When parasites were treated for 48 h with 2 μM F1792-0016, 51% ± 1.0% of the parasites were in the S/M phase of the cell cycle, compared to 32% ± 2.4% in untreated controls (P = 0.0002) (Fig. 9). This suggests that a possible mechanism of action for F1792-0016 may be to arrest parasites in the S/M phase of the cell cycle, as has been seen with other inhibitors of T. gondii growth, such as monensin (36). Previously, mutants resistant to the antibiotic monensin were identified, and we also examined whether one such mutant (MSH-1) was also resistant to F1792-0016 and found that it was equally susceptible to F1792-0016 as a wild-type strain (for both strains, 48 h of F1792-0016 exposure resulted in 40% S/M parasites, compared to 29% in controls; P > 0.05).
Fig. 9.
Effects of treatment with 2 μM F1792-0016 on the cell cycle in Toxoplasma. (A to C) Parasites were treated with vehicle (A) or with compound for 24 h (B) or 48 h (C), and DNA content was assessed using flow cytometry. (D) F1792-0016 treatment causes a significant increase in the percentage of parasites in the S/M phase of the cell cycle compared to controls (P = 0.0002).
DISCUSSION
In this study, we used chemoinformatics approaches to identify compounds that may have affinities for protein kinases and/or other ATP binding proteins in the human pathogen Toxoplasma gondii, and we tested in vitro the effects of these compounds on parasite growth. Members of the ROP2 superfamily of secreted protein kinases are known to play key roles in parasite pathogenesis (10), including the secreted kinase ROP18, which has moderate effects on parasite growth in vitro (25) and potent effects on pathogenesis in the mouse (44). To do this, we generated a homology model of type II ROP18 based on the most closely related homologs with solved structures and used virtual docking to identify compounds that docked with comparatively low energy into the ATP binding site of the homology model. Simultaneously, we performed morphological similarity searches using two different commercially available libraries to identify compounds that are highly similar to the ATP. In principle, we aimed to identify ATP-like compounds that could act as inhibitors of ROP18. Through this process, and based on previous work demonstrating that expression of the appropriate ROP18 allele can increase parasite growth in vitro, we examined the effects of these compounds on parasite growth. This compound may not target ROP18, since there is no difference in sensitivity to this compound in strains that express high levels of ROP18 and those that do not. Regardless, the homology model of the ROP18 kinase domain, which was built based on similarity to two Toxoplasma proteins with kinase folds (ROP2 and ROP8) (34), served as a starting point for the identification of compounds that may interact with parasite kinases. Future work will focus on identifying the targets of this class of compounds. The importance of our study is that we were able to identify benzodioxole-containing compounds that halt parasite growth in vitro.
The active compound F1792-0016 strongly inhibits parasite growth in vitro. This compound contains both a quinoxaline group and a benzodioxole group, and by testing closely related analogs (F1656-0035, F1792-0007, and F1792-0008; structures are shown in Fig. 3) for their activity against Toxoplasma, the benzodioxole moiety appears to be important for the inhibitory activity of this compound. F1656-0035 has an identical structure except for the addition of a methyl group on C-1 of the quinoxaline moiety, while F1792-0007 and F1792-0008 both have altered forms of the benzodioxole. F1656-0035 inhibited parasite growth in vitro in a fashion similar to that for F1792-0016 (albeit at reduced potency), while both F1792-0007 and F1792-0008 enhanced parasite growth in vitro. While the mechanism for the observed growth enhancement by these two analogs is not known (i.e., whether the analogs interact with the same target[s] as F1792-0016 or others), these restricted structure-activity studies suggest that the benzodioxole moiety is important for the inhibitory activity of F1792-0016.
Benzodioxole-containing compounds have been implicated as inhibitors of eukaryotic protein kinases, and their effects on the receptor tyrosine kinase EphB4 have been studied in great detail (4–6). Benzodioxole-containing compounds were found to be highly effective at inhibiting EphB4 activity in vitro, and cocrystals with EphB4 reveal that the benzodioxole moiety interacts with the gatekeeper threonine residue in the kinase (6). When the benzodioxole is replaced with other moieties, this dramatically reduces effectiveness against EphB4 (4). The only exception to this was when the benzodioxole was replaced with an indazole, as this compound retained activity against the kinase in vitro (4). In the Toxoplasma genome, most predicted active kinases have bulky amino acids (e.g., Met, Leu, or Ile) at the gatekeeper position, and only a smaller subset of protein kinases from Toxoplasma have smaller residues such as threonine at this position (www.toxodb.org; unpublished data). The nature of the gatekeeper residue, and in particular the presence of smaller amino acids (such as threonine) at this position, allows for alteration of the putative target for the kinase inhibitor and subsequent testing of the parasites for resistance to the compound. In a bioinformatic survey of the Toxoplasma genome, a gene coding for a cyclin-dependent protein kinase homolog (TGME49_019100) does have a threonine residue at the gatekeeper position (www.toxodb.org). Based on the fact that transcript levels of genes involved in transitions between different phases of the cell cycle are altered by exposure to F1792-0016 and the fact that other benzodioxole-containing compounds interact with the gatekeeper threonine in kinases such as EphB4 (4–6), the Toxoplasma cyclin-dependent kinase may be an excellent candidate for the target of F1729-0016.
