Abstract
Lysobactor enzymogenes strain OH11 is an emerging biological control agent of fungal and bacterial diseases. We recently completed its genome sequence and found it contains a large number of gene clusters putatively responsible for the biosynthesis of nonribosomal peptides and polyketides, including the previously identified antifungal dihydromaltophilin (HSAF). One of the gene clusters contains two huge open reading frames, together encoding 12 modules of nonribosomal peptide synthetases (NRPS). Gene disruption of one of the NRPS led to the disappearance of a metabolite produced in the wild type and the elimination of its antibacterial activity. The metabolite and antibacterial activity were also affected by the disruption of some of the flanking genes. We subsequently isolated this metabolite and subjected it to spectroscopic analysis. The mass spectrometry and nuclear magnetic resonance data showed that its chemical structure is identical to WAP-8294A2, a cyclic lipodepsipeptide with potent anti-methicillin-resistant Staphylococcus aureus (MRSA) activity and currently in phase I/II clinical trials. The WAP-8294A2 biosynthetic genes had not been described previously. So far, the Gram-positive Streptomyces have been the primary source of anti-infectives. Lysobacter are Gram-negative soil/water bacteria that are genetically amendable and have not been well exploited. The WAP-8294A2 synthetase represents one of the largest NRPS complexes, consisting of 45 functional domains. The identification of these genes sets the foundation for the study of the WAP-8294A2 biosynthetic mechanism and opens the door for producing new anti-MRSA antibiotics through biosynthetic engineering in this new source of Lysobacter.
INTRODUCTION
The discovery of new anti-infectives is a pressing and continual need for human health due to the constant emergence of drug-resistant pathogens (50, 51). Natural products (NPs) have been a very prolific source of anti-infective agents. Traditionally, soil bacteria, especially the Gram-positive Streptomyces, have been the primary source for anti-infectives. However, many “user-friendly,” prolific NP producers, such as the Gram-negative soil/water bacteria Lysobacter spp., have not been well explored.
Lysobacter belongs to the Xanthomonadaceae family within the Gammaproteobacteria (10, 48). They are ubiquitous inhabitants of soil and water. These Gram-negative gliding bacteria have drawn increased attention for their ecological diversity, industrial applications and, most notably, unique biotic interactions. Lysobacter species display a number of unique traits that distinguish them from other taxonomically and ecologically related microbes (10, 48). These include a high-GC genome (65 to 72%), gliding motility but without flagella, and prolific production of extracellular lytic enzymes. Within the genus, Lysobacter enzymogenes is the best-known species (54). We have been using L. enzymogenes strains C3 and OH11 as new sources for bioactive NP discovery and as new biocontrol agents of crop diseases (29–31, 33, 39, 52). Strain C3 was originally isolated from grass foliage and exhibits field efficacy against a wide range of fungal pathogens (14, 25, 26, 53, 55, 57). Strain OH11 was isolated from the rhizosphere soil of pepper plants (39). Both strains are emerging as novel agricultural biocontrol agents with a broad spectrum of antifungal and antibacterial activities.
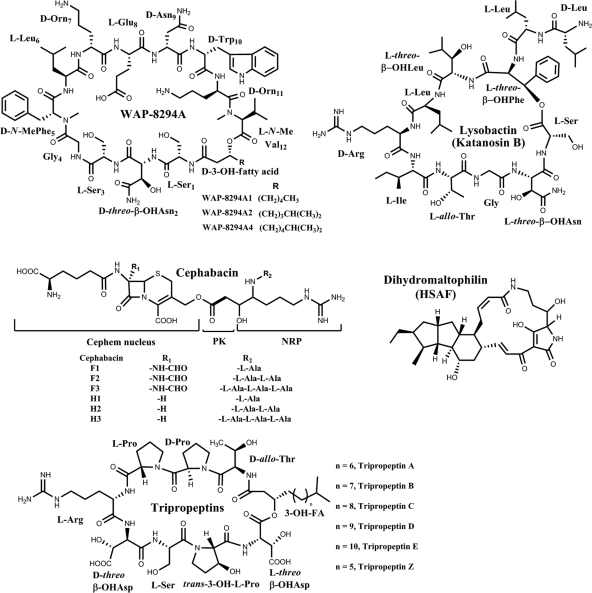
Producing bioactive small-molecule NPs is a characteristic of the biocontrol strains. Shown in Fig. 1 are examples of NPs isolated from various Lysobacter species. Lysobactin (katanosin B) was isolated from Lysobacter sp. SC14067 (7, 20, 37); tripropeptins were isolated from Lysobacter sp. BMK-333-48F3 (18, 19). Both lysobactin and tripropeptins are cyclic peptides with strong anti-methicillin-resistant Staphylococcus aureus (MRSA) and anti-vancomycin-resistant Enterococcus (VRE) activities. Cephabacins have been isolated from Lysobacter lactamgenus (17, 36, 45). These compounds belong to the cephem-type β-lactams, composed of a cephem nucleus, an acetate unit, and an oligopeptide side chain. Dihydromaltophilin (HSAF) was isolated from Streptomyces sp. (15) and L. enzymogenes (29–31, 33, 52). This compound belongs to polycyclic tetramate macrolactams, which are emerging as a new class of NPs with distinct structures and a new mode of action (6, 8). Dihydromaltophilin exhibits potent inhibitory activities against a wide range of fungi and shows a novel mode of action, as it disrupts the biosynthesis of sphingolipids (30). WAP-8294As are cyclic depsipeptides originally isolated from Lysobacter staphylocidin (16, 22–24). They are potent anti-MRSA compounds (50% effetive dose [ED50]14 times more active than vancomycin) and, among them, WAP-8294A2 is in phase I/II clinical studies by aRigen Pharmaceuticals (1). However, the WAP-8294A2 biosynthetic genes have not been described previously, and the molecular mechanism by which Lysobacter synthesizes WAP-8294A2 remains unclear.
Fig. 1.
Chemical structures of representative bioactive natural products isolated from various species of Lysobacter.
We obtained the genome sequence of L. enzymogenes OH11. The genome contains a large number of genes that are predicted for NP biosynthesis. Among them, at least nine clusters contain polyketide synthase and nonribosomal peptide synthetase (PKS-NRPS) genes, including the HSAF cluster (33, 52). The products of the remaining clusters are not clear. Here, we report the identification and characterization of the WAP-8294A2 biosynthetic gene cluster from L. enzymogenes OH11.
