Abstract
Recent studies have demonstrated that some morphologically atypical Aspergillus fumigatus strains are different species belonging to the section Fumigati. Aspergillus lentulus, one of these sibling species, is increasingly reported in patients under corticosteroid treatment. MICs of most antifungals in clinical use are elevated against A. lentulus, and it shows primary resistance to azole drugs. Two A. lentulus cytochrome P450 14-α sterol demethylases, encoded by A. lentulus cyp51A (Alcyp51A) and Alcyp51B genes, were identified. Targeted cyp51A gene knockout in A. lentulus showed that the intrinsic azole resistance of this species is cyp51A dependent. The Δcyp51A strain was morphologically indistinguishable from the A. lentulus wild-type strain, retaining the ability to cause pulmonary disease in neutropenic mice. The heterologous expression of A. lentulus cyp51A was performed in an A. fumigatus cyp51A-deficient strain, confirming that Cyp51A is responsible for the differences in A. lentulus-azole drug interaction.
INTRODUCTION
Invasive aspergillosis (IA) commonly develops in recipients of allogeneic hematopoietic stem cell transplantation (HSCT) and in patients with persistent neutropenia (22). However, some studies indicate that the population at risk for pulmonary or disseminated IA can be expanded to patients with chronic obstructive pulmonary disease (COPD) as well as to nontransplant intensive care patients (11, 17). Aspergillus fumigatus is the main causative agent of IA, although the number of other Aspergillus species able to produce fungal disease is on the rise, increasing the complexity of this infection (24, 45).
Several studies have demonstrated that some strains morphologically identified as A. fumigatus are different species belonging to the section Fumigati (5, 7, 8). Aspergillus lentulus, one of these sibling species of A. fumigatus, is increasingly reported in hematological and cystic fibrosis patients and in those under corticosteroid treatment (4, 6, 33, 42). The risk factors associated with these cryptic species still are undefined, but it is known that they show a different antifungal drug susceptibility profile than A. fumigatus (3). Strains of A. fumigatus are intrinsically susceptible to the expanded-spectrum triazole drugs, such as itraconazole, voriconazole, and posaconazole, and to the polyene drug amphotericin B (12). However, the development of secondary resistance to azole drugs in clinical strains is well documented (10, 15, 21, 40, 47). For triazole drugs, secondary resistance involving the acquisition of resistance in a susceptible strain accounts for all of the resistance described in A. fumigatus. On the other hand, the MICs of azole drugs for A. lentulus clinical samples are high, as are those of echinocandins and amphotericin B (3, 9, 41).
The azole-derived antifungal agents inhibit the ergosterol biosynthesis pathway via the inhibition of 14-α sterol demethylase (Cyp51) (18). Analyses of A. fumigatus azole-resistant strains at the molecular level have already identified many underlying bases for A. fumigatus azole resistance. Resistance due to modifications of the azole target enzyme 14-α sterol demethylase (Cyp51A) or its overexpression are the main mechanisms involved in A. fumigatus azole resistance (13, 16, 26, 30, 32, 34). However, the molecular mechanism of the intrinsic resistance of A. lentulus remains unknown. Since A. lentulus isolates cause invasive disease, its primary resistance to azole drugs is a matter of concern that may have both epidemiological and clinical significance (3, 25, 28).
Similarly to what happens in A. fumigatus, two different cyp51-related genes encoding 14-α sterol demethylase-like enzymes were identified in A. lentulus (Alcyp51A and Alcyp51B) (29). Since the cytochrome P450 14-α sterol demethylase, encoded by the cyp51A gene, is responsible for the azole drug affinity in A. fumigatus, here we describe the characterization of A. lentulus Cyp51A and its role in azole drug susceptibility and in virulence by the disruption of the azole target (Cyp51A). Finally, its function was confirmed by the heterologous expression of A. lentulus cyp51A in A. fumigatus cyp51A knockout strains.
MATERIALS AND METHODS
Strains, media, and growth conditions.
Fungal strains used in this study were (i) Aspergillus lentulus strain CM-1290 (CM refers to the fungal collection of the Mycology Reference Laboratory), with a clinical origin; (ii) A. fumigatus strain CM-237, previously used to describe the cyp51A and cyp51B gene sequences and also used as a control strain (29); (iii) A. fumigatus strain akuBKU80 (14), used as the recipient strain for some experiments of fungal transformation; and (iv) A. fumigatus ATCC 2004305 and Aspergillus flavus ATCC 2004304, used as quality-control strains for EUCAST antifungal susceptibility testing (37).
Aspergillus strains were grown at 37°C in either GYEP (2% glucose, 0.3% yeast extract, 1% peptone) or Sabouraud (2% glucose, 1% mycopeptone) medium. Escherichia coli JM109 was grown in Luria-Bertani (LB) medium (38) supplemented with ampicillin (100 μg/ml) for the propagation of plasmids for DNA purification. For standard cloning and subcloning procedures, the vector pGEM-T Easy vector (Promega, Madrid, Spain) was used.
Aspergillus lentulus cyp51A and cyp51B gene sequences.
A. fumigatus primers previously used for cyp51A and cyp51B gene amplification were used to amplify each of the genes from A. lentulus (29). Once the full sequence for the A. lentulus cyp51A gene was obtained, specific oligonucleotides were designed for A. lentulus (Table 1) when needed. All of the primers used in the present work were synthesized by Sigma Genosys (Madrid, Spain) (Table 1). The PCRs were carried out in a 50-μl volume containing 10 mM (NH4)2SO4; 10 mM KCl; 20 mM Tris-Cl (pH 8.8); 2 mM MgSO4; 10 ng bovine serum albumin (BSA); 0.1% Triton X-100; 250 μM (each) dATP, dGTP, dCTP, and dTTP (Applied Biosystem, Madrid, Spain); 1 μM each primer; 2.5 U of Titanium or Advantage 2 Taq polymerase (Clontech, Madrid, Spain); and 50 ng of genomic DNA. Amplification was performed in a thermal cycler (Amersham Biosciences, Madrid, Spain) for 1 cycle of 5 min at 94°C, 45 s at 56°C, and 2 min at 72°C; 30 cycles of 30 s at 94°C, 45 s at 56°C, and 2 min at 72°C; and then a final extension for 10 min at 72°C. PCR products were analyzed by electrophoresis on agarose gels and visualized by transillumination after staining them with ethidium bromide.
Table 1.