The importance of the quinoxaline-containing moiety in F1792-0016 cannot be ruled out, and other compounds with this core structure have been shown to have antimicrobial effects. In particular, N-oxide-containing quinoxaline derivatives were shown to reduce the growth of Mycobacterium tuberculosis both in vitro and in vivo (43, 50, 52, 53) and also that of the human malaria parasite Plasmodium falciparum (51). In the present study, we have found that a quinoxaline-containing compound without 1,4-N-oxides is also effective against an apicomplexan parasite, in this case Toxoplasma gondii.
Since T. gondii is an obligate intracellular parasite, determining whether effective compounds are targeting the host cell, the parasite, or both can be difficult to address. In this study, we used an MTS assay and visual observation to address host cell cytotoxicity of the compounds, and this led us to rule out compound F0920-5744. Based on these assays, F1792-0016-exposed cells are healthy. However, the compounds could be altering parasite growth or survival by altering certain key (and as-yet-unknown) signaling pathways within the host cell that are crucial for Toxoplasma survival. When we treated uninfected host cells with F1792-0016 for 48 h, we found that very few host cell transcripts were significantly altered in abundance, except for a short list of genes involved in responding to stress and/or chemical insults (such those for as P450 and TIPARP). Moreover, host cell treatment with an enhancer of parasite growth (F1792-0007) during the same experiment altered levels of 11 of the 12 transcripts altered by F1792-0016 treatment. This provides strong evidence that the inhibitory effects of F1792-0016 are due to effects on the parasite rather than on the host cell.
As might be expected from an inhibitor that results in the death of parasites within the host cell, a large number of parasite transcripts were altered by exposure to the drug. At 48 h posttreatment, approximately 30% of the parasite vacuoles exhibit the disrupted phenotype, while the rest of the vacuoles appear normal. Some of these normal vacuoles do undergo subsequent rounds of division, although all of them eventually succumb to the effects of the drug. The specificity of the parasite transcriptional profile may shed light on possible mechanisms of action of F1792-0016. Given the significantly enriched GO categories related to nucleosomes and nucleosome assembly and DNA content analyses, F1792-0016 may halt parasites at a particular phase of the cell cycle. Recently, transcriptional analyses were conducted on cell cycle-synchronized T. gondii parasites (RH strain) (8). The authors found that certain genes in the Toxoplasma transcriptome could be divided into either an S/M-phase subtranscriptome or a G1-phase subtranscriptome. Interestingly, histone components, such as histone H3 (gene model TGME49_061240), were from 7- to 54-fold more abundant in F1792-0016-treated parasites, and this gene is more highly expressed during the S/M phase than during G1 (8; www.toxodb.org). In contrast, proteasome components are a part of the G1 transcriptome (8). While not all components of these subtranscriptomes were of different abundance after exposure to F1792-0016, it is possible that a larger percentage of F1792-0016-treated parasites are in the S/M phase of the cell cycle. The effect of F1792-0016 on histone-related transcript levels is very similar to that observed previously for monensin, which was found to have potent anti-Toxoplasma activity. Microarray analyses of monensin-treated parasites resulted in increased abundances of some of the same transcripts that are upregulated in the presence of F1792-0016, particularly members of the histone family (H2, H3, and H4) (36). Consistent with this observation, monensin also increases the number of parasites that are in the S/M phase of the cell cycle (36). When we tested whether F1792-0016 was acting in a similar fashion, we found that a larger percentage of compound-treated parasites than of vehicle-treated controls were in the S/M phase of the cell cycle, suggesting that cell cycle disruption may be a mechanism of action and/or a downstream effect of F1792-0016 treatment. While the effects on the percentage of parasites in S/M is clear, it is difficult to know if F1792-0016 specifically blocks exit from S/M. Compared to monensin, this is a difficult hypothesis to test with F1792-0016, since treatment with 2 μM always results in the eventual destruction of the parasites within the host cell, possibly due to a lack of production of key effectors that aid in the prevention of recognition of the parasites and lysosomal fusion.