MATERIALS AND METHODS
Bacterial strains, plasmids, and general DNA manipulations.
Escherichia coli DH5α strain was used as the host for general DNA propagation. E. coli S17-1 was used as the conjugal strain. L. enzymogenes and other bacterial strains were grown in Luria-Bertani (LB) broth medium or 1/10-strength tryptic soy broth (TSB; Sigma). Genomic DNA of L. enzymogenes was prepared as previously described (25). The pGEM-zf series from Promega (Madison, WI) and pANT841 (41) were used for cloning and sequencing. Plasmid preparation and DNA gel extraction were carried out using kits from Qiagen. PCR primers were synthesized by Integrated DNA Technologies (Coralville, IA). All other manipulations were performed according to standard methods (44).
Generation of gene disruption mutants.
To construct the plasmid vectors for gene disruption, an internal fragment was amplified from each of the open reading frames (ORFs) by using the primer pairs described in the supplemental material. Genomic DNA from the wild-type L. enzymogenes OH11 served as the PCR template. Primers waps1-F and waps1-R were used to amplify a 442-bp fragment from WAPS1 (ORF3, encoding the 7-module NRPS). The fragment was digested with XhoI/BamHI and then cloned into the conjugation vector pJQ200SK (40) to produce pJQ200SK-waps1. Primers orf4-F and orf4-R were used to amplify a 437-bp fragment from ORF4 (taurine catabolism dioxygenase). Primers orf8-F and orf8-R were used to amplify a 575-bp fragment from ORF8 (TonB-dependent receptor). Both fragments were digested with BamHI/XbaI and then cloned into the conjugation vector pJQ200SK to produce pJQ200SK-orf4 and pJQ200SK-orf8, respectively. Each of the pJQ200SK constructs was transformed into E. coli S17-1, which was mated with L. enzymogenes OH11 for conjugal transfer of the vectors. The positive colonies grown on LB plates containing gentamicin (20 μg/ml) were picked and inoculated into liquid cultures containing gentamicin. Genomic DNA was prepared from each of the cultures, and diagnostic PCR was performed to identify mutants that resulted from a homologous recombination. To screen the WAPS1 gene disruption mutants, PCR was performed using the primers waps1-V-F and pJQ-R, which amplified a 584-bp fragment when mutants resulted from a homologous recombination, but not from the wild type or mutants that resulted from a random insertion of the construct (see Fig. S1A in the supplemental material). To screen the ORF4 gene disruption mutants, PCR was performed using the primers orf4-V-F and pJQ-R, which amplified an 826-bp fragment when mutants resulted from a homologous recombination (see Fig. S1B). To screen the ORF8 mutants, primers orf8-V-F and pJQ-R were used to amplify a 1,074-bp fragment from homologous recombination mutants (see Fig. S1C). Because the signal was weak, the mutants were further checked using primers orf8-F and pJQ-R to amplify the 654-bp product (see Fig. S1C).
Production and analysis of the metabolites in mutants.
OH11 or its mutant was grown in 1/10 TSB for 1 day, and an aliquot of 2 ml was transferred to a 250-ml flask containing 50 ml of fermentation medium (2.5% glucose, 2% soybean flour, 0.4% soybean oil, 0.25% NaCl, 0.5% CaCO3; pH 7.2). The culture was incubated at 28°C for 3 days with shaking at 200 rpm. To extract the metabolites, the 50-ml broth culture was collected, and the pH was adjusted to 2.5 with 1 N HCl. The supernatant was extracted with n-butanol–ethyl acetate (1/1, vol/vol) containing 0.05% trifluoroacetic acid (TFA). The organic phase was dried with a rotavapor (R-200; Buchi) to obtain the crude extract. The extract was dissolved in 2 ml methanol containing 0.05% TFA. A 50-μl aliquot of each extract was analyzed by high-pressure liquid chromatography (HPLC; ProStar 210; Varian) using a reverse-phase column (Alltima C18LL; 5 μm; 4.6 mm by 250 mm). Water–0.025% TFA (solvent A) and acetonitrile–0.025% TFA (solvent B) were used as the mobile phases with a flow rate of 1.0 ml/min. The HPLC program was as follows: 5 to 25% solvent B in solvent A for the first 0 to 10 min, with an increase to 80% B at 25 min, to 100% at 26 min, and back to 5% B at 30 min. The metabolites were detected at 220 nm on a UV detector (ProStar 310; Varian).
Antibacterial activity test.
Bacillus subtilis was used as the indicator strain for anti-Gram-positive bacterial activity of various strains of Lysobacter. B. subtilis was mixed with LB medium (20 ml) and poured into a plate. After solidification of the agar plate, sterilized stainless steel tubes were put on the surface of the LB plate, and aliquots (50 μl) of various Lysobacter extracts were added into each of the tubes. The plates were incubated at 37°C overnight until clear inhibition zones appeared.
Preparation of WAP-8294A2 and MS and NMR analysis.
To prepare WAP-8294A2, the fermentation broth (8 liters) was centrifuged, and the cell mass was discarded. The supernatant was collected, evaporated in vacuo to 1 liter, and partitioned with ethyl acetate (EtOAc) until the EtOAc layer was colorless. The aqueous phase was partitioned with n-butanol as TFA was added to the solvent system at 0.005 M. The n-butanol layer was concentrated, and a crude extract was precipitated by addition of acetone. The precipitate was collected through filtration and dissolved in methanol (MeOH) to obtain MeOH extract (15 g). The MeOH extract (15 g) was divided into two parts and subjected to medium-pressure LC (MPLC; 80 g RP-18 silica gel; 60% and 100% MeOH containing 0.005 M TFA; 800 ml for each gradient) to obtain two fractions (a and b). Fraction a (440 mg) was separated by column chromatography (CC) on Sephadex LH-20 (in MeOH containing 0.005 M TFA) to obtain fraction a1 (58.3 mg). Fraction b (1.5 g) was subjected to CC on Sephadex LH-20 (in MeOH containing 0.005 M TFA) and further fractionated on MPLC (30 g RP-18 silica gel; 60% and 100% MeOH containing 0.005 M TFA; 300 ml for each gradient) to afford fraction b1. Fraction a1 and fraction b1 were combined and applied to CC (100 g macroporous resin D101; 70%, 100% MeOH, and 100% MeOH containing 0.005 M TFA) to obtain fraction ab1. Fraction ab1 (32.6 mg) was further purified on a semipreparative HPLC column of ZORBAX Eclipse XDB-C18 (9.4 by 250 nm; Agilent). The mobile phases were water (phase A) and MeOH (phase B) containing 0.005 M TFA, respectively, with a gradient of 70% mobile phase B in mobile phase A in the first 3 min, 70% to 85% mobile B in mobile A from 3 to 16 min, 85% to 70% mobile B in mobile A from 16 to 17 min, and 70% mobile B in mobile A from 17 to 20 min. The flow rate was 4 ml/min. The peaks were detected at 280 nm on a UV-visible detector. The peak with a retention time of 12 min was collected and dried to obtain WAP-8294A2 (6 mg). To analyze the structure of the compound, high-resolution quadrupole time of flight mass spectrometry (HR-Q-TOF-MS) data were acquired by using Agilent Q-TOF 6520 mass spectrometer. Nuclear magnetic resonance (NMR) spectra (1H and 13C NMR and heteronuclear single quantum correlation [HSQC]) were recorded on a Bruker DRX-600 spectrometer, at 600/150 MHz, respectively, in DMSO-d6, δ in ppm relative to Me4Si.