Oligonucleotides used throughout this work
| Name and group | Orientation | Sequence (5′-3′) | Purpose |
|---|---|---|---|
| A. lentuluscyp51A deletion | |||
| L-1 | Sense | CACTGTTTCTCATACAGGGAAACCTCATGG | 1st PCR, 5′-end vector construction; 2nd PCR, fusion vector |
| LH1 | Sense | GTTTTCGGATCTGACGTGGTGTATGATTGTTCGTCGACGTTAACTGATATTGAAGGAGCA | Hygromycin cassette amplification |
| LH2 | Antisense | TGCTCCTTCAATATCAGTTAACGTCGACGAACAATCATACACCACGTCAGATCCGAAAAC | 5′-End vector construction |
| HL3 | Sense | CAGTTAACGTCGACGAATTCGATATCAAGCGACCTCTATCATGATCTGGACAAGGGCTTT | Hygromycin cassette amplification |
| HL2 | Antisense | AAAGCCCTTGTCCAGATCATGATAGAGGTCGCTTGATATCGAATTCGTCGACGTTAACTG | Hygromycin cassette amplification |
| L-2 | Antisense | CCCTGAGAAGAGCGATGAATAGTCGGTATC | 1st PCR, 3′-end vector construction; 2nd PCR, fusion vector |
| L-3 | Sense | TTGTGCCTAGCAAGGAAAAAGACAAGAAA | PCR gene deletion verification |
| A. lentuluscyp51A expression in A. fumigatus | |||
| A | Sense | TAGAATGAGTGAGCTGATTTGCCATGGATT | 1st PCR, 5′-end vector construction; 2nd PCR, fusion vector |
| B2 | Antisense | GTAGGCCGTGAGCAATAGCATCGACACCATTTCGAGGAGACACAGGGAGGGTGAGCCCGA | 5′-End vector construction |
| C2 | Sense | TCGGGCTCACCCTCCCTGTGTCTCCTCGAAATGGTGTCGATGCTATTGCTCACGGCCTAC | Coding Cyp51A A. lentulus insertion |
| D3 | Antisense | CCTTTGAAGTCCTCGATGGTTACAACAGTCTCACTTGGATGTGTCTTTAGAACGCTTCTC | Coding Cyp51A A. lentulus insertion |
| E3 | Sense | GAGAAGCGTTCTAAAGACACATCCAAGTGAGACTGTTGTAACCATCGAGGACTTCAAAGG | A. fumigatus terminator insertion |
| F3 | Antisense | TGCTCCTTCAATATCAGTTAACGTCGACGAGCGTGCCAAGGCCAAGGCTTGATTAAGTAT | A. fumigatus terminator insertion |
| G3 | Sense | ATACTTAATCAAGCCTTGGCCTTGGCACGCTCGTCGACGTTAACTGATATTGAAGGAGCA | Hygromycin resistance cassette |
| F4 | Antisense | CTGTTCAGTCAGGGCATGTCCTTGGAGACTAAGCTTGATATCGAATTCGTCGACGTTAAC | Hygromycin resistance cassette |
| G4 | Sense | GTTAACGTCGACGAATTCGATATCAAGCTTAGTCTCCAAGGACATGCCCTGACTGAACAG | 3′-End vector construction |
| H2 | Antisense | AGAGATAGAACCTTGGAGTCTGCTTGCCTC | 1st PCR, 3′-end vector construction; 2nd PCR, fusion vector |
| A. lentuluscyp51A expression assessment | |||
| Len-A4 | Sense | CAGACATGATTTGGAACC | cDNA amplification |
| Len-Cypa2 | Antisense | TTAGAACGCTTCTCCCAG | cDNA amplification |
| A. fumigatuscyp51A expression analysis | |||
| A4 | Sense | CAGACATGATATGGAACC | cDNA amplification |
| Cypa2 | Antisense | TTCGACCGCTTCTCCCAG | cDNA amplification |
| Tub5 | Sense | TGACCCAGCAGATGTT | House-keeping gene used as reference for normalization |
| Tub6 | Antisense | GTTGTTGGGAATCCACTC | House-keeping gene used as reference for normalization |
The full coding sequences of cyp51A and cyp51B from A. lentulus were amplified by PCR, and both strands were sequenced by the BigDye Terminator cycle sequencing ready reaction system (Applied Biosystems, Madrid, Spain) according to the manufacturer's instructions. Sequence analysis was performed on an ABI prism 377 DNA sequencer (Applied Biosystem) using the sequencing facilities available at the Genomic Department at Instituto de Salud Carlos III, Majadahonda, Madrid, Spain.
Construction of the deletion and heterologous expression cassettes.
Two different fusion cassettes were constructed for Aspergillus transformation by PCR overlap as previously described (49). Basically, the first step is a conventional PCR in which oligonucleotide primers are partially complementary at their 5′ ends to the adjacent fragments that are fused to create the chimera. The second PCR step consists of the PCR amplification of the fusion of the PCR fragments generated in the first step by using the complementary extremities of the primers. The final PCR product is a chimeric vector built up with the different amplified PCR fragments (49). Oligonucleotides used for the construction of both fusion cassettes are enumerated in Table 1. Primer location and fusion vector design are described in Fig. 1 and 2. The PCR conditions for the amplification of the deletion and expression cassettes were previously described (23).
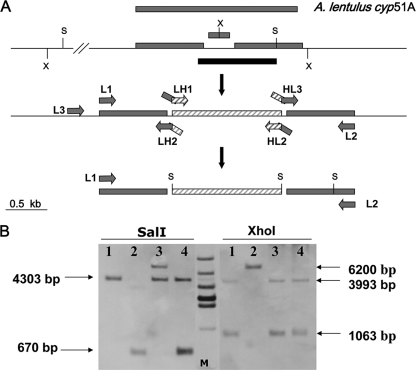
Fig. 1.
Construction of the A. lentulus cyp51A-defective mutant strain. (A) Design of the fusion vector for A. lentulus cyp51A gene deletion. The location of restriction sites used for the Southern analysis are indicated before (upper section) and after (lower section) the cyp51A locus. The transformants were selected using the hyg gene (1.4 kb) flanked by SalI (S) restriction sites included in the deletion cassette (striped bars). The excised cyp51A coding fragment and the probe used for hybridization are indicated in gray and black bars, respectively. The location of the primers for fusion vector construction is indicated in the middle section of the diagram. (B) Southern hybridization of genomic DNA of A. lentulus digested with SalI and XhoI (X). Lane 1, wild-type strain; lane 2, A. lentulus cyp51A-deficient T38.18 strain. Lanes 3 and 4 show two ectopic transformants that maintained the wild-type cyp51A copy. Sizes of the expected fragment are indicated in bp on the right side. Lane M, 1-kb molecular size marker.
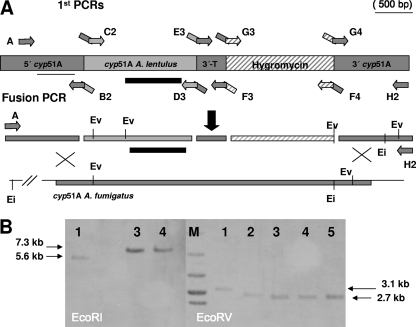
Fig. 2.
Construction of the A. fumigatus cyp51A-defective mutant strain expressing the A. lentulus cyp51A gene (Alcyp51A). (A) Design of the fusion vector for A. fumigatus cyp51A gene deletion and target integration of the Alcyp51A gene driven by the A. fumigatus cyp51A promoter (5′cyp51A) and terminator (3′ T) and including the hygromycin cassette (hyg) for transformant selection. At least 1 kb of A. fumigatus sequence outside the cyp51A coding region was included on both sides of the fusion construction to facilitate the homologous recombination. The location of restriction sites used for the Southern analysis are indicated before (upper section) and after (lower section) the cyp51A locus. The transformants were selected using the hyg gene (1.4 kb) included in the deletion cassette (striped bars). The location of the primers for fusion vector construction is indicated in this section of the diagram. (B) Southern hybridization of genomic DNA with EcoRV (Ev) and EcoRI (Ei) restriction enzyme digested with A. fumigatus wild type (lanes 1), A. lentulus CM-1290 wild type (lanes 2), and three hygromycin-resistant transformants with the wild-type cyp51A deleted and replaced by A. lentulus cyp51A copy T51.7 (lanes 3), T51.8 (lanes 4), and T51.9 (lane 5). Sizes of the expected fragment are indicated in bp on the right side. Lane M, 1-kb molecular size marker. The A. lentulus cyp51A probe used for hybridization is indicated by a black bar.
