Abstract
Gallium ions have previously been shown to exhibit antibacterial and antibiofilm properties. In this study, we report differential bactericidal activities of two gallium complexes, gallium desferrioxamine B (Ga-DFOB) and gallium citrate (Ga-Cit). Modeling of gallium speciation in growth medium showed that DFOB and citrate both can prevent precipitation of Ga(OH)3, but some precipitation can occur above pH 7 with citrate. Despite this, Ga-Cit 90% inhibitory concentrations (IC90) were lower than those of Ga-DFOB for clinical isolates of Pseudomonas aeruginosa and several reference strains of other bacterial species. Treatment with Ga compounds mitigated damage inflicted on murine J774 macrophage-like cells infected with P. aeruginosa PAO1. Again, Ga-Cit showed more potent mitigation than did Ga-DFOB. Ga was also taken up more efficiently by P. aeruginosa in the form of Ga-Cit than in the form of Ga-DFOB. Neither Ga-Cit nor Ga-DFOB was toxic to several human cell lines tested, and no proinflammatory activity was detected in human lung epithelial cells after exposure in vitro. Metabolomic analysis was used to delineate the effects of Ga-Cit on the bacterial cell. Exposure to Ga resulted in lower concentrations of glutamate, a key metabolite for P. aeruginosa, and of many amino acids, indicating that Ga affects various biosynthesis pathways. An altered protein expression profile in the presence of Ga-Cit suggested that some compensatory mechanisms were activated in the bacterium. Furthermore, the antibacterial effect of Ga was shown to vary depending on the carbon source, which has importance in the context of medical applications of gallium.
INTRODUCTION
Resistance of pathogenic bacteria to antibiotics is an escalating problem worldwide, and therefore, development of alternative antibacterial drugs is of tremendous relevance. Unlike classical antibiotics, which target a specific reaction or process, metals and metaloorganic compounds often affect several different groups of biomolecules, and therefore, development of resistance is unlikely, which makes these compounds attractive as antibacterial agents (25, 39). While classical antibiotics are less efficient against bacterial biofilms than planktonic cells, there is evidence that this is not the case for metal ions (24). Gallium was recently identified as a potential antibacterial drug and shown to be taken up by both Gram-positive and Gram-negative bacteria (9, 15, 30, 37, 41). It has been hypothesized that bacteria sequester Ga through their iron uptake systems (5), since Ga has been shown to bind to iron siderophores (9, 17). A siderophore is a small secreted iron-binding molecule that together with a siderophore receptor protein is a part of bacterial iron uptake systems. Each siderophore has its own receptor, and bacteria often express receptors for siderophores produced by other species. In several groups of bacteria, a number of siderophores have been well characterized and studied. Pseudomonas aeruginosa secures its iron acquisition by the endogenous siderophores pyoverdine and pyochelin in combination with the ability to utilize exogenous iron chelators like citrate and desferrioxamine B (DFOB), which is a hydroxamate-based siderophore. Proteins involved in uptake of iron citrate and iron DFOB are induced in iron-starved P. aeruginosa (34, 36). Ferric citrate was also shown to be an iron source for Escherichia coli along with the siderophores enterobactin and ferric hydroxamate (18, 46). Staphylococcus aureus uses siderophores such as staphyloferrins A and B and staphylobactin and additionally possesses the capacity for ferric hydroxamate uptake. Staphylococcus epidermidis produces only staphyloferrin B (6, 10).
Recently, it was found that planktonic P. aeruginosa cells are killed by the addition of Ga in micromolar concentrations, and even more importantly, the metal appeared to be especially efficient in reducing biofilms (30). The antibacterial properties of Ga are generally attributed to its ability to substitute for Fe3+ in bacterial metabolism. Unlike Fe, Ga cannot redox cycle, and as a consequence, it blocks multiple processes in bacteria where the shift between reduced and oxidized iron (Fe2+ and Fe3+) is crucial (7, 12). It was suggested that in P. aeruginosa, Ga decreases the uptake of Fe by disrupting Fe signaling through repression of pvdS, which encodes a sigma factor that enables transcription of the genes for pyoverdine synthesis. Gallium's antibiofilm properties are less well understood, though it has been proposed that bacteria in the inner layers of biofilms are more susceptible to Ga due to the Fe limitation in this region (30). Nevertheless, this hypothesis does not explain why subinhibitory concentrations of Ga prevent biofilm formation even during the early stages of attachment (30).