RESULTS
The 12-module NRPS genes.
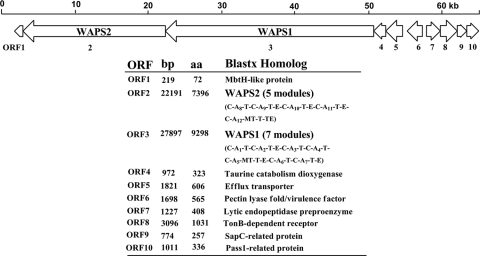
We initially found a strong antifungal activity in L. enzymogenes and subsequently identified the antifungal dihydromaltophilin (HSAF) and its biosynthetic genes (33, 52). During this process, we noticed that strain OH11 also has a potent activity against bacteria. To determine the component(s) responsible for this antibacterial activity, we first analyzed the recently completed genome draft (5.6 Mb, putatively for 4,043 genes, with a GC content of 70.03%) of L. enzymogenes OH11. Our focus was on large gene clusters in the genome that presumably are dedicated to complex natural product (NP) biosynthesis. This revealed at least nine gene clusters, including those for multimodular polyketide synthase (PKS) and nonribosomal peptide synthetase (NRPS). Among them, a cluster (designated WAPS; accession number JN596952) appeared particularly interesting because it hosts two huge NRPSs that are next to each other. One NRPS (WAPS1) contains 7 modules, and the other (WAPS2) contains 5 modules (Fig. 2). Together, there are a total of 45 domains that make up the 12 NRPS modules, which represent one of the largest NRPS complexes. The complex is rich in epimerase (E) domains, implying that its product(s) is rich in d-amino acids. Additionally, the cluster hosts genes predicted to encode enzymes putatively involved in tailoring, resistance, and regulation (Fig. 2).
Fig. 2.
Map of the 63,187-bp WAPS gene cluster from L. enzymogenes OH11. This gene cluster contains two giant NRPSs (WAPS2 and WAPS1), made up of a total of 12 modules. The domain compositions of the NRPSs are indicated. Abbreviations: A, adenylation domain, C, condensation domain; E, epimerase domain; T, thiolation (PCP) domain; TE, thioesterase domain; MT, methyltransferase domain.
Antibacterial activity and metabolites in NRPS disruption mutants.
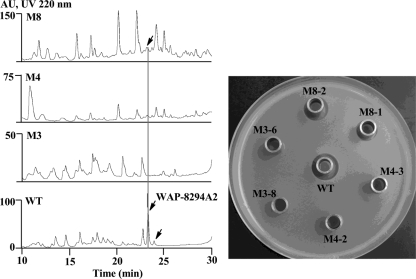
To find out the product(s) synthesized by this large NRPS cluster, we first disrupted ORF3, which encodes the 7-module WAPS1. The first adenylation domain of this NRPS was specifically disrupted by homologous recombination. The resulting mutants (M3-6 and M3-8 are shown) did not have any obvious differences from the wild type except for the loss of the inhibitory activity against B. subtilis (Fig. 3). This suggests that the WAPS cluster is likely to be a major contributor, if not the only one, to the activity against Gram-positive bacteria. We then set out to identify the compound(s) that is responsible for this activity. The total metabolites from six independent mutant strains were compared to that from the wild type by HPLC. All strains showed the absence of a major peak (retention time, 23.27 min), as well as a few minor peaks, compared to the metabolite profile of the wild type (Fig. 3). We then purified this major metabolite and found that it indeed had anti-Bacillus activity.
Fig. 3.
HPLC analysis and antibacterial activity test of the gene disruption strains. M8, mutant of ORF8; M4, mutant of ORF4; M3, mutant of ORF3; WT, wild type. The arrows indicate WAP-8294A2 or a putative analog. In the activity test, Bacillus subtilis was used as the testing organism, and two independent strains of each of the mutants were tested.
Chemical structure of the main active compound.
We prepared ∼6 mg of the compound from 8 liters of OH11 culture. The compound was isolated as a white powder. HR-electrospray ionization (ESI)-MS gave an m/z of 1,562.8255 for [M(C73H111N17O21) + H]+ (calculated, 1,562.8219). Its 1H and 13C NMR data are summarized in Table 1 (1H and 13C NMR spectra and selected HSQC correlations are provided in the supplemental material). A comparison of these data to those in the literature readily revealed that this compound is WAP-8294A2, a cyclic lipodepsipeptide originally isolated from L. staphylocidin (16, 22–24). WAP-8294A2 consists of 12 amino acid residues and a fatty acid side chain (3-hydroxy-7-methyloctanoic acid) (Fig. 1). The amino group of the first amino acid (l-Ser1) is capped with the fatty acid side chain by forming an amide bond with the carboxyl group of the fatty acid. The carboxyl group of the last amino acid (N-Me-Val12) is condensed with the 3-hydroxy group of the fatty acid side chain to form the only ester bond of the cyclic depsipeptide. Six of the 12 amino acids are in a d-configuration, the α-amino nitrogens of three amino acids are methylated, and both Asn2 and the fatty acid side chain contain a β-hydroxyl group. The results demonstrated that the WAPS cluster is responsible for the biosynthesis of WAP-8294A2. Since the disruption of the WASP1 gene also led to the disappearance of at least one other compound with similar retention time as WAP-8294A2, it is likely that this gene cluster is also responsible for other members in the WAP-8294A family that differ only in the acyl side chain length (Fig. 1) (16, 22–24).