Aspergillus transformation.
The transformation of A. fumigatus strains was achieved by electroporation as previously described (31), with minor modifications to adjust the differences of growth between A. fumigatus and A. lentulus. After electroporation, transformants were selected on minimal medium plates containing hygromycin B (130 μg/ml; Sigma). Mutants were named with a letter followed by two numbers (i.e., T38.18 and T52.7), indicating the transformation experiment number and the transformant number.
Deletion verification by PCR and Southern blot analysis.
Gene targeting was first verified by PCR using primers selected outside the flanking regions (Table 1 and Fig. 1) used for cyp51A deletion fusion vector construction, in combination with primers used for resistance marker amplification. Homologous recombination at the correct locus of A. lentulus (Fig. 1) or A. fumigatus (Fig. 2) was confirmed by Southern blot analysis. Genomic DNA from hygromycin-resistant PCR-positive transformants was isolated using a rapid extraction procedure (43), digested with different restriction enzymes, and fractioned by electrophoresis through 0.8% agarose gels in Tris-acetate-EDTA (TAE) buffer. Southern analysis was performed as previously described (20). A probe for the A. lentulus cyp51A gene was obtained by the PCR amplification of the desired fragment, fractionation in 0.7% low-melting-point agarose gels, and further gel excision for labeling. A random-prime DNA labeling system (ECL; Amersham Pharmacia Biotech, Madrid, Spain) was used to label DNA probes according to the manufacturer's instructions.
RNA isolation and real-time reverse transcription-PCR (RT-PCR).
Mycelial mats were blot dried, frozen with liquid nitrogen, and then ground to powder using a pestle and mortar. RNA was isolated from mycelial powder by using an RNeasy plant minikit (Qiagen, Madrid, Spain) by following the manufacturer's instructions. Reverse transcription was carried out in a 20-μl reaction volume containing 10 mM Tris-HCl, pH 8.8, 50 mM KCl, 0.1% Triton X-100, 5 mM MgCl2, 1 mM each deoxynucleoside triphosphate, 0.5 μg of specific primer [oligo(dT)15 primer], 20 U rRNasin RNase inhibitor, and 15 U of avian myeloblastosis virus reverse transcriptase (AMV-RT) (reverse transcription system; Promega, Madrid, Spain) on 2.5 μg of total A. fumigatus RNA. The reaction conditions were 1 h at 42°C. A tube containing all of the reaction components without the AMV-RT enzyme, to check for the presence of contaminating DNA in the RNA sample, was always included as a negative control.
Two μl of cDNA products was used as target DNA for amplification using Light Cycler Fast Start (Roche). The primers Len-A4 and Len-CypA2 were used to amplify the cDNA from A. lentulus cyp51A, and the primers A4 and CypA2 were used for A. fumigatus cyp51A (Table 1).
Fold changes in expression were calculated using the 2−ΔΔCT method (39) and normalized to data for the β-tubulin housekeeping gene (GenBank accession number AY048754).
The RT-PCR products were sequenced to confirm the identity of the expressed DNA. The cDNA from A. lentulus also was used to check for the absence of an intron, which is present in the DNA fragment, and to delimitate its size and boundaries.
Antifungal susceptibility testing.
Inoculum preparations were performed by means of counting spores in a hematocytometer chamber (1) and were adjusted to 105 conidia per ml. For the Etest, gradient strips of voriconazole, posaconazole, itraconazole, and fluconazole, obtained from AB Biodisk (bioMérieux, Madrid, Spain), were used. The MICs were read after 48 h of incubation at 35°C. MICs were read as the lowest concentration at which the border of the elliptical inhibition zone intersected the scale on the strip. Microcolonies inside the inhibition zone were ignored. The strips of fluconazole, voriconazole, itraconazole, and posaconazole contained concentration gradients of 0.002 to 32 mg/liter. Antifungal susceptibility testing was repeated at least twice on different days. To define susceptibility and resistance in vitro, the criteria used were according to the epidemiological cutoffs (designated ec-off) recently published for A. fumigatus. The wild-type populations were defined as isolates against which the itraconazole and voriconazole MICs were ≤1 μg/ml and against which the posaconazole MIC was ≤0.25 μg/ml (36).
Itraconazole, voriconazole, and posaconazole (0.015 to 8 μg/ml) also were tested by microdilution by following EUCAST procedures (37). Since amphotericin B testing is not validated by Etest testing, amphotericin B (Sigma-Aldrich Química) was tested only by microdilution (range, 0.03 to 16 μg/ml).
Experimental model of infection.
The pathogenicity of the A. lentulus Δcyp51A (T38.18) mutant strain was compared to that of its A. lentulus parental strain and that of A. fumigatus CM-237 (inoculum size of 105 spores/mice). Strain virulence was assessed in a neutropenic mice model (ICR; specific pathogen free; 6 weeks old; CRIFFA, Barcelona, Spain) as previously described (31, 44). Immunosuppression was induced with 175 mg/kg of body weight of cyclophosphamide (Pras-Farma, Barcelona, Spain) administered intraperitoneally (i.p.) and 112 mg/kg of cortisone 21-acetate (C-3130; Sigma) administered subcutaneously, both given 3 and 1 day before the start of the experiment. Afterwards only cyclophosphamide (175 mg/kg) was used every 3 days until the completion of the experiment. After immunosuppression, mice were infected with a 30-μl drop of a fresh suspension of A. fumigatus or A. lentulus conidia. Two inoculum sizes were tested for A. lentulus wild-type and mutant strains (105 and 106 spores/mice). Survival was monitored at least twice a day for a time period of 14 days, and moribund animals were sacrificed. The experiments were carried out with eight mice per group. Lungs of mice infected with conidia of different Aspergillus strains were analyzed to check for the recovery of the infected strain: A. fumigatus, the A. lentulus parental strain, or the A. lentulus Δcyp51A mutant strain.
Statistical analysis.
Kaplan-Meier survival analysis was used to determine differences in the pathogenicity test. Statistical analysis was done with the SPSS package (version 12.0; SPSS S.L., Madrid, Spain).
Computer analysis.
The amino acid sequences of putative 14-α sterol demethylase genes cyp51A and cyp51B were deduced from nucleotide sequences and analyzed using the MegAlign software package (DNAstar, Inc., Lasergene, Madison, WI) run on a personal computer. The amino acid alignments were derived by CLUSTAL analysis (19).
Nucleotide sequence accession numbers.
The full nucleotide sequences of the cyp51A and cyp51B genes from A. lentulus and the deduced amino acid sequences determined in this work appear in the GenBank nucleotide sequence database under accession numbers GU479991 for Aspergillus lentulus cyp51A and JF276895 for Aspergillus lentulus cyp51B. A. fumigatus AF338659 and AF33860 were the reference sequences used for Cyp51A and Cyp51B, respectively.