It has been shown that Ga and another Fe mimetic, Al, have severe toxicity for Pseudomonas fluorescens, impeding the functionality of the electron transport chain and enzymes containing Fe-S clusters (12). Chenier et al. found that as a consequence of the pronounced stress in the presence of Ga or Al, P. fluorescens responds with active reorganization of its metabolic pathways to use the tricarboxylic acid (TCA) cycle (12, 32). At present, it is not known whether pathogenic bacteria possess similar mechanisms and how the presence or absence of such mechanisms can affect their virulence. Elucidation of the metabolic effects of Ga in pathogens is a key to understanding the differential toxicity of Ga for a variety of bacterial species. Such an understanding may allow more efficient usage of Ga in the treatment of infectious diseases.
This work illustrates how solution chemistry and the versatility of bacterial metabolic networks can increase or decrease the toxicity of Ga for a pathogenic bacterium. We show that the chemical speciation of Ga influences its uptake by bacteria as well as the outcome of bacterial infections in a mammalian cell model.
MATERIALS AND METHODS
Strains and chemicals.
Gallium citrate (Ga-Cit) was prepared by mixing an aqueous solution of Ga(NO3)3 with Na citrate, and the total concentration of dissolved Ga was verified by atomic absorption spectroscopy. Ga-Cit was mixed with an equimolar aqueous solution of desferrioxamine B (DFOB) to yield Ga-DFOB. Ga-DFOB was always prepared fresh using a stock solution of Ga-Cit to avoid changes in Ga concentration due to precipitation. [If Ga(NO3)3 solution is used as a stock, it needs to have a much lower pH (<2) to maintain Ga ions in solution.] Iso-Sensitest broth was purchased from Oxoid (Cambridge, United Kingdom), and Hussain, Hastings and White (HHW) medium was prepared as described in reference 27 but without iron. All chemicals were purchased from Sigma-Aldrich if not stated otherwise. The strains used in this study (Table 1) were clinical isolates of P. aeruginosa, including cystic fibrosis isolates (40), Staphylococcus epidermidis, including strains obtained from patients with infections associated with catheters and prostheses, methicillin-resistant S. aureus (MRSA), and reference strains of different species known to form biofilms.
Table 1.
Strains and IC90 of Ga-Cit and Ga-DFOB
| Strain | Origin | IC90 (μM) of: |
|||
|---|---|---|---|---|---|
| Ga-Cit (Iso-Sensitest) | Ga-DFOB (Iso-Sensitest) | Ga-Cit (HHW) | Ga-DFOB (HHW) | ||
| Clinical isolates of Pseudomonas aeruginosa | |||||
| 9806/93 | W, NUH | 160 | 640 | 20 | 80 |
| 9791/93 | W, NUH | 80 | 320 | 20 | 40 |
| 9704/93 | W, NUH | 160 | 640 | 40 | 640 |
| 48/94 | W, NUH | 80 | 320 | 20 | 80 |
| 10243/93 | W, NUH | 160 | 640 | 20 | 80 |
| 10031/93 | W, NUH | 160 | 640 | 40 | 80 |
| 9962/93 | C/P, NUH | 80 | 320 | 20 | 40 |
| 9960/93 | C/P, NUH | 80 | 320 | 20 | 40 |
| 10140/93 | W, NUH | 160 | 640 | 20 | 160 |
| 7444/93 | W, NUH | 80 | 160 | 20 | 40 |