Support for the activation of host innate immune processes after compound treatment is found in the altered host cell transcript levels for parasitized cells in the presence of F1792-0016. A large number of the human immune-related genes queried by the Toxoplasma expression array were significantly upregulated in the presence of parasites. By comparing our data from the Illumina WG-6 chips on uninfected host cells, it is clear that these transcripts are not changing in abundance due to direct effects of F1792-0016 on the host cells but rather are doing so due to indirect effects of the compound on the parasite. Many key innate immune mediators, including 2 chemokines (CXCL12 and SCYB10), Toll-like receptor 4, and STAT2, are of increased abundance in infected, F1792-0016-treated cells. What is interesting about these data is that they appear to provide a snapshot of the transcriptional profile of host cells that are in the process of targeting Toxoplasma-containing vacuoles for destruction. This would be expected for a compound that is somehow incapacitating the parasites such that key effector molecules required for maintenance of the vacuole, and permitting continued growth of the parasites, are no longer produced or have been rendered ineffective.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yaw Adomako-Ankomah and Greg Wier for critical reading of the manuscript and George Bondar (Life Chemicals) for helpful discussions about compound libraries.
This work was supported by NIH scholar development award K22 AI080977 and a Pew Scholarship in the Biomedical Sciences to J.P.B., University of Pittsburgh Biological Sciences summer and academic-year fellowships funded by the Howard Hughes Medical Institute to T.M., and NIH grant R01 AI 89808-01 and American Cancer Society grant RSG-08-19-01-MBC to G.A.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 26 September 2011.
REFERENCES
- 1. ACD Labs 2009, posting date. ADME/TOX boxes. http://www.acdlabs.com
- 2. Austeng M. E., et al. 2010. Maternal infection with Toxoplasma gondii in pregnancy and the risk of hearing loss in the offspring. Int. J. Audiol. 49:65–68 [DOI] [PubMed] [Google Scholar]
- 3. Bahl A., et al. 2010. A novel multifunctional oligonucleotide microarray for Toxoplasma gondii. BMC Genomics 11:603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bardelle C., et al. 2010. Inhibitors of the tyrosine kinase EphB4. 3. Identification of non-benzodioxole-based kinase inhibitors. Bioorg. Med. Chem. Lett. 20:6242–6245 [DOI] [PubMed] [Google Scholar]
- 5. Bardelle C., et al. 2008. Inhibitors of the tyrosine kinase EphB4. 2. Structure-based discovery and optimisation of 3,5-bis substituted anilinopyrimidines. Bioorg. Med. Chem. Lett. 18:5717–5721 [DOI] [PubMed] [Google Scholar]
- 6. Bardelle C., et al. 2008. Inhibitors of the tyrosine kinase EphB4. 1. Structure-based design and optimization of a series of 2,4-bis-anilinopyrimidines. Bioorg. Med. Chem. Lett. 18:2776–2780 [DOI] [PubMed] [Google Scholar]
- 7. Becker K., Jablonowski H., Haussinger D. 1996. Sulfadiazine-associated nephrotoxicity in patients with the acquired immunodeficiency syndrome. Medicine 75:185–194 [DOI] [PubMed] [Google Scholar]
- 8. Behnke M. S., et al. 2010. Coordinated progression through two subtranscriptomes underlies the tachyzoite cycle of Toxoplasma gondii. PLoS One 5:e12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bertschy S., et al. 2006. Discontinuation of maintenance therapy against Toxoplasma encephalitis in AIDS patients with sustained response to anti-retroviral therapy. Clin. Microbiol. Infect. 12:666–671 [DOI] [PubMed] [Google Scholar]
- 10. Boothroyd J. C., Dubremetz J. F. 2008. Kiss and spit: the dual roles of Toxoplasma rhoptries. Nat. Rev. Microbiol. 6:79–88 [DOI] [PubMed] [Google Scholar]
- 11. Bordoli L., et al. 2009. Protein structure homology modeling using SWISS-MODEL workspace. Nat. Protoc. 4:1–13 [DOI] [PubMed] [Google Scholar]
- 12. Boutros P. C., et al. 2011. Hepatic transcriptomic responses to TCDD in dioxin-sensitive and dioxin-resistant rats during the onset of toxicity. Toxicol. Appl. Pharmacol. 251:119–129 [DOI] [PubMed] [Google Scholar]