RESULTS
Identification of the Aspergillus lentulus cyp51A and cyp51B genes.
PCR amplification based on A. fumigatus oligonucleotides allowed for the cloning and sequencing of two different cyp51 homologue genes in A. lentulus (31). The two cyp51-related genes encoding 14-α sterol demethylases-like enzymes were named A. lentulus cyp51A (AlCyp51A) and A. lentulus cyp51B (AlCyp51B) by following A. fumigatus nomenclature. There was a 92% identity match between the Alcyp51A sequence and that of A. fumigatus cyp51A (Afcyp51A). The alignment of the two DNA sequences revealed that both have exactly the same length (1,619 bp), including the size and location of an intron of 71 bp. The second gene, cyp51B, has a length of 1,733 bp, including three predicted introns at the same location as Afcyp51B but with different lengths: 54 bp (58 bp in A. fumigatus), 51 bp (45 bp in A. fumigatus), and a third intron of 53 bp in both species. There was an overall 96% identity match between Alcyp51B and Afcyp51B.
The comparison of the deduced AlCyp51A (515 amino acids) and AlCyp51B (524 amino acids) proteins to Cyp51A and Cyp51B of A. fumigatus showed that the amino acid sequences of these newly described proteins had percent identity matches of 95 and 98%, respectively. Sequence alignments clearly demonstrated that these two enzymes (AlCyp51A and AlCyp51B) are the corresponding A. fumigatus Cyp51 homologues. Amino acid differences between both Cyp51 proteins in A. fumigatus and A. lentulus are listed in Tables 2 and 3.
Table 2.
Amino acid differences between Cyp51A from A. fumigatus and A. lentulusb
| Amino acid | A. fumigatus | A. lentulus |
|---|---|---|
| Pro3 | P3 (CCG) | P3S (TCG) |
| Trp6 | W6 (TGG) | W6L (TTG) |
| Met11 | M11 (ATG)a | M11T (ACG)a |
| Val15 | V15 (GTG) | V15M (ATG) |
| Ala18 | A18 (GCA) | A18V (GCA) |
| Fhe29 | F29 (TTT)a | F29Y (TAC)a |
| Ser49 | S49 (AGT) | S49N (AAT) |
| Leu67 | L67 (AAG) | K67R (AGG) |
| Asp161 | D161 (GAT) | D161N (AAT) |
| Arg171 | R171 (CGG) | R171Q (CAG) |
| Met172 | M172 (ATG) | M172V (GTG) |
| Asp255 | D255 (GAC) | D255G (GGA) |
| Cys270 | C270 (TGC) | C270S (AGC) |
| Lys314 | K314 (AAA) | K314Q (CAG) |
| Ala330 | A330 (GCC) | A330I (ATT) |
| Ser335 | S335 (AGT) | S335N (AAT) |
| Phe337 | P337 (CCT)a | P337S (TCT)a |
| His352 | H352 (CAT)a | H352Q (CAG)a |
| Ile354 | I354 (ATT) | I354V (GTT) |
| Ile360 | I360 (ATT) | I360L (CTT) |
| Ile367 | I367 (ATC) | I367L (CTC) |
| Met383 | M383 (ATG) | M383V (GTG) |
| Thr420 | T420 (ACT) | T420A(GCC) |
| Leu464 | L464 (CTT) | L464I (ATT) |
| Glu488 | E488 (GAA) | E488D (GAT) |
| Gly505 | G505 (GGC) | G505R (CGC) |
Polymorphisms between strains of A. lentulus.
Amino acid numbers are the same for both species.
Table 3.
Amino acid differences between Cyp51B of A. fumigatus and A. lentulusa
| Amino acid | A. fumigatus | A. lentulus |
|---|---|---|
| Ile26 | I26 (ATA) | I26V (GTC) |
| Asn48 | N48 (AAT) | N48S (TCT) |
| Lys175 | K175 (ATG) | K175R (ATG) |
| Ala179 | A179 (GTG) | A179S (ATG) |
| Phe180 | F180 (GCA) | F180L (GCA) |
| Lys263 | K263 (TTT) | K263R (TTT) |
| Ala268 | A268 (AGT) | A268T (AGT) |
| Ala385 | A385 (AGT) | A385V (AGT) |
| Asn420 | N420 (GAT) | N420D (GAT) |
| Ser446 | S446 (CGG) | S446N (CGG) |
| Arg485 | R485 (ATG) | R485K (CGG) |
| Ile495 | I495 (GAC) | I495V (GAC) |
Amino acid numbers are the same for both species.
Generation of the Δcyp51A Aspergillus lentulus strain.
To analyze the role of the azole target designated 14-α sterol demethylase A (AlCyp51A) in A. lentulus, the corresponding cyp51A gene was deleted from the A. lentulus wild-type strain CM-1290. For this purpose, a fusion vector containing the cyp51A knockout construct with the hygromycin (hyg) resistance marker (Fig. 1A) was generated (see Materials and Methods). A full cyp51A-deleted mutant was not pursued, since there is not enough A. lentulus sequence knowledge to allow for the excision of the full Alcyp51A coding sequence.
After the transformation of A. lentulus strain CM-1290 with the fusion vector, the integration of the Δcyp51A-Hyg construct of the cyp51A gene in the resulting transformants was detected by PCR. To verify the results obtained by PCR, Southern blot analysis after digestion with two different restriction enzymes was performed. As shown in Fig. 1B, using SalI, the characteristic wild-type band (4.3 kb) for the Afcyp51A gene was absent from the Δcyp51A A. lentulus mutant and was replaced by a 670-bp band because of the inclusion of an SalI site corresponding to the Hyg cassette (the only remaining DNA segment able to hybridize with the probe). To double check, the use of XhoI clearly detected that the two characteristic bands for the cyp51A gene of the wild type (3.9 and 1 kb) had disappeared from the Δcyp51A mutant. Instead, there was a single band of 6.2 kb, indicative of cyp51A gene replacement as a consequence of the inclusion of the hyg resistance gene, and the disappearance of an XhoI restriction site located in the deleted section of the cyp51A gene (Fig. 1A and B).
Phenotype of the A. lentulus Δcyp51A strain.
The Alcyp51A-disrupted strain was morphologically indistinguishable from the A. lentulus wild-type strain CM-1290. However, antifungal susceptibility testing clearly showed that the absence of Cyp51A renders A. lentulus significantly more susceptible to all azole drugs, with decreased azole MICs that varied from 5- to 100-fold depending on the azole tested (Table 4 and Fig. 3). Azole MICs for the A. lentulus Δcyp51A mutant resemble those of A. fumigatus Δcyp51A mutants (31). Moreover, since azole MICs for A. lentulus strains are higher than those for A. fumigatus, the decrease in azole MICs was even more remarkable (Table 4). Differences in antifungal susceptibility values for itraconazole, voriconazole, and posaconazole between the A. lentulus wild type and the Δcyp51A mutant strain (T18.38) were statistically significant (P < 0.05), while there were no differences in susceptibility to amphotericin B.
Table 4.