Table 1.
1H and 13C NMR spectroscopic data for WAP-8294A2 measured in DMSO-d6 at 600 and 150 MHz, respectively
| Structural moiety | Position(s) | δH (ppm) | δC (ppm) |
|---|---|---|---|
| 3-OH-7-Me-octanoic acid | CO | 169.3 | |
| C-2 | 2.31, 2.56 | 38.1 | |
| C-3 | 4.92 | 70.9 | |
| C-4 | 1.50 | 33.0 | |
| C-5 | 1.22 | 21.9 | |
| C-6 | 1.12 | 38.0 | |
| C-7 | 1.48 | 27.2 | |
| C-8, C-8′ | 0.82 | 22.2, 22.3 | |
| Ser1 | CO | 169.5 | |
| C-α | 4.85 | 53.8 | |
| C-β | 3.47, 3.54 | 62.8 | |
| NH | 7.96 | ||
| OHAsn | CO | 169.1 | |
| C-α | 4.98 | 54.5 | |
| C-β | 4.24 | 71.2 | |
| CO-γ | 173.5 | ||
| NH-γ | 7.38 | ||
| NH-γ′ | 7.44 | ||
| NH | 8.10 | ||
| Ser2 | CO | 169.8 | |
| C-α | 4.85 | 54.7 | |
| C-β | 3.62 | 62.0 | |
| NH | 8.34 | ||
| Gly | CO | 169.0 | |
| C-α | 3.87 | 42.1 | |
| NH | 8.23 | ||
| NMePhe | CO | 169.4 | |
| C-α | 4.42 | 63.5 | |
| C-β | 2.94, 3.26 | 33.8 | |
| C-1 | 138.2 | ||
| C-2, C-6 | 7.16 | 129.0 | |
| C-3, C-5 | 7.27 | 128.3 | |
| C-4 | 7.21 | 126.3 | |
| N-Me | 2.58 | 35.3 | |
| Leu | CO | 171.5 | |
| C-α | 4.24 | 51.2 | |
| C-β | 1.36, 1.52 | 39.2a | |
| C-γ | 1.38 | 24.0 | |
| C-δ | 0.65 | 20.7 | |
| C-δ′ | 0.69 | 22.8 | |
| NH | 7.96 | ||
| Orn1 | CO | 170.9 | |
| C-α | 4.65 | 50.6 | |
| C-β | 1.66 | 30.2 | |
| C-γ | 1.55 | 22.9 | |
| C-δ | 2.84 | 38.4 | |
| NH2 | 7.77 | ||
| NH | 7.49 | ||
| Glu | CO | 169.8 | |
| C-α | 4.61 | 51.2 | |
| C-β | 1.66, 1.80 | 28.5 | |
| C-γ | 2.18 | 29.6 | |
| CO-δ | 174.0 | ||
| NH | 8.55 | ||
| Asn | CO | 170.4 | |
| C-α | 4.78 | 49.1 | |
| C-β | 2.42, 2.61 | 38.0 | |
| C-γ | 171.8 | ||
| NH-γ | 6.95 | ||
| NH-γ′ | 7.35 | ||
| NH | 8.21 | ||
| Trp | CO | 170.6 | |
| C-α | 4.72 | 52.6 | |
| C-β | 2.78, 3.12 | 27.3 | |
| C-2 | 7.14 | 123.2 | |
| C-3 | 109.2 | ||
| C-4 | 7.46 | 118.0 | |
| C-5 | 6.87 | 117.9 | |
| C-6 | 6.98 | 120.6 | |
| C-7 | 7.31 | 111.3 | |
| C-8 | 127.1 | ||
| C-9 | 135.7 | ||
| NH-1 | 10.27 | ||
| NH | 8.80 | ||
| Orn2 | CO | 170.1 | |
| Cα | 4.56 | 48.2 | |
| Cβ | 1.55, 1.66 | 28.0 | |
| Cγ | 1.46 | 23.1 | |
| Cδ | 2.78 | 38.6 | |
| NH2 | 7.77 | ||
| NH | 8.00 | ||
| NMeVal | CO | 169.0 | |
| C-α | 4.60 | 60.8 | |
| C-β | 2.06 | 25.4 | |
| C-γ | 0.69 | 17.9 | |
| C-γ′ | 0.86 | 19.0 | |
| N-Me | 2.52 | 29.0 |
Overlapped with the carbon signal of dimethyl sulfoxide (DMSO)-d6 and was recognized by assignment of HSQC correlations.
Predicted substrate specificity of NRPS adenylation domains.
The substrate specificity of the adenylation (A) domains of the 12 NRPS modules were predicted based on sequence alignments of the 10-amino-acid “nonribosomal peptide codes” defined by Stachelhaus et al. (42, 46). Table S1 in the supplemental material shows the predicted “nonribosomal peptide codes” from the A domains of the 7-module WAPS1, which would activate and incorporate Ser1, Asn2, Ser3, Gly4, Phe5, Leu6, and Orn7, and of the 5-module WAPS2, which would activate and incorporate Glu8, Asn9, Trp10, Orn11, and Val12. The results were in good agreement with the amino acid sequence in the chemical structure of WAP-8492A2, suggesting a colinear assembly line of the modular template for peptide biosynthesis (34).
The WAPS gene cluster.