- 13. Bowie J. U., Luthy R., Eisenberg D. 1991. A method to identify protein sequences that fold into a known three-dimensional structure. Science 253:164–170 [DOI] [PubMed] [Google Scholar]
- 14. Castillo-Davis C. I., Hartl D. L. 2003. GeneMerge—post-genomic analysis, data mining, and hypothesis testing. Bioinformatics 19:891–892 [DOI] [PubMed] [Google Scholar]
- 15. Catalano-Pons C., et al. 2004. Sulfadiazine-induced nephrolithiasis in children. Pediatr. Nephrol. 19:928–931 [DOI] [PubMed] [Google Scholar]
- 16. Cory A. H., Owen T. C., Barltrop J. A., Cory J. G. 1991. Use of an aqueous soluble tetrazolium/formazan assay for cell growth assays in culture. Cancer Commun. 3:207–212 [DOI] [PubMed] [Google Scholar]
- 17. Darde M. L. 2008. Toxoplasma gondii, “new” genotypes and virulence. Parasite 15:366–371 [DOI] [PubMed] [Google Scholar]
- 18. Demar M., et al. 2007. Fatal outbreak of human toxoplasmosis along the Maroni River: epidemiological, clinical, and parasitological aspects. Clin. Infect. Dis. 45:e88–e95 [DOI] [PubMed] [Google Scholar]
- 19. Derouin F., Pelloux H., and ESCMID Study Group on Clinical Parasitology 2008. Prevention of toxoplasmosis in transplant patients. Clin. Microbiol. Infect. 14:1089–1101 [DOI] [PubMed] [Google Scholar]
- 20. Diani-Moore S., et al. 2010. Identification of the aryl hydrocarbon receptor target gene TIPARP as a mediator of suppression of hepatic gluconeogenesis by 2,3,7,8-tetrachlorodibenzo-p-dioxin and of nicotinamide as a corrective agent for this effect. J. Biol. Chem. 285:38801–38810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Di Carlo P., Romano A., Schimmenti M. G., Mazzola A., Titone L. 2008. Materno-fetal Toxoplasma gondii infection: critical review of available diagnostic methods. Infezioni Med. 16:28–32 [PubMed] [Google Scholar]
- 22. Donald R. G., Carter D., Ullman B., Roos D. S. 1996. Insertional tagging, cloning, and expression of the Toxoplasma gondii hypoxanthine-xanthine-guanine phosphoribosyltransferase gene. Use as a selectable marker for stable transformation. J. Biol. Chem. 271:14010–14019 [DOI] [PubMed] [Google Scholar]
- 23. Dunning M. J., Barbosa-Morais N. L., Lynch A. G., Tavare S., Ritchie M. E. 2008. Statistical issues in the analysis of Illumina data. BMC Bioinformatics 9:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. El Hajj H., et al. 2006. The ROP2 family of Toxoplasma gondii rhoptry proteins: proteomic and genomic characterization and molecular modeling. Proteomics 6:5773–5784 [DOI] [PubMed] [Google Scholar]
- 25. El Hajj H., et al. 2007. ROP18 is a rhoptry kinase controlling the intracellular proliferation of Toxoplasma gondii. PLoS Pathog. 3:e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gautier L., Cope L., Bolstad B. M., Irizarry R. A. 2004. Affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20:307–315 [DOI] [PubMed] [Google Scholar]
- 27. Guha R., et al. 2006. The Blue Obelisk—interoperability in chemical informatics. J. Chem. Infect. Model. 46:991–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Holliday J. D., Hu C. Y., Willett P. 2002. Grouping of coefficients for the calculation of inter-molecular similarity and dissimilarity using 2D fragment bit-strings. Comb. Chem. High Throughput Screen. 5:155–166 [DOI] [PubMed] [Google Scholar]
- 29. Irwin J. J., Shoichet B. K. 2005. ZINC—a free database of commercially available compounds for virtual screening. J. Chem. Infect. Model. 45:177–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jain A. N. 2000. Morphological similarity: a 3D molecular similarity method correlated with protein-ligand recognition. J. Comput. Aided Mol. Des. 14:199–213 [DOI] [PubMed] [Google Scholar]
- 31. Japertas P., Didziapetris R., Petrauskas A. 2003. Fragmental methods in the analysis of biological activities of diverse compound sets. Mini Rev. Med. Chem. 3:797–808 [DOI] [PubMed] [Google Scholar]
- 32. Kiefer F., Arnold K., Kunzli M., Bordoli L., Schwede T. 2009. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 37:D387–D392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kongsaengdao S., Samintarapanya K., Oranratnachai K., Prapakarn W., Apichartpiyakul C. 2008. Randomized controlled trial of pyrimethamine plus sulfadiazine versus trimethoprim plus sulfamethoxazole for treatment of toxoplasmic encephalitis in AIDS patients. J. Int. Assoc. Physicians AIDS Care 7:11–16 [DOI] [PubMed] [Google Scholar]