MICs of different antifungals against Aspergillus fumigatus and A. lentulus isolates and their derived mutant strains
| Isolate | Origin and/or background | Species | cyp5A gene copy origin | MIC (μg/ml) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AmB |
ITC |
VCZ |
POS |
|||||||
| EUCAST | EUCAST | Etest | EUCAST | Etest | EUCAST | Etest | ||||
| CM-1290 | Sputum | A. lentulus | A. lentulus | 1–4 | 0.5–8 | 8–32 | 4 | 4–6 | 012–0.25 | 1 |
| ATCC 2004305 | Reference strain | A. fumigatus | A. fumigatus | 0.25–0.5 | 0.12–0.25 | 2 | 0.25–0.5 | 0.25 | 0.03–0.12 | 0.19 |
| akuBKU80 | BALa | A. fumigatus | A. fumigatus | 0.25–0.5 | 0.12–0.25 | 1.5–4 | 0.25 | 0.38–0.25 | 0.03–006 | 0.012–0.38 |
| CM-237 | Sputum | A. fumigatus | A. fumigatus | 0.25–0.5 | 0.25 | 0.75–3 | 0.25–0.5 | 012–0.19 | 0.06–012 | 0.012–0.38 |
| CM237-A-8 | Mutant; CM237 | A. fumigatus | Δcyp51A | 0.5 | 0.03 | 0.047 | 0.12 | 0.064 | 0.015 | <0.02 |
| CM1290-T29.18 | Mutant; CM1290 | A. lentulus | Δcyp51A | 2–4 | 0.03–0.06 | 0.032–0.064 | 0.25 | 0.064 | 0.015 | 0.064 |
| Mutant T52.7 | Mutant; akuBKU80 | A. fumigatus | A. lentulus | 0.25 | 0.5–8 | 8–32 | 2 | 2–4 | 0.25 | 0.25–1.5 |
| Mutant T52.8 | Mutant; akuBKU80 | A. fumigatus | A. lentulus | 0.25 | 0.25 | 4 | 1 | 1–2 | 0.06–0.12 | 1 |
| Mutant T52.9 | Mutant; akuBKU80 | A. fumigatus | A. lentulus | 0.25 | 0.5 | 8 | 1 | 1–3 | 0–12 | 0.5–1 |
BAL, bronchoalveolar lavage.
Fig. 3.
Etest susceptibility testing of itraconazole (ITC), voriconazole (VCZ), fluconazole (FCZ), and posaconazole (POS) for the A. lentulus wild-type strain (CM-1290) and an A. lentulus cyp51A-deficient strain (T38.18).
Aspergillus lentulus Alcyp51A gene expression in an A. fumigatus Δcyp51A deletion strain.
To confirm the implication of AlCy51A in azole resistance, the Alcyp51A gene copy was expressed in an A. fumigatus strain, generating a cyp51A-deleted strain. A PCR fusion vector was constructed for this purpose, containing 1,237 bp of homology at the 5′ upstream sequence of A. fumigatus, followed by the full cyp51A gene sequence of A. lentulus and 270 bp of the 3′ Afcyp51A gene terminator, which was followed by the 1.4-kb hygromycin-selectable marker cassette and 1,000 bp of cyp51A sequence homologous to the 3′ downstream sequence of A. fumigatus. The constructed vector favors homologous recombination at the Afcyp51A locus, replacing its endogenous gene with the construct which includes the wild-type A. lentulus cyp51A gene copy expressed under the A. fumigatus cyp51A promoter and terminator and including the hygromycin resistance cassette for transformant selection (Fig. 2A).
After A. fumigatus transformation, several hygromycin-resistant mutants were analyzed by PCR. The single integration of the fusion vector at the right locus was confirmed in three transformants (T52.7, T52.8, and T52.9) by Southern blot analysis (Fig. 2B). Also, a PCR fragment containing the whole integrated construction was sequenced in each transformant to verify the replacement of the A. fumigatus cyp51A sequence by cyp51A of A. lentulus and its correct nucleotide sequence.
The effect on the susceptibility to azoles in the A. fumigatus Δcyp51A gene-replaced strains was investigated by Etest and microdilution EUCAST methods, which showed that the A. lentulus azole-resistant phenotype was reproduced in the Δcyp51A A. fumigatus strain (Fig. 4). Accordingly, there were no changes in amphotericin B susceptibility values for any of the transformants (Table 4).
Fig. 4.
Etest susceptibility testing of voriconazole (VCZ) and posaconazole (POS) for the A. lentulus wild-type strain (CM-1290), the A. fumigatus parental strain (akuBKU80), and three mutant strains (T51.7, T51.8, and T51.9).
To verify that the MIC change was not due to a modification of cyp51A expression, we quantified the cyp51A mRNA levels of these mutants by real-time PCR. As is shown in Table 5, the A. fumigatus transformants expressed Alcyp51A at the same level as the homologous Afcyp51A in the wild-type strain (akuBKU80) that was used as the reference for gene expression.
Table 5.
A. lentulus cyp51A mRNA transcription levels with respect to A. fumigatus wild-type reference strain akuBKU80
| Mutant strain |
cyp51A mRNA transcription levela |
||
|---|---|---|---|
| Geometric mean | SD | VC (%) | |
| akuBKU80 T52.7 | 1.2 | 0.10 | 6.9 |
| akuBKU80 T52.8 | 1.1 | 1.61 | 22.1 |
| akuBKU80 T52.9 | 1.1 | 0.65 | 15.2 |
Geometric means are from at least two different RNAs and six cDNAs. SD, standard deviation; VC, variation coefficient.
Virulence in an experimental model of pulmonary aspergillosis with Aspergillus lentulus wild-type and cyp51A-deleted strains.
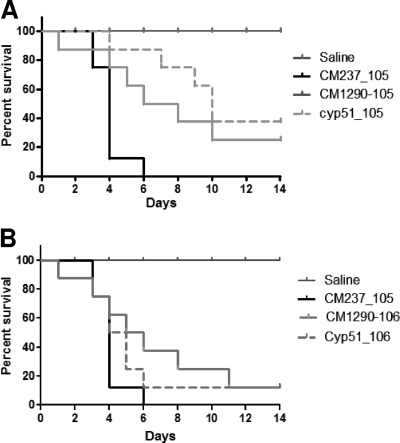
To determine whether the lack of 14-α sterol demethylase A influenced fungal pathogenicity, the A. lentulus Δcyp51A mutant was tested in a neutropenic mouse model of invasive aspergillosis by intranasal infection. The results of the course of the infections are shown in Fig. 5. A control group remained uninfected (they were given an inhalation of saline) to monitor the influence of the immunosuppression procedure on survival. In the control groups, mortality rates were 0% in the saline group and 100% in the A. fumigatus group (with an inoculum size of 105 spores/mice). The A. lentulus wild-type strain was pathogenic, although there was a significant difference in the duration of the course of the infection compared to that of A. fumigatus. Therefore, two different inoculum sizes were tested for A. lentulus (105 and 106 spores/mice). Higher doses of A. lentulus (106 spores/mice) showed a survival profile similar to that of A. fumigatus (105 spores/mice). However, there were no differences, with either of the inocula sizes, between the A. lentulus parental and the Δcyp51A mutant strains (Fig. 5). Fourteen days after infection with inocula of 105 or 106 conidia, the overall mortality rates for the A. lentulus mutant strain and for the parental wild-type strain CM-1290 were 10 and 20%, respectively.