| 9870/93 | W, NUH | 160 | 320 | 20 | 40 |
| 8046/93 | W, NUH | 320 | 640 | 40 | 80 |
| 76/94 | C/P, NUH | 160 | 640 | 40 | 80 |
| 71/94 | C/P, NUH | 160 | 1,280 | 40 | 320 |
| 1824 | CF, DH(39) | 640 | 640 | 40 | 80 |
| 1956 | CF, DH(39) | 80 | 160 | 20 | 40 |
| 3308 | CF, DH(39) | 80 | 640 | 20 | 80 |
| 2988 | CF, DH(39) | 160 | 320 | 20 | 40 |
| 2152 | CF, DH(39) | 320 | 1,280 | 40 | 160 |
| 1949 | CF, DH(39) | 160 | 320 | 40 | 80 |
| Clinical isolates of coagulase-negative Staphylococcus epidermidis | |||||
| 38. 04-77651 | BB, NUH | >5,120 | 5,120 | 2,560 | 1,280 |
| 39. 04-139122 | BB, NUH | 2,560 | 1,280 | 1,280 | 640 |
| 40. 05-95325 | BB, NUH | 2,560 | 1,280 | 1,280 | 640 |
| 41. 04-26895 | BB, NUH | 5,120 | 5,120 | 2,560 | 2,560 |
| 42. 0691964 | BB, NUH | 5,120 | 5,120 | 2,560 | 1,280 |
| 45. 06-87106 | BB, NUH | 2,560 | 1,280 | 1,280 | 1,280 |
| 132. 2008-02-3902 | C/P, NUH | 5,120 | 2,560 | 2,560 | 640 |
| 137. 2008-02-2068 | C/P, NUH | 2,560 | 2,560 | 2,560 | 640 |
| 135. 2008- 03-7703 | C/P, NUH | 2,560 | 2,560 | 2,560 | 640 |
| Reference strains | |||||
| P. aeruginosa PAO1 | DSMZ | 160 | 320 | 40 | 80 |
| P. aeruginosa 14 | MW | 160 | 320 | 20 | 40 |
| P. aeruginosa 17619 | CCUG | 80 | 320 | 20 | 40 |
| S. epidermidis 1621 | CCUG | 2,560 | 2,560 | 2,560 | 1,280 |
| S. epidermidis 43038 | CCUG | >5,120 | 2,560 | 1,280 | 640 |
| S. aureus Newman | ES | 5,120 | 2,560 | 320 | 640 |
| S. aureus Newman Δfur | ES | 5,120 | 5,120 | NG | NG |
| S. aureus 1800 | CCUG | 5,120 | 5,120 | 640 | 320 |
| S. aureus 1828 | CCUG | 2,560 | 2,560 | 5,120 | >5,120 |
| S. aureus 10778 | CCUG | 2,560 | 2,560 | 5,120 | 5,120 |
| S. aureus 15915 | CCUG | 5,120 | 5,120 | 2,560 | 2,560 |
| S. aureus 42765 | CCUG | 5,120 | 5,120 | 5,120 | 5,120 |
| S. aureus MRSA16 | MW | >5,120 | 2,560 | 5,120 | 2,560 |
| S. aureus MRSA15 | MW | 5,120 | 5,120 | 5,120 | 5,120 |
| E. faecalis 9997 | CCUG | 1,280 | 1,280 | NG | NG |
| E. faecalis ATCC29212 | MW | 2,560 | 5,120 | NG | NG |
| K. pneumoniae 225 | CCUG | 1,280 | 2,560 | 1,280 | 2,560 |
| K. pneumoniae 10785 | CCUG | 2,560 | 2,560 | 1,280 | 1,280 |
| K. pneumoniae 38027 | CCUG | >5,120 | 5,120 | 5,120 | 2,560 |
| P. mirabilis 26767 | CCUG | 1,280 | 1,280 | 1,280 | 1,280 |
| E. coli 10979 | CCUG | >5,120 | >5,120 | 1,280 | 1,280 |
| E. coli 17620 | CCUG | 5,120 | >5,120 | 2,560 | 5,120 |
| E. coli UPEC 536 MP10:24 | SNW | 2,560 | 2,560 | 2,560 | 2,560 |
| V. cholerae A1 552 O1 ELTOR CTX+ | SNW | 1,280 | 1,280 | 2,560 | 1,280 |
| V. cholerae V:5 NONO1 0139 | SNW | 1,280 | 1,280 | 1,280 | 1,280 |
W, wound; BB, bone biopsy; CF, cystic fibrosis; C/P, catheter/prosthesis; NG, no growth; CCUG, culture collection, University of Gothenburg; SNW, Sun Nyunt Wai collection, Umeå University; MW, Martin Welch collection, University of Cambridge; DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen; ES, Erik Scaar collection, Vanderbilt University; NUH, Norrland University Hospital; DH, Didier Hocquet collection, Addenbrooke's Hospital.