- 34. Labesse G., et al. 2009. ROP2 from Toxoplasma gondii: a virulence factor with a protein-kinase fold and no enzymatic activity. Structure 17:139–146 [DOI] [PubMed] [Google Scholar]
- 35. Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. 1996. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8:477–486 [DOI] [PubMed] [Google Scholar]
- 36. Lavine M. D., Arrizabalaga G. 2011. The antibiotic monensin causes cell cycle disruption of Toxoplasma gondii mediated through the DNA repair enzyme TgMSH-1. Antimicrob. Agents Chemother. 55:745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lipinski C. A. 2000. Drug-like properties and the causes of poor solubility and poor permeability. J. Pharmacol. Toxicol. Methods 44:235–249 [DOI] [PubMed] [Google Scholar]
- 38. Martin Y. C., Kofron J. L., Traphagen L. M. 2002. Do structurally similar molecules have similar biological activity? J. Med. Chem. 45:4350–4358 [DOI] [PubMed] [Google Scholar]
- 39. Ojo K. K., et al. 2010. Toxoplasma gondii calcium-dependent protein kinase 1 is a target for selective kinase inhibitors. Nat. Struct. Mol. Biol. 17:602–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Peitsch M. C. 1995. Protein modeling by email. Biotechnology 13:658–660 [Google Scholar]
- 41. Qiu W., et al. 2009. Novel structural and regulatory features of rhoptry secretory kinases in Toxoplasma gondii. EMBO J. 28:969–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ramachandran G. N., Ramakrishnan C., Sasisekharan V. 1963. Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 7:95–99 [DOI] [PubMed] [Google Scholar]
- 43. Ramalingam P., Ganapaty S., Rao Ch B. 2010. In vitro antitubercular and antimicrobial activities of 1-substituted quinoxaline-2,3(1H,4H)-diones. Bioorg. Med. Chem. Lett. 20:406–408 [DOI] [PubMed] [Google Scholar]
- 44. Saeij J. P., et al. 2006. Polymorphic secreted kinases are key virulence factors in toxoplasmosis. Science 314:1780–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schwede T., Kopp J., Guex N., Peitsch M. C. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Smyth G. K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:Article 3 [DOI] [PubMed] [Google Scholar]
- 47. Taylor S., et al. 2006. A secreted serine-threonine kinase determines virulence in the eukaryotic pathogen Toxoplasma gondii. Science 314:1776–1780 [DOI] [PubMed] [Google Scholar]
- 48. Thomsen R., Christensen M. H. 2006. MolDock: a new technique for high-accuracy molecular docking. J. Med. Chem. 49:3315–3321 [DOI] [PubMed] [Google Scholar]
- 49. Tsai H. C., et al. 2002. Treatment of Toxoplasma brain abscess with clindamycin and sulfadiazine in an AIDS patient with concurrent atypical Pneumocystis carinii pneumonia. J. Formos. Med. Assoc. 101:646–649 [PubMed] [Google Scholar]
- 50. Vicente E., Duchowicz P. R., Castro E. A., Monge A. 2009. QSAR analysis for quinoxaline-2-carboxylate 1,4-di-N-oxides as anti-mycobacterial agents. J. Mol. Graph. Model. 28:28–36 [DOI] [PubMed] [Google Scholar]
- 51. Vicente E., et al. 2008. Synthesis and structure-activity relationship of 3-phenylquinoxaline 1,4-di-N-oxide derivatives as antimalarial agents. Eur. J. Med. Chem. 43:1903–1910 [DOI] [PubMed] [Google Scholar]
- 52. Vicente E., et al. 2009. Selective activity against Mycobacterium tuberculosis of new quinoxaline 1,4-di-N-oxides. Bioorg. Med. Chem. 17:385–389 [DOI] [PubMed] [Google Scholar]
- 53. Vicente E., et al. 2008. Efficacy of quinoxaline-2-carboxylate 1,4-di-N-oxide derivatives in experimental tuberculosis. Antimicrob. Agents Chemother. 52:3321–3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wallqvist A., Huang R., Thanki N., Covell D. G. 2006. Evaluating chemical structure similarity as an indicator of cellular growth inhibition. J. Chem. Infect. Model. 46:430–437 [DOI] [PubMed] [Google Scholar]
- 55. Zheng J., et al. 1993. A refined crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MnATP and a peptide inhibitor. Acta Crystallogr. D Biol. Crystallogr. 49:362–365 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.