Fig. 5.
Comparative analysis of parental A. lentulus wild-type and cyp51A-deficient strains in a neutropenic murine model of pulmonary aspergillosis. Survival of CD-1 immunocompromised mice infected with the A. fumigatus wild-type CM-237 strain (105 spores/mice) and the A. lentulus wild-type (CM-1290) and cyp51A-deficient (Cyp51) strains. Two inoculum sizes, 105 (A) and 106 (B) spores/mouse, were used for both A. lentulus strains. A control group with mice immunocompromised and inoculated only with saline also was included. Survival was monitored for a time period of 14 days, and moribund animals were sacrificed.
DISCUSSION
The use of azoles is increasing in the management of invasive aspergillosis. Voriconazole is the recommended drug for the primary therapy of invasive aspergillosis (35). Another azole drug, posaconazole, is used prophylactically, as it has been reported to reduce the number of invasive fungal infections in neutropenic patients (48). Azole resistance in Aspergillus species is not yet a frequently reported event. As the number of patients exposed to azole therapy broadens, the probability of resistance is expected to increase. Unfortunately, widespread azole usage could lead to both the development of acquired resistance (especially among A. fumigatus strains) and a shift toward less azole-susceptible fungal species, as could be happening with A. lentulus.
A. lentulus prevalence is unknown, although since its identification in 2005 it is being reported more frequently in some countries (4, 7, 28, 33, 42, 45). Although this could be due to improved identification and therefore does not reflect increasing incidence, this fact is of relevance, since A. lentulus has been proven to be responsible for invasive aspergillosis (6). Currently, A. lentulus infection in susceptible patients is being described but is mostly associated with A. fumigatus infections (4, 33, 42). The main importance of this species is that A. lentulus could be considered intrinsically azole resistant if we follow the proposed breakpoints for azole resistance in A. fumigatus (46). Since A. fumigatus Cyp51A and Cyp51B were described in 2001 (29), the search for the resistance mechanism operating in A. fumigatus isolates has revealed at least four different mechanisms, and all of them are related to cyp51A alterations (13, 16, 26, 30, 32, 34). To date, A. fumigatus azole resistance has always been defined as secondary or acquired resistance. However, all A. lentulus strains have a similar antifungal susceptibility pattern (3, 28), which would indicate primary resistance operating in A. lentulus. It is well known that the modification of some Cyp51A amino acid residues is the most important azole resistance mechanism in A. fumigatus (15, 26, 28, 30, 31, 34). However, no changes have been identified in cyp51B in relation to azole susceptibility, suggesting that this enzyme has a different function or acts in alternative growth conditions. Backing this up, the alignment between A. fumigatus and A. lentulus Cyp51A proteins showed that there were 26 different amino acids, while both Cyp51B proteins were more conserved, with only 12 different residues.
A. lentulus is macro- and microscopically indistinguishable from A. fumigatus, although some differences in growth rate and sporulation (6, 8) could account for differences in azole drug interaction or drug uptake. In this study, we generated a cyp51A knockout strain of A. lentulus. The lack of morphological defects compared to the A. lentulus wild type, together with its unchanged pathogenicity, indicates that Cyp51A is not essential for A. lentulus viability. However, the susceptibility testing of the mutant strain showed significantly decreased MICs for all azoles, with the A. lentulus Δcyp51A mutants being severalfold more susceptible to all azole drugs than the parental wild type. This increased susceptibility was observed even with fluconazole (Fig. 3), an azole drug to which all Aspergillus spp. are intrinsically resistant. The azole susceptibility profile resembled that of A. fumigatus Δcyp51A mutants in susceptible and resistant strains (31), suggesting that the intrinsic azole resistance shown by A. lentulus is dependent on Cyp51A function. In addition, the replacement of the A. fumigatus cyp51A gene with the A. lentulus cyp51A gene restored the susceptibility phenotype of the A. lentulus wild-type strain, transforming an A. fumigatus (cyp51A-deleted) azole-hypersusceptible strain into an azole-resistant one. A. fumigatus strains expressing A. lentulus cyp51A were significantly less susceptible to azole drugs than when they were expressing their constitutive cyp51A gene. Some discrepancies were observed between Etest and EUCAST testing, especially with itraconazole, where the Etest showed generally higher values. Etest was used to show the results, because differences in susceptibility values are clearer with this methodology, showing clear endpoints, as well as providing clear visual reading which can be easily photographed. In any case, since we are using modified strains with the same background strain (isogenic strains), the differences in susceptibility should be considered significant because we can exclude strain variability.
However, the azole MICs for A. lentulus are slightly higher for almost every wild-type strain tested. Therefore, it seems that an additional factor, other than protein-azole interaction, contributes to the decreased azole susceptibility shown by A. lentulus. Two major circumstances could be responsible for this: (i) differences in the intracellular azole concentrations achieved within A. lentulus because of differences in azole drug uptake or an increased expression of ABC transporters (among other efflux pumps), or (ii) due to the design of the experiment, only the coding region of the A. lentulus cyp51A was expressed in A. fumigatus. Therefore, differences in the A. lentulus cyp51A promoter could allow for transcription factor interaction, leading to increased cyp51A expression. Alternatively, differences in the A. lentulus 3′-untranslated region could allow for higher cyp51A mRNA stability, increasing the possibilities of resistance expression. Both of these possibilities have been shown to be potential resistance determinants on their own or as contributing factors (32, 27).
The results obtained in this study proved that the A. lentulus wild-type strain was pathogenic in immunocompromised mice, although there was a significant difference in the duration of the course of the infection compared to that for A. fumigatus. This indicates a reduced virulence capability for A. lentulus and might explain why both species are found mainly in mixed infections (4, 33, 42). Given the differences in antifungal susceptibility between both species, this could represent a threat for patients under long-term azole treatment.
As the sequences of AfCyp51A and AlCyp51A are nearly identical (95%), it is difficult to base the different azole susceptibility profiles on single-amino-acid differences, especially since none of the different residues are located in important conserved regions of the protein (2, 50). However, the study of molecular dynamics applied to three-dimensional protein models of the molecular docking of the A. lentulus cyp51A and Afcyp51A, in combination with voriconazole, has provided information about some critical differences found in the putative closed form adopted by the proteins upon voriconazole binding. These results suggest that some major differences in the protein's BC loop differently affect voriconazole lock up and in turn correlate with A. lentulus differences in voriconazole susceptibility (2). The results derived could be of use for rational azole drug design or azole drug improvement in general. In addition, the A. lentulus Δcyp51A mutant could be used in the screening for the identification of inhibitory compounds that specifically target Cyp51B activity. These findings could be confirmed by testing antifungal compounds in a murine model of invasive aspergillosis.