Having identified the product of the WAPS gene, we then tried to define the gene cluster required for WAP-8294A2 biosynthesis. Immediately upstream of the 5-module WAPS2 is an MbtH-like protein (ORF1). This is a small protein known to be associated with certain NRPS and required for biosynthesis (13, 56). Interestingly, there is no apparent ORF in the 4.3-kb upstream region of ORF1. The further upstream region contains ORFs predicted to encode a putative DNA mismatch repair protein, endonuclease subunit, and other products that do not appear to be relevant to WAP-8294A2 biosynthesis. Downstream of the 7-module WAPS1 are a series of tightly clustered ORFs (Fig. 2). ORF4 is homologous to taurine catabolism dioxygenase, which is a nonheme iron, 2-ketoglutarate-dependent oxygenase (49). We subsequently disrupted this gene and found that the mutants lost antibacterial activity as well as the WAP-8294A2 peak on HPLC (Fig. 3). The result showed that ORF4 is essential for WAP-8294A2 biosynthesis. WAP-8294A2 contains two hydroxyl groups, one on β-hydroxy Asn2 and the other on 3-hydroxy fatty acyl chain. This dioxygenase is likely involved in one or both of the hydroxylations. ORF5 is homologous to efflux transporters. The ORF5 disruption mutant still exhibited the antibacterial activity and produced WAP-8294A2 (data not shown). ORF6 is homologous to pectin lyase fold/virulence factor, which is related to peptidases. The disruption of this gene led to the loss of antibacterial activity and the disappearance of the WAP-8294A2 peak on HPLC (data not shown). ORF7 is homologous to lytic endopeptidase preproenzyme, which is related to a family of bacterial extracellular metalloendopeptidases. ORF5, -6, and -7 may be involved in self-protection. ORF8 is homologous to the TonB-dependent receptor, part of a family of outer membrane β-barrel proteins that sense extracellular signals (35). The disruption of this gene led to a partial loss of the antibacterial activity and a reduced WAP-8294A2 peak on HPLC (Fig. 3). The result suggests that ORF8 is involved but not essential for WAP-8294A2 production, consisting with the predicted function of this gene as a regulator. Interestingly, the WAP-8294A2 peak was significantly smaller than that in the wild type, while the antibacterial activity was only partly reduced. One possibility is that the inactivation of this receptor gene could have led to activation of biosynthesis of other antibacterial metabolites, as evidenced by HPLC with multiple new peaks that resulted from this mutation. ORF9 is homologous to SapC-related protein, which is related to peptide transport (38), and ORF10 is homologous to Pass1-related protein (32). These two ORFs and ORF5 may be involved in WAP-8294A transport. It is notable that the WAPS gene cluster contains an unusually high number of resistance and transport-related genes. The downstream region of ORF10 contains ORFs homologous to cellulase, glucokinase, and other sugar-related products, which do not appear to be relevant to WAP-8294A biosynthesis. Together, the data suggest that the region between ORF1 and ORF10 is the WAPS gene cluster.
All WAP-8294A compounds contain a 3-hydroxy fatty acyl chain. However, there is not a candidate for acyl coenzyme A (CoA) ligase (ACL) present in this gene cluster or its flanking regions. Such an enzyme is needed to activate and introduce the 3-hydroxy fatty acid chain into the peptide. The activated acyl-S-CoA can directly serve as substrate to be incorporated into the peptide (21) or is transferred to a dedicated acyl carrier protein (ACP) as acyl-S-ACP, which is then incorporated into the peptide (4). To test if the WAP assembly could use an ACL in other metabolic pathways, we disrupted an ACL-encoding ORF (accession number JN596953) present in one of the uncharacterized PKS/NRPS gene clusters found in the genome of OH11. The mutants showed a partial loss of the antibacterial activity and produced approximately 50% of the WAP-8492A yield of the wild type (data not shown). The results suggested that the WAP-8492A biosynthetic pathway likely recruits an ACL from other pathways. This type of in trans ACL generally exhibits a broad specificity and is functionally redundant (2), which is consistent with the fact that all WAP-8492A compounds have the same amino acid composition but vary in the length of the fatty acid side chain (16, 22–24).
DISCUSSION
A huge number of gene clusters discovered by genome sequencing have been predicted for secondary metabolite biosynthesis in numerous microbes. In many cases, the actual products of the gene clusters remain unclear. In this work, we identified the product of a previously uncharacterized NRPS gene cluster from the recently completed genome sequence of the biocontrol agent L. enzymogenes OH11. This strain exhibited a strong antibacterial activity, but the active component(s) for this activity was not known. Through genome mining, target-specific gene disruption, metabolite isolation, and structural elucidation, we were able to link this new NRPS gene cluster to a group of potent anti-MRSA antibiotics, the WAP-8294A family. Although these compounds have been known for many years and WAP-8294A2 is currently in clinical studies, their biosynthetic genes and mechanism have not been reported.
The core of the WAPS cluster is the two huge NRPS genes (WAPS1 and WAPS2). A total of 45 domains were identified from the two genes, representing one of the largest NRPS systems. The domains make up the 12 modules responsible for the recognition, activation, thiolation, condensation, and modification of the 12 amino acids found in WAP-8294A2. The domain organization of WAPS1 and WAPS2 reveals a “canonical” modular assembly line. The predicted adenylation (A) domain specificity, based on the “nonribosomal peptide codes” (46), matches well with the amino acid sequence in the structure of WAP-8492A2 (Fig. 1 and 2). In addition, the positions of the epimerase (E) and methytransferase (MT) domains in WAPS1 and WAPS2 are coincident with the occurrence of the d-amino acids and N-methyl amino acids in WAP-8492A2. The 12-module organization contains six E domains, matching the six d-amino acids. There is a perfect “colinearity” between the 12 modules and the 12 amino acid residues of WAP-8294A2. WAPS1 starts with a C domain (type CIII), which is a common feature found in NRPSs for cyclic lipopeptides, such as surfactin (27), daptomycin (3), and calcium-dependent antibiotic (CDA) of Streptomyces coelicolor A3(2) (9). The N terminus of surfactin is also acylated with a 3-hydroxy fatty acid that forms a lactone with the C-terminal carboxyl of the peptide, whereas in daptomycin and CDA the N terminus is acylated with a nonhydroxylated fatty acid and the cyclization takes place between the C-terminal carboxyl and the hydroxyl group of an internal amino acid residue (threonine).
While the daptomycin and CDA gene clusters host a dedicated acyl-CoA ligase and an ACP for the activation and incorporation of the fatty acyl side chain (3, 9), the surfactin gene cluster (27) and the WAPS cluster do not contain an ACL. In the case of surfactin, the B. subtilis genome contains four putative ACL genes. An in vitro study showed that two of the ACLs can catalyze the activation and transfer of the 3-hydroxy fatty acids to the N terminus of the peptide (27). When each of the four ACL genes was disrupted, the yield of surfactin was decreased by 38 to 55%. However, even deletion of all four genes did not abolish surfactin production. Similarly, a disruption of an ACL gene (accession number JN596953) found in the genome of L. enzymogenes OH11 lowered the antibacterial activity and the yield of WAP-8294A2. The data suggest that WAP-8492A2 is biosynthetically more close to surfactin than daptomycin and CDA.