Determination of inhibitory concentrations.
Concentrations that inhibit 90% of bacterial growth (IC90) were determined as described in reference 2 with small modifications. Briefly, the number of bacteria per ml in a suspension with an optical density at 600 nm (OD600) of 1 was determined for each strain, and approximately 5 × 106 to 1 × 107 bacteria were added to either Iso-Sensitest or HHW medium containing 2-fold-dilution series of Ga citrate or Ga-DFOB in 24-well plates. Plates were incubated with shaking at 37°C for 10 h, and OD600 was determined using a plate reader.
Biofilm studies.
Biofilm formation by P. aeruginosa PAO1 expressing the pJBA129 plasmid, encoding green fluorescence protein (GFP), was analyzed by diluting an overnight culture of fresh 20% Iso-Sensitest supplemented with tetracycline at 10 μg/ml (to maintain the GFP-containing plasmid) and subinhibitory concentrations of Ga citrate (0, 2.5, 10, or 20 μM). Aliquots (1 ml each) were dispensed into 24-well plates; corner wells and the inner volume of the plates were filled with water to prevent evaporation. The plates were incubated with shaking at 37°C. GFP fluorescence in four independent wells was measured after 4 h (corresponds to late exponential phase) using an Infinite series 200 plate reader (Tecan). Growth curves and viable counts were measured in parallel experiments.
Speciation modeling.
The theoretical speciation of gallium in the presence of DFOB or citrate in 20% Iso-Sensitest was predicted through calculations using the WinSGW program based on the SolGasWater algorithm (19) and equilibrium constants described in the literature (48).
A total concentration of 5 or 20 μM Ga3+ was used in the calculations. The metal-to-ligand ratio was 1:1 for DFOB and 1:2.5 for citrate. These ratios were chosen to match the experimental ratios used. The media contained large numbers of ingredients, some of which lack reported equilibrium constants. In order to perform the calculations, the structure of each ingredient (see Table S1 in the supplemental material) was examined for possible association with gallium ions. Ligands with the ability to form complexes were compared to known ligands and ranked. For example, most amino acids were estimated to have a weaker complexation than cysteine, and to simplify, the binding constant for cysteine was used for all amino acids. Consequently, the calculations might overestimate the binding of gallium ions to components in defined media such as HHW medium. Iso-Sensitest is a semidefined medium and contains some hydrolyzed casein as well as peptones. Equilibrium constants for the undefined components could not be estimated, and consequently their effects were not taken into account in the calculations.
Preparation of iron-free medium.
Iron-depleted medium was prepared by adding 0.1% Chelex-100 chelating resin (Bio-Rad) to 20% Iso-Sensitest and kept overnight with stirring at 4°C. Next, medium was filtered through a 0.2-μm filter (Millipore) to remove Chelex-100 with bound cations. Fresh Chelex-100 was added, and the medium was treated in the same way once again. After the second filtration, medium was complemented with a trace element solution to produce the following final concentrations: MgCl2·6H2O, 197 μM; CaCl2·H2O, 155 μM; CuSO4·5 H2O, 0.8 μM; ZnSO4·H2O, 1.1 μM; MnSO4·H2O, 2.3 μM. The Fe-free medium was kept in a plastic bottle pretreated with 1% nitric acid and dry sterilized.
Ga and Fe uptake.
To study how the presence of citrate or DFOB influences Ga and Fe uptake by P. aeruginosa, PAO1 was grown to an OD600 of 1.1 in Fe-depleted 20% Iso-Sensitest. Bacteria from 200 ml of culture were collected by centrifugation and washed twice with 20 ml phosphate-buffered saline (PBS). The washed bacteria were added to obtain an OD600 of 1.1 in each of the following media: (i) iron-depleted 20% Iso-Sensitest; (ii) iron-depleted 20% Iso-Sensitest with 20 μM Ga and 50 μM citrate; (iii) iron-depleted 20% Iso-Sensitest with 20 μM Ga, 50 μM citrate, and 20 μM DFOB; (iv) iron-depleted 20% Iso-Sensitest with 10 μM Ga, 10 μM Fe, and 25 μM citrate; (v) iron-depleted 20% Iso-Sensitest with 10 μM Ga, 10 μM Fe, 25 μM citrate, and 20 μM DFOB.
After 1 h of incubation at 37°C with shaking, 2 ml of the medium was centrifuged, and bacteria were washed twice in 2 ml PBS and resuspended in 1 ml 0.1% sodium dodecyl sulfate (SDS) plus 1 mM NaOH. Bacteria were pipetted and vortexed until they dissolved (approximately 10 min). The resulting liquid was filtered through a 0.2-μm filter. All samples were stored, handled, and pipetted with plastic equipment to avoid iron contamination.
Ga concentrations in dilutions of the samples were measured with an Elan DRC-e ICP-MS (Perkin Elmer SCIEX, Ontario, Canada). Gallium was quantified by external calibration with matrix-matched standards using the isotope 71Ga+, with indium (115In+) as the internal standard.
Determination of total intracellular Fe was performed using a ferrozine assay as described by Lindgren et al. (33), with the exception that bacterial viable counts were done for every sample.
J774 murine macrophage infection.
A total of 2 × 105 J774 macrophages were seeded in 24-well plates in 1 ml warm GIBCO Dulbecco's modified Eagle medium plus GlutaMax-l (Invitrogen) with 10% fetal bovine serum. After 24 h, the culture medium was exchanged for fresh medium containing P. aeruginosa PAO1 at a multiplicity of infection (MOI) of 0.5 or 5. Tetracycline (15 μg/ml), Ga-Cit (150 μM), Ga-DFOB (150 μM), Hoechst (2 μg/ml), propidium iodide (30 μM), and apo-transferrin (1,000 and 250 μg/ml) were added together with the bacteria.
Bacterial toxicity for macrophages was judged by the release of lactate dehydrogenase (LDH) into the culture medium using a CytoTox96 nonradioactive cytotoxicity assay (Promega) according to the manufacturer's recommendations. In short, 50-μl aliquots of culture medium were withdrawn from each well at 0, 1.5, 3, 4.5, 6, and 7 h postinfection. The absorbance of each sample was determined at 490 nm using an Infinite series 200 plate reader (Tecan). All experiments were performed in triplicate.
To monitor the progression of infection of J774 macrophages using microscopy, cells were cultured and infected in petri dishes with optical glass bottoms (MatTek Corporation, MA). A Nikon Eclipse Ti-E inverted microscope equipped with an Andor iXon+ EMCCD (electron multiplying charge-coupled device) camera was used to capture differential interference contrast (DIC) images and fluorescence from propidium iodide, Hoechst, and GFP at 4.5 h postinfection. In each petri dish, infection was started with a 10-min interval, and pictures were taken at 10-min intervals to reach equal time postinfection for every sample. Viability was monitored using propidium iodide. Propidium iodide stains only cells with compromised viability, and hence red cells were considered damaged. The toxicity was judged by the average number of damaged macrophages counted in seven different fields.
Exposure of mammalian cell lines to gallium complexes.
The human type II alveolar epithelial cell line A549 (ATCC CCL-185; American Type Culture collection), the human bronchial epithelial cell line BEAS-2B (ATCC CRL-9609), the mouse fibroblast cell line L929 (ATCC CCL1), and the mouse monocyte-macrophage cell line J774 (ATCC TIB 67) were cultured in Dulbecco's modified Eagle medium (Gibco BRL, Paisley, United Kingdom), with and without 10% fetal calf serum (FCS; HyClone, Perbio Science, Aalst, Belgium) and 50 μg/ml gentamicin. All cells were maintained at 37°C in a humidified atmosphere with 5% CO2. For experiments, cells were seeded in 24- or 96-well culture plates at 5 × 104 and 4 × 103 cells/well, respectively, and allowed to attach overnight before being exposed to Ga-Cit or Ga-DFOB.
Cell stress was analyzed by monitoring the release of cytokines from the cells using enzyme-linked immunosorbent assay (ELISA). Cells cultured in 24-well culture plates were exposed to gallium citrate or gallium DFOB at concentrations of 2.5, 10, 80, 160, and 640 μM for 24 h. The supernatants were separated from the cells by centrifugation, and interleukin 8 (IL-8), IL-6, and monocyte-chemotactic protein 1 (MCP-1) were measured in the cell-free fluid using a DuoSet ELISA development kit (R&D Systems, Abingdon, United Kingdom) according to the manufacturer's protocol.
Cell viability was measured using an alamarBlue assay (Serotec Scandinavia, Kidlington, United Kingdom) (42). Cells cultured in flat-bottomed 96-well culture plates were exposed to Ga-Cit or Ga-DFOB for 24 h at concentrations ranging from 0.6 μM to 320 μM and in cell culture medium with or without FCS. Thereafter, 20 μl of alamarBlue was added to each well, and plates were further incubated for 6 to 20 h before the fluorescence was measured by fluorescence spectroscopy (excitation at 560 nm and emission at 590 nm) using a Fluorostar plate reader (BMG Labtechnologies GmbH, Hilden, Germany). Unstimulated cells served as a control.
Metabolomic analysis.
Bacteria for metabolomic analysis were grown in 100% Iso-Sensitest with addition of either 20 μM Ga-Cit or 20 μM Na citrate. This medium composition was chosen to avoid restricting the bacterial metabolism of certain pathways by using a medium containing only a limited number of components. When cultures reached an OD600 of 1.0 (still in log phase), bacteria were harvested by centrifugation at 4°C, washed once in 20 ml of ice-cold PBS, and frozen in liquid nitrogen. Two independent liquid cultures of P. aeruginosa PAO1 were used to start the 20 cultures used in the experiment. Of 10 cultures derived from the same starting culture, 5 were grown with Ga-Cit and 5 with Na citrate. Metabolites were extracted using a water-methanol mixture (1:9) containing 11 internal standards, each at 3.5 ng μl−1. Samples were normalized to the number of cells so that 500 μl of the extraction mixture was added to 1010 cells. Thereafter, the cells were broken using a tungsten bead in a vibration mill (30 Hz for 3 min). After removal of the bead, the samples were centrifuged for 10 min at 14,000 rpm and 4°C. A volume of 200 μl of the supernatant was transferred to microvials for gas chromatography-mass spectroscopy (GC-MS) and dried in a Speed-Vac concentrator. Derivatization was carried out the day before GC-MS analysis. A volume of 15 μl of methoxyamine was added to each sample, and samples were shaken for 10 min in a shaker and incubated at 25°C for 16 h. Thereafter, 15 μl of N-methyl-trimethylsilyltrifluoroacetamide (MSTFA) (1% trimethylchlorosilane [TMCS]) was added, vortexed, and incubated at 25°C for 1 h. Finally, 15 μl heptane, including 500 ng of methyl stearate, was added before samples were loaded onto the GC-MS instrument.
GC-MS and data collection were performed as described in reference 28a. Briefly, an 1-μl aliquot of derivatized sample was injected (splitless mode) into an Agilent 6980 GC equipped with a 10 m × 0.18 mm i.d. fused-silica capillary column chemically bonded with 0.18 μm DB5-MS stationary phase (J&W Scientific, Folsom, CA). The column effluent was introduced into the ion source of a Pegasus III TOFMS instrument (Leco Corp., St Joseph, MI). Masses were recorded from m/z 50 to 800 at a rate of 30 spectra s−1, and files of acquired samples were exported to MATLAB 7.3 (R2006b) (Mathworks, Natick, MA) in NetCDF format for further data processing and analysis.
All data pretreatment procedures, including baseline correction, chromatogram alignment, setting of time windows, and hierarchical multivariate curve resolution (H-MCR) were performed in MATLAB using custom scripts (29). Alignment and smoothing using a moving average were carried out before H-MCR was used to resolve pure chromatographic and spectral profiles. Before multivariate modeling, all peak areas were normalized to the peak areas of 11 internal standards eluting over the entire chromatographic time range. Mass spectra of all detected compounds were compared with spectra in the NIST 2.0 library (as of 31 January 2001), using the in-house mass spectrum library database established by Umeå Plant Science Center (UPSC) or the mass spectrum library maintained by the Max Planck Institute in Golm (http://csbdb.mpimp-golm.mpg.de/csbdb/gmd/gmd.html).
The H-MCR-processed GC-MS data were mean-centered and scaled to unit variance prior to multivariate data analysis. Principal component analysis (PCA) was used initially to study the main variation in the data. In a second step, orthogonal partial least-squares discriminant analysis (OPLS-DA) was performed to model the systematic variation in the metabolomic data related to and orthogonal to predefined sample classes among the samples, here considering Ga citrate versus Na citrate. Cross validation was used to determine the predictive ability of the models. The OPLS-DA model loadings combined with univariate P values (Student's t test) were used to highlight significant metabolites associated with detected class differences. All multivariate analyses were carried out with SIMCA-P+ software (version 12.0; Umetrics AB, Umeå, Sweden).
Polyacrylamide gel electrophoresis (PAGE) and protein analysis.
15% SDS-PAGE was done according to a standard procedure using a 20 cm Protean II xi cell (Bio-Rad). The run time was 12 h at 24 mA. Three bands that differed visually between the Ga-containing sample and the control were cut out and sent for analysis (Alphalyse A/S, Odense, Denmark). The protein samples were reduced and alkylated with iodoacetamide (i.e., carbamidomethylated) and subsequently digested with trypsin. The resulting peptides were concentrated on a ZipTip micropurification column and eluted onto an anchor chip target for analysis on a Bruker Autoflex III matrix-assisted laser desorption ionization–time of flight (MALDI TOF/TOF) instrument. The peptide mixture was analyzed in positive reflector mode for accurate peptide mass determination. MALDI tandem mass spectrometry (MS/MS) was performed on selected peptides for peptide fragmentation analysis, i.e., partial sequencing. The MS and MS/MS spectra were combined and used for database searching using Mascot software (Matrix Science Ltd.). The Mascot score is defined as −10 × log10(P), where P is the absolute probability, and a score is significant if it is above 67, i.e., P is <0.05. The data were searched against in-house protein databases downloaded from NCBI, including the NRDB database, which contains more than 12 million known nonredundant protein sequences.
Effect of carbon source.
P. aeruginosa PAO1 was grown in 10% Iso-Sensitest with the addition of 4 mg/ml of one of the following compounds: Casamino Acids, glucose, succinate, pyruvate, glutamate, acetate, and citrate. Bacteria (∼5 × 106/ml) were added to 24-well plates, and optical density at 600 nm was measured to monitor growth (Infinite series 200 plate reader; Tecan). To these mixtures either 20 μM Ga citrate or 20 μM FeCl3 was added; 10% Iso-Sensitest medium was used as a control. Pyoverdine production was assessed in cell-free supernatants by measuring absorbance at 405 nm.
X-ray photoelectron spectroscopy analysis.
P. aeruginosa PAO1 was grown to the mid-exponential and late exponential phases in 100 and 20% Iso-Sensitest. Cells from 50 ml liquid culture were collected by centrifugation and washed twice in 20 ml of PBS. For each sample, a volume of 20 μl of the cell pellet was quickly frozen using liquid nitrogen inside the loading chamber of the spectrometer. X-ray photoelectron spectroscopy (XPS) spectra of the frozen sample were collected using a Kratos Axis Ultra DLD spectrometer with a monochromated Al Kα source operated at 150 W. Analyzer pass energies of 160 eV and 20 eV were used for acquiring survey spectra and for acquiring spectra of individual photoelectron lines, respectively. The analysis area was 0.3 by 0.7 mm. The spectrometer charge neutralizing system was used to compensate for sample charging during the measurement, and the binding energy scale was referenced to the C1s aliphatic carbon peak at 285.0 eV. The analysis gives the near-surface (<10 nm) chemical composition of the bacterial surface.
RESULTS
Chemical speciation of gallium in media.
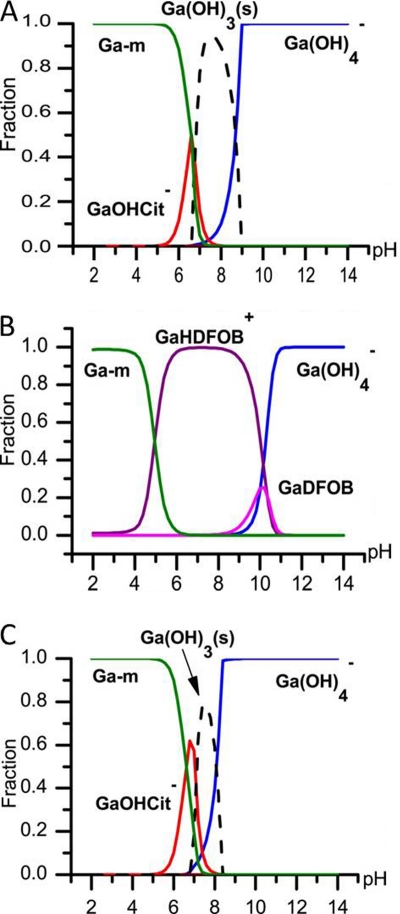
The biological effect of metal ions is greatly influenced by their complexation to ligands in solution. Theoretical calculations of the equilibrium speciation of gallium ions show that in pure water solutions at neutral pH, gallium ions precipitate as gallium hydroxide, whereas citrate keeps gallium in solution (see Fig. S1 in the supplemental material). Consequently, aqueous solutions of noncomplexed gallium ions can change composition with age unless they are kept at a pH of <2 or have ligands present that can form soluble complexes with gallium ions. Furthermore, the addition of growth medium can influence speciation and biological effect of metal ions (44) by ligand exchange. Figure 1 shows results from equilibrium calculations of the speciation for 20 μM gallium in 20% Iso-Sensitest for the two complexes investigated in this study, Ga-Cit and Ga-DFOB. The calculations show that the gallium complexes can be expected to persist in solution around neutral pH even in the presence of medium components. With time, a precipitate from Ga(OH)3 can be expected in Ga-Cit solutions at a pH above 7 in a concentration-dependent manner (Fig. 1A and C). In solutions with Ga-DFOB this precipitation is completely prevented. However, the equilibrium calculations do not take into account the undefined ingredients in the Iso-Sensitest media that could possibly affect the bioavailability of gallium.
Fig. 1.
Results from the calculation of gallium speciation in 20% Iso-Sensitest medium. Each line represents the fraction of the total amount of gallium in a specific complex in solution. Broken lines represent precipitation of Ga(OH)3. According to the calculations, Ga ions are expected to bind to medium components (Ga-m) only at low pH. At high pH, Ga exists in form of soluble Ga(OH)4−. (A) Twenty micromolar Ga3+ and 50 μM citrate; (B) 20 μM Ga3+ and 20 μM DFOB. (C) At lower total concentrations of Ga3+ (5 μM), the relative precipitation of Ga(OH)3(s) is lower than that at higher concentrations.
Ga citrate has an antibiofilm effect and is more bactericidal than Ga-DFOB.
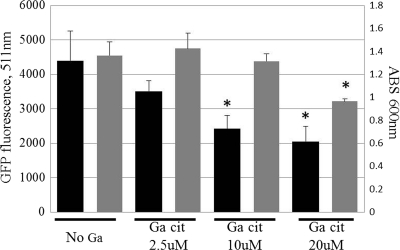
The Ga-DFOB complex was shown previously to display an antibacterial effect on P. aeruginosa (6). However, little is known regarding its effect in solutions with different compositions and regarding its effect against a large range of bacteria. To investigate this, the 90% inhibitory concentration (IC90) of Ga-DFOB and Ga-Cit were determined in two different growth media for a broad spectrum of Gram-negative and Gram-positive bacteria, including clinical isolates (Table 1). Ga(NO3)3 was not used as a control for uncomplexed Ga3+, since precipitation lowers the soluble concentration of