In summary, PCR based on A. fumigatus oligonucleotides allowed for the amplification and sequencing of two different cyp51 homologue genes in A. lentulus. The molecular mechanisms of high azole MICs for A. lentulus are Cyp51A dependent but are different from what has been described previously for A. fumigatus. The intrinsic azole resistance shown by A. lentulus may represent a problem if colonization by this species becomes more frequent in the susceptible host. As the role of A. lentulus in invasive aspergillosis remains unclear, surveillance networks of clinical isolates are needed to determine the species distribution, their implication in clinical disease, and the outcomes of patients with invasive aspergillosis.
ACKNOWLEDGMENTS
This work was supported by SAF2008-04143 from the Ministerio de Ciencia e Innovacion (MICINN). L.A.-F. held a postdoctoral contract from the EU-STREP project (LSHM-CT-2005-518199).
E.M. thanks the ESF Research Networking Programme FUMINOMICS, Ref. 06-RNP-132 (EMRC-LESC). We thank Christophe d′Enfert for helpful discussion of vector design for Aspergillus lentulus cyp51A expression. We are also grateful to Frank Hodgkins for correcting the English.
E.M. and L.A.-F. do not have a financial relationship with any commercial entities that have an interest in the subject of the manuscript. In the past 5 years, M.C.E. has received grant support from Astellas Pharma, bioMerieux, Gilead Sciences, Merck Sharp and Dohme, Pfizer, Schering Plough, Soria Melguizo SA, the European Union, the ALBAN program, the Spanish Agency for International Cooperation, the Spanish Ministry of Culture and Education, The Spanish Health Research Fund, The Instituto de Salud Carlos III, The Ramon Areces Foundation, and The Mutua Madrileña Foundation. He has been an advisor/consultant to the Panamerican Health Organization, Gilead Sciences, Merck Sharp and Dohme, Pfizer, and Schering Plough. He has been paid for talks on behalf of Gilead Sciences, Merck Sharp and Dohme, Pfizer, and Schering Plough In the past 5 years, J.L.R.-T. has received grant support from Astellas Pharma, Gilead Sciences, Merck Sharp and Dohme, Pfizer, Schering Plough, Soria Melguizo SA, the European Union, the Spanish Agency for International Cooperation, the Spanish Ministry of Culture and Education, The Spanish Health Research Fund, The Instituto de Salud Carlos III, The Ramon Areces Foundation, and The Mutua Madrileña Foundation. He has been an advisor/consultant to the Panamerican Health Organization, Gilead Sciences, Merck Sharp and Dohme, Mycognostica, Pfizer, and Schering Plough. He has been paid for talks on behalf of Gilead Sciences, Merck Sharp and Dohme, Pfizer, and Schering Plough.
Footnotes
Published ahead of print on 26 September 2011.
REFERENCES
- 1. Aberkane A., et al. 2002. Comparative evaluation of two different methods of inoculum preparation for antifungal susceptibility testing of filamentous fungi. J. Antimicrob. Chemother. 50: 719–722 [DOI] [PubMed] [Google Scholar]
- 2. Alcazar-Fuoli L., Cuesta I., Cuenca-Estrella M., Rodriguez-Tudela J. L., Mellado E. 2011. Three dimensional models of 14-α sterol demethylase (Cyp51A) from Aspergillus lentulus and A. fumigatus: An insight into differences in voriconazole interaction. Int. J. Antimicrob. Agents 38: 426–434 [DOI] [PubMed] [Google Scholar]
- 3. Alcazar-Fuoli L., Mellado E., Alastruey-Izquierdo A., Cuenca-Estrella M., Rodriguez-Tudela J. L. 2008. Aspergillus section Fumigati: antifungal susceptibility patterns and sequence-based identification. Antimicrob. Agents Chemother. 52: 1244–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alhambra A., et al. 2008. Isolation of Aspergillus lentulus in Spain from a critically ill patient with chronic obstructive pulmonary disease. Rev. Iberoam. Micol. 25: 246–249 [DOI] [PubMed] [Google Scholar]
- 5. Balajee S. A., et al. 2005. Mistaken identity: Neosartorya pseudofischeri and its anamorph masquerading as Aspergillus fumigatus. J. Clin. Microbiol. 43: 5996–5999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Balajee S. A., Gribskov J. L., Hanley E., Nickle D., Marr K. A. 2005. Aspergillus lentulus sp. nov., a new sibling species of A. fumigatus. Eukaryot. Cell 4: 625–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Balajee S. A., et al. 2009. Molecular identification of Aspergillus species collected for the transplant-associated infection surveillance network. J. Clin. Microbiol. 47: 3138–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Balajee S. A., Nickle D., Varga J., Marr K. A. 2006. Molecular studies reveal frequent misidentification of Aspergillus fumigatus by morphotyping. Eukaryot. Cell 5: 1705–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Balajee S. A., Weaver M., Imhof A., Gribskov J., Marr K. A. 2004. Aspergillus fumigatus variant with decreased susceptibility to multiple antifungals. Antimicrob. Agents Chemother. 48: 1197–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bueid A., et al. 2010. Azole antifungal resistance in Aspergillus fumigatus: 2008 and 2009. J. Antimicrob. Chemother. 65: 2116–2118 [DOI] [PubMed] [Google Scholar]
- 11. Bulpa P., Dive A., Sibille Y. 2007. Invasive pulmonary aspergillosis in patients with chronic obstructive pulmonary disease. Eur. Respir. J. 30: 782–800 [DOI] [PubMed] [Google Scholar]
- 12. Cuenca-Estrella M., Gomez-Lopez A., Mellado E., Buitrago M. J., Monzon A., Rodriguez-Tudela J. L. 2006. Head-to-head comparison of the activities of currently available antifungal agents against 3,378 Spanish clinical isolates of yeasts and filamentous fungi. Antimicrob. Agents Chemother. 50: 917–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. da Silva Ferreira M. E., et al. 2004. In vitro evolution of itraconazole resistance in Aspergillus fumigatus involves multiple mechanisms of resistance. Antimicrob. Agents Chemother. 48: 4405–4413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. da Silva Ferreira M. E., et al. 2006. The akuBKU80 mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot. Cell 5: 207–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Denning D. W., et al. 2011. High-frequency triazole resistance found in nonculturable Aspergillus fumigatus from lungs of patients with chronic fungal disease. Clin. Infect. Dis. 52: 1123–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Diaz-Guerra T. M., Mellado E., Cuenca-Estrella M., Rodriguez-Tudela J. L. 2003. A point mutation in the 14alpha-sterol demethylase gene cyp51A contributes to itraconazole resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 47: 1120–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dimopoulos G., et al. 2003. Disseminated aspergillosis in intensive care unit patients: an autopsy study. J. Chemother. 15: 71–75 [DOI] [PubMed] [Google Scholar]
- 18. Ghannoum M. A., Rice L. B. 1999. Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin. Microbiol. Rev. 12: 501–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Higgins D. G., Sharp P. M. 1988. CLUSTAL: a package for performing multiple sequence alignments on a microcomputer. Gene 73: 237–244 [DOI] [PubMed] [Google Scholar]