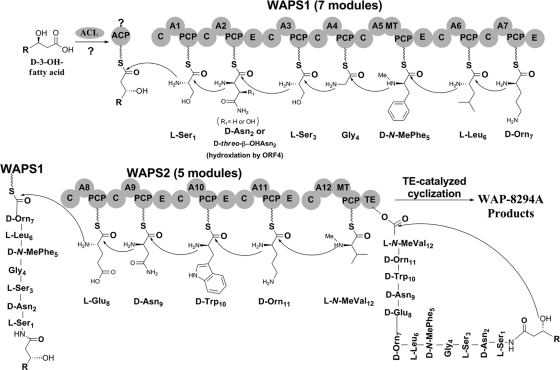
Based on the results, we propose a biosynthetic pathway for WAP-8294A2 (Fig. 4). The pathway starts with the activation of the 3-hydroxy fatty acid by the action of a generic ACL. To incorporate the acyl chain into the peptide, a transferase type of enzyme is required. This is likely to be accomplished by the first C domain of WAPS1. This type of so-called type CIII domain is commonly found in the beginning of the NRPS for cyclic lipopeptides (3, 4, 27). Such an enzyme can either use the fatty acyl-S-CoA directly as substrate (21) or use an intermediate acyl-S-ACP (Fig. 4) (4). The 7 modules of WAPS1 are predicted to activate, respectively, l-Ser1, l-Asn2, l-Ser3, Gly4, l-Phe5, l-Leu6, and l-Orn7. The E domain in the second, fifth, and seventh modules is expected to epimerize the l-amino acid to the corresponding d-form. The MT domain in the fifth module is responsible for adding the methyl group on the amino nitrogen of the phenylalanine residue. The second amino acid residue, Asn2, is also hydroxylated at the β position. This is likely carried out by the 2-ketoglutarate-dependent dioxygenase (ORF4), which is required for WAP-8294A2 production. The timing of this hydroxylation can be prior to or after the condensation between this residue and the following residue (Ser3), while the intermediates are WAPS1 bound. Like WAPS1, WAPS2 also starts with a condensation (C) domain. This C domain is responsible for transferring the heptapeptidyl chain on the last PCP domain of WAPS1 to WAPS2 by catalyzing the condensation between this peptidyl chain and the eighth residue (Glu8) on the first PCP domain of WAPS2. The following three modules all contain an E domain, corresponding to the d-configuration of Asn9, Trp10, and Orn11. The last module of the 5-module WAPS2 contains an MT, which is expected to transfer the methyl group onto the amino nitrogen of the 12th residue (Val12) of WAP-8294A2. Finally, the fatty acyl-capped dodecapeptidyl chain is transferred to the serine active site of the thioesterase (TE) domain located at the end of WAPS2. This TE is predicted to catalyze the intramolecular macrocyclization by using the 3-hydroxy group of the fatty acyl chain as the nucleophile to attack the carbonyl carbon of the peptidyl-O-TE (Fig. 4) (12). This would release the lipopeptide as a lactone. The TE domain is expected to have a relax substrate specificity for the length of the fatty acyl chain, since WAP-8294A analogs differ mainly in this side chain length. The relaxed substrate specificity may provide an opportunity to generate new WAP-8294A analogs through precursor feeding or overexpression of an ACL that activates a particular type of acyl chains.
Fig. 4.
Proposed biosynthetic pathway for WAP-8294A2 products from L. enzymogenes.
A number of cyclic lipopeptides, including surfactin, daptomcyin, and CDA, have been extensively studied (3, 4, 27, 43, 47). So far, daptomycin is the only one that has been incorporated into the market (Cubicin). In light of the enormous success of daptomcyin and the clinical trials of WAP-8294A2, the identification and characterization of the WAP-8492A2 biosynthetic gene cluster will set the foundation for future genetic engineering to produce new “surfactin-type” anti-MRSA antibiotics. One particularly attractive point of this gene cluster is the producer organism, Lysobacter, which is a ubiquitous soil/water microorganism but has not been well exploited. Lysobacter is a Gram-negative gliding bacterium distinct from Streptomyces. The majority of the currently in-use or under-study cyclic peptides (including daptomcyin) or polyketide anti-infectives were isolated from Streptomyces or other Gram-positive bacteria. Lysobacter is fast-growing, simple to use and maintain, and readily accessible to genetic tools, with capacities for posttranslational modifications and availability of the substrate pool for polyketides and nonribosomal peptide biosynthesis. Furthermore, Lysobacter is known as a prolific producer of bioactive compounds, such as cephabacins (11, 28), HSAF (33, 52), lysobactin (5, 7, 20, 37), and tripropeptins (18, 19). So far, none of these pathways has been genetically manipulated for new antibiotic production. The availability of the WAP-8294A2 biosynthetic genes, the development of the genetic system for L. enzymogenes, and an understanding of the biosynthetic mechanism will open a new area of antibiotic research of the Gram-negative producers.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by NSFC (31028019), the NIH (AI073510), Nebraska Research Council, and National High Technology Research and Development Program of China (2011AA10A210). W.Z. (Xiamen University) and Y.W. (Shandong University) were supported by a scholarship from the China Scholarship Council. Y.L. was supported by the Postdoctoral Innovation Foundation of Shandong Province (201003070).
We thank Ron Cerny and Kurt Wulser, Department of Chemistry, University of Nebraska—Lincoln, for technical assistance.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 19 September 2011.
REFERENCES
- 1. aRigen-Pharmaceuticals 2009. WAP-8294A2, a first-line anti-MRSA product candidate, funded by NEDO (New Energy and Industrial Technology Development Organization) in Japan. Press release. http://www.evaluatepharma.com/Universal/View.aspx?type=Story&id=192830
- 2. Arora P., Vats A., Saxena P., Mohanty D., Gokhale R. S. 2005. Promiscuous fatty acyl CoA ligases produce acyl-CoA and acyl-SNAC precursors for polyketide biosynthesis. J. Am. Chem. Soc. 127: 9388–9389 [DOI] [PubMed] [Google Scholar]
- 3. Baltz R. H. 2009. Daptomycin: mechanisms of action and resistance, and biosynthetic engineering. Curr. Opin. Chem. Biol. 13: 144–151 [DOI] [PubMed] [Google Scholar]
- 4. Baltz R. H., Miao V., Wrigley S. K. 2005. Natural products to drugs: daptomycin and related lipopeptide antibiotics. Nat. Prod. Rep. 22: 717–741 [DOI] [PubMed] [Google Scholar]
- 5. Bernhard F., Demel G., Soltani K., Dohren H. V., Blinov V. 1996. Identification of genes encoding for peptide synthetases in the gram-negative bacterium Lysobacter sp. ATCC 53042 and the fungus Cylindrotrichum oligospermum. DNA Seq. 6: 319–330 [DOI] [PubMed] [Google Scholar]
- 6. Blodgett J. A., et al. 2010. Common biosynthetic origins for polycyclic tetramate macrolactams from phylogenetically diverse bacteria. Proc. Natl. Acad. Sci. U. S. A. 107: 11692–16927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bonner D. P., O'Sullivan J., Tanaka S. K., Clark J. M., Whitney R. R. 1988. Lysobactin, a novel antibacterial agent produced by Lysobacter sp. II. Biological properties. J. Antibiot. (Tokyo) 41: 1745–1751 [DOI] [PubMed] [Google Scholar]
- 8. Cao S., Blodgett J. A., Clardy J. 2010. Targeted discovery of polycyclic tetramate macrolactams from an environmental Streptomyces strain. Org. Lett. 12: 4652–4654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chong P. P., et al. 1998. Physical identification of a chromosomal locus encoding biosynthetic genes for the lipopeptide calcium-dependent antibiotic (CDA) of Streptomyces coelicolor A3(2). Microbiology 144: 193–199 [DOI] [PubMed] [Google Scholar]
- 10. Christensen P., Cook F. D. 1978. Lysobacter, a New genus of non-fruiting, gliding bacteria with a high base ratio. Int. J. Syst. Bacteriol. 28: 367–393 [Google Scholar]
- 11. Demirev A. V., Lee C. H., Jaishy B. P., Nam D. H., Ryu D. D. 2006. Substrate specificity of nonribosomal peptide synthetase modules responsible for the biosynthesis of the oligopeptide moiety of cephabacin in Lysobacter lactamgenus. FEMS Microbiol. Lett. 255: 121–128 [DOI] [PubMed] [Google Scholar]
- 12. Du L., Lou L. 2010. PKS and NRPS release mechanisms. Nat. Prod. Rep. 27: 255–278 [DOI] [PubMed] [Google Scholar]
- 13. Felnagle E. A., et al. 2010. MbtH-like proteins as integral components of bacterial nonribosomal peptide synthetases. Biochemistry 49: 8815–8817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Giesler L. J., Yuen G. Y. 1998. Evaluation of Stenotrophomonas maltophilia strain C3 for biocontrol of brown patch disease. Crop Protect. 17: 509–513 [Google Scholar]
- 15. Graupner P. R., et al. 1997. Dihydromaltophilin: a novel fungicidal tetramic acid-containing metabolite from Streptomyces sp. J. Antibiot. (Tokyo) 50: 1014–1019 [DOI] [PubMed] [Google Scholar]
- 16. Harad K. I., et al. 2001. Separation of WAP-8294A components, a novel anti-methicillin-resistant staphylococcus aureus antibiotic, using high-speed counter-current chromatography. J. Chromatogr. A 932: 75–81 [DOI] [PubMed] [Google Scholar]
- 17. Harada S., Tsubotani S., Ono H., Okazaki H. 1984. Cephabacins, new cephem antibiotics of bacterial origin. II. Isolation and characterization. J. Antibiot. (Tokyo) 37: 1536–1545 [DOI] [PubMed] [Google Scholar]
- 18. Hashizume H., et al. 2004. Tripropeptins, novel antimicrobial agents produced by Lysobacter sp. J. Antibiot. (Tokyo) 57: 52–58 [DOI] [PubMed] [Google Scholar]
- 19. Hashizume H., et al. 2001. Tripropeptins, novel antimicrobial agents produced by Lysobacter sp. I. Taxonomy, isolation and biological activities. J. Antibiot. (Tokyo) 54: 1054–1059 [DOI] [PubMed] [Google Scholar]
- 20. Hou J., Robbel L., Marahiel M. A. 2011. Identification and characterization of the lysobactin biosynthetic gene cluster reveals mechanistic insights into an unusual termination module architecture. Chem. Biol. 18: 655–664 [DOI] [PubMed] [Google Scholar]
- 21. Imker H. J., Krahn D., Clerc J., Kaiser M., Walsh C. T. 2010. N-acylation during glidobactin biosynthesis by the tridomain nonribosomal peptide synthetase module GlbF. Chem. Biol. 17: 1077–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kato A., Hirata H., Ohashi Y., Fujii K., Mori K., Harada K. A new anti-MRSA antibiotic complex, WAP-8294A. II. Structure characterization of minor components by ESI LCMS and MS/MS. J. Antibiot. (Tokyo) 64: 373–379 [DOI] [PubMed] [Google Scholar]
- 23. Kato A., et al. 1998. A new anti-MRSA antibiotic complex, WAP-8294A. I. Taxonomy, isolation and biological activities. J. Antibiot. (Tokyo) 51: 929–935 [DOI] [PubMed] [Google Scholar]
- 24. Kato A., Nakaya S., Ohashi Y., Hirata H. 1997. WAP-8294A(2), a novel anti-MRSA antibiotic produced by Lysobacter sp. J. Am. Chem. Soc. 119: 6680–6681 [Google Scholar]
- 25. Kobayashi D. Y., Reedy R. M., Palumbo J. D., Zhou J. M., Yuen G. Y. 2005. A clp gene homologue belonging to the Crp gene family globally regulates lytic enzyme production, antimicrobial activity, and biological control activity expressed by Lysobacter enzymogenes strain C3. Appl. Environ. Microbiol. 71: 261–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kobayashi D. Y., Yuen G. Y. 2005. The role of clp-regulated factors in antagonism against Magnaporthe poae and biological control of summer patch disease of Kentucky bluegrass by Lysobacter enzymogenes C3. Can. J. Microbiol. 51: 719–723 [DOI] [PubMed] [Google Scholar]
- 27. Kraas F. I., Helmetag V., Wittmann M., Strieker M., Marahiel M. A. 2010. Functional dissection of surfactin synthetase initiation module reveals insights into the mechanism of lipoinitiation. Chem. Biol. 17: 872–880 [DOI] [PubMed] [Google Scholar]