- 20. Holden D. W., Kronstad J. W., Leong S. A. 1989. Mutation in a heat regulated hsp70 gene of Ustilago maydis. EMBO J. 8: 1927–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Howard S. J., et al. 2009. Frequency and evolution of Azole resistance in Aspergillus fumigatus associated with treatment failure. Emerg. Infect. Dis. 15: 1068–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kontoyiannis D. P., et al. 2010. Prospective surveillance for invasive fungal infections in hematopoietic stem cell transplant recipients, 2001–2006: overview of the Transplant-Associated Infection Surveillance Network (TRANSNET) database. Clin. Infect. Dis. 50: 1091–1100 [DOI] [PubMed] [Google Scholar]
- 23. Lamarre C., Ibrahim-Granet O., Du C., Calderone R., Latgé J. P. 2007. Characterization of the SKN7 ortholog of Aspergillus fumigatus. Fungal Genet. Biol. 44: 682–690 [DOI] [PubMed] [Google Scholar]
- 24. Lionakis M. S., et al. 2005. Increased frequency of non-fumigatus Aspergillus species in amphotericin B- or triazole-pre-exposed cancer patients with positive cultures for aspergilli. Diagn. Microbiol. Infect. Dis. 52: 15–20 [DOI] [PubMed] [Google Scholar]
- 25. Malani A. N., Kauffman C. A. 2007. Changing epidemiology of rare mould infections: implications for therapy. Drugs 67: 1803–1812 [DOI] [PubMed] [Google Scholar]
- 26. Mann P. A., et al. 2003. Mutations in Aspergillus fumigatus resulting in reduced susceptibility to posaconazole appear to be restricted to a single amino acid in the cytochrome P450 14alpha-demethylase. Antimicrob. Agents Chemother. 47: 577–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Manoharlal R., Gaur N. A., Panwar S. L., Morschhäuser J., Prasad R. 2008. Transcriptional activation and increased mRNA stability contribute to overexpression of CDR1 in azole-resistant Candida albicans. Antimicrob. Agents Chemother. 52: 1481–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mellado E., et al. 2006. New resistance mechanisms to azole drugs in Aspergillus fumigatus and emergence of antifungal drugs-resistant A. fumigatus atypical strains. Med. Mycol. 44(Suppl.): 367–371 [DOI] [PubMed] [Google Scholar]
- 29. Mellado E., Diaz-Guerra T. M., Cuenca-Estrella M., Rodriguez-Tudela J. L. 2001. Identification of two different 14-alpha sterol demethylase-related genes (cyp51A and cyp51B) in Aspergillus fumigatus and other Aspergillus species. J. Clin. Microbiol. 39: 2431–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mellado E., Garcia-Effron G., Alcazar-Fuoli L., Cuenca-Estrella M., Rodriguez-Tudela J. L. 2004. Substitutions at methionine 220 in the 14alpha-sterol demethylase (Cyp51A) of Aspergillus fumigatus are responsible for resistance in vitro to azole antifungal drugs. Antimicrob. Agents Chemother. 48: 2747–2750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mellado E., et al. 2005. Targeted gene disruption of the 14-alpha sterol demethylase (cyp51A) in Aspergillus fumigatus and its role in azole drug susceptibility. Antimicrob. Agents Chemother. 49: 2536–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mellado E., et al. 2007. A new Aspergillus fumigatus resistance mechanism conferring in vitro cross-resistance to azole antifungals involves a combination of cyp51A alterations. Antimicrob. Agents Chemother. 51: 1897–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Montenegro G., et al. 2009. Phenotypic and genotypic characterization of Aspergillus lentulus and Aspergillus fumigatus isolates in a patient with probable invasive aspergillosis. J. Med. Microbiol. 58: 391–395 [DOI] [PubMed] [Google Scholar]
- 34. Nascimento A. M., et al. 2003. Multiple resistance mechanisms among Aspergillus fumigatus mutants with high-level resistance to itraconazole. Antimicrob. Agents Chemother. 47: 1719–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pasqualotto A. C., Xavier M. O., Andreolla H. F., Linden R. 2010. Voriconazole therapeutic drug monitoring: focus on safety. Expert Opin. Drug Safety 9: 125–137 [DOI] [PubMed] [Google Scholar]
- 36. Rodriguez-Tudela J. L., et al. 2008. Epidemiological cut-offs and cross resistance to azole drugs in Aspergillus fumigatus. Antimicrob. Agents Chemother. 52: 2468–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rodriguez-Tudela J. L., et al. 2008. EUCAST technical note on the method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for conidia-forming moulds. Subcommittee on Antifungal Susceptibility Testing (AFST) of the ESCMID European Committee for Antimicrobial Susceptibility Testing (EUCAST) Clin. Microbiol. Infect. 14: 982–984 [DOI] [PubMed] [Google Scholar]
- 38. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning. A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 39. Schmittgen T. D., Livak K. J. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3: 1101–1118 [DOI] [PubMed] [Google Scholar]
- 40. Snelders E., et al. 2008. Emergence of azole resistance in Aspergillus fumigatus and spread of a single resistance mechanism. PLoS Med. 5: e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Staab J. F., Kahn J. N., Marr K. A. 2010. Differential Aspergillus lentulus echinocandin susceptibilities are Fksp independent. Antimicrob. Agents Chemother. 54: 4992–4998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Symoens F., et al. 2010. Unusual Aspergillus species in patients with cystic fibrosis. Med. Mycol. 48: S10–S16 [DOI] [PubMed] [Google Scholar]
- 43. Tang C. M., Cohen J., Holden D. W. 1992. An Aspergillus fumigatus alkaline protease mutant constructed by gene disruption is deficient in extracellular elastase activity. Mol. Microbiol. 6: 1663–1671 [DOI] [PubMed] [Google Scholar]
- 44. Tang C. M., Cohen J., Krausz T., Van Noorden S., Holden D. W. 1993. The alkaline protease of Aspergillus fumigatus is not a virulence determinant in two murine models of invasive pulmonary aspergillosis. Infect. Immun. 61: 1650–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Van Der Linden J. W., Warris A., Verweij P. E. 2011. Aspergillus species intrinsically resistant to antifungal agents. Med. Mycol. 49: S82–S89 [DOI] [PubMed] [Google Scholar]
- 46. Verweij P. E., Howard S. J., Melchers W. J., Denning D. W. 2009. Azole-resistance in Aspergillus: proposed nomenclature and breakpoints. Drug Resist. Updat. 12: 141–147 [DOI] [PubMed] [Google Scholar]
- 47. Verweij P. E., Mellado E., Melchers W. J. 2007. Multiple-triazole-resistant aspergillosis. N. Engl. J. Med. 356: 1481–1483 [DOI] [PubMed] [Google Scholar]
- 48. Walsh T. J., et al. 2007. Treatment of invasive aspergillosis with posaconazole in patients who are refractory to or intolerant of conventional therapy: an externally controlled trial. Clin. Infect. Dis. 44: 2–12 [DOI] [PubMed] [Google Scholar]
- 49. Wurch T., Lestienne F., Pauwels P. J. 1998. A modified overlap extension PCR method to create chimeric genes in the absence of restriction enzymes. Biotechnol. Techniques 12: 653–657 [Google Scholar]
- 50. Xiao L., et al. 2004. Three-dimensional models of wild-type and mutated forms of cytochrome P450 14alpha-sterol demethylases from Aspergillus fumigatus and Candida albicans provide insights into posaconazole binding. Antimicrob. Agents Chemother. 48: 568–574 [DOI] [PMC free article] [PubMed] [Google Scholar]