- 28. Lee J. S., et al. 2008. Expression and characterization of polyketide synthase module involved in the late step of cephabacin biosynthesis from Lysobacter lactamgenus. J. Microbiol. Biotechnol. 18: 427–433 [PubMed] [Google Scholar]
- 29. Li S., Calvo A. M., Yuen G. Y., Du L., Harris S. D. 2009. Induction of cell wall thickening by the antifungal compound HSAF disrupts fungal growth and is mediated by sphingolipid biosynthesis. J. Eukaryot. Microbiol. 56: 182–187 [DOI] [PubMed] [Google Scholar]
- 30. Li S., Du L., Yuen G., Harris S. D. 2006. Distinct ceramide synthases regulate polarized growth in the filamentous fungus Aspergillus nidulans. Mol. Biol. Cell 17: 1218–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li S., et al. 2008. An antibiotic complex from Lysobacter enzymogenes strain C3: antimicrobial activity and role in plant disease control. Phytopathology 98: 695–701 [DOI] [PubMed] [Google Scholar]
- 32. Liu C., Gilmont R. R., Benndorf R., Welsh M. J. 2000. Identification and characterization of a novel protein from Sertoli cells, PASS1, that associates with mammalian small stress protein hsp27. J. Biol. Chem. 275: 18724–18731 [DOI] [PubMed] [Google Scholar]
- 33. Lou L., et al. 2011. Biosynthesis of HSAF, a tetramic acid-containing macrolactam from Lysobacter enzymogenes. J. Am. Chem. Soc. 133: 643–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Marahiel M. A., Stachelhaus T., Mootz H. D. 1997. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 97: 2651–2674 [DOI] [PubMed] [Google Scholar]
- 35. Nau C. D., Konisky J. 1989. Evolutionary relationship between the TonB-dependent outer membrane transport proteins: nucleotide and amino acid sequences of the Escherichia coli colicin I receptor gene. J. Bacteriol. 171: 1041–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ono H., Nozaki Y., Katayama N., Okazaki H. 1984. Cephabacins, new cephem antibiotics of bacterial origin. I. Discovery and taxonomy of the producing organisms and fermentation. J. Antibiot. (Tokyo) 37: 1528–1535 [DOI] [PubMed] [Google Scholar]
- 37. O'Sullivan J., et al. 1988. Lysobactin, a novel antibacterial agent produced by Lysobacter sp. I. Taxonomy, isolation and partial characterization. J. Antibiot. (Tokyo) 41: 1740–1744 [DOI] [PubMed] [Google Scholar]
- 38. Parra-Lopez C., Baer M. T., Groisman E. A. 1993. Molecular genetic analysis of a locus required for resistance to antimicrobial peptides in Salmonella typhimurium. EMBO J. 12: 4053–4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Qian G. L., Hu B. S., Jiang Y. H., Liu F. Q. 2009. Identification and characterization of Lysobacter enzymogenes as a biological control agent against some fungal pathogens. Agric. Sci. China 8: 68–75 [Google Scholar]
- 40. Quandt J., Hynes M. F. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127: 15–21 [DOI] [PubMed] [Google Scholar]
- 41. Rajgarhia V. B., Strohl W. R. 1997. Minimal Streptomyces sp. strain C5 daunorubicin polyketide biosynthesis genes required for aklanonic acid biosynthesis. J. Bacteriol. 179: 2690–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rausch C., Weber T., Kohlbacher O., Wohlleben W., Huson D. H. 2005. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs). Nucleic Acids Res. 33: 5799–5808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Robbel L., Marahiel M. A. 2010. Daptomycin, a bacterial lipopeptide synthesized by a nonribosomal machinery. J. Biol. Chem. 285: 27501–27508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 45. Sohn Y. S., Nam D. H., Ryu D. D. 2001. Biosynthetic pathway of cephabacins in Lysobacter lactamgenus: molecular and biochemical characterization of the upstream region of the gene clusters for engineering of novel antibiotics. Metab. Eng. 3: 380–392 [DOI] [PubMed] [Google Scholar]
- 46. Stachelhaus T., Mootz H. D., Marahiel M. A. 1999. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6: 493–505 [DOI] [PubMed] [Google Scholar]
- 47. Strieker M., Marahiel M. A. 2009. The structural diversity of acidic lipopeptide antibiotics. ChemBioChem 10: 607–616 [DOI] [PubMed] [Google Scholar]
- 48. Sullivan R. F., Holtman M. A., Zylstra G. J., White J. F., Kobayashi D. Y. 2003. Taxonomic positioning of two biological control agents for plant diseases as Lysobacter enzymogenes based on phylogenetic analysis of 16S rDNA, fatty acid composition and phenotypic characteristics. J. Appl. Microbiol. 94: 1079–1086 [DOI] [PubMed] [Google Scholar]
- 49. Ueki I., et al. 21 June 2011, posting date Knockout of the murine cysteine dioxygenase gene results in severe impairment in ability to synthesize taurine and an increased catabolism of cysteine to hydrogen sulfide. Am. J. Physiol. Endocrinol. Metab. 301: E668–E684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Walsh C. T. 2003. Antibiotics: actions, origins, resistance. ASM Press, Washington, DC [Google Scholar]
- 51. Walsh C. T. 2003. Where will new antibiotics come from. Nat. Rev. Microbiol. 1: 65–70 [DOI] [PubMed] [Google Scholar]
- 52. Yu F., et al. 2007. Structure and biosynthesis of heat-stable antifungal factor (HSAF), a broad-spectrum antimycotic with a novel mode of action. Antimicrob. Agents Chemother. 51: 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yuen G. Y., Jochum C. C., Osborne L. E., Jin Y. 2003. Biocontrol of Fusarium head blight in wheat by Lysobacter enzymogenes C3. Phytopathology 93: S93 [Google Scholar]
- 54. Yuen G. Y., Kobayashi D. K., Caswell-Chen E. P. 2006. Ecology and biological control of plant pathogens by Lysobacter enzymogenes. Phytopathlogy 96:S154 [Google Scholar]
- 55. Yuen G. Y., Steadman J. R., Lindgren D. T., Schaff D., Jochum C. C. 2001. Bean rust biological control using bacterial agents. Crop Protect. 20:395–402 [Google Scholar]
- 56. Zhang W., Heemstra J. R., Jr., Walsh C. T., Imker H. J. 2010. Activation of the pacidamycin PacL adenylation domain by MbtH-like proteins. Biochemistry 49: 9946–9947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang Z., Yuen G. Y. 1999. Biological control of Bipolaris sorokiniana on tall fescue by Stenotrophomonas maltophilia C3. Phytopathology 89: 817–822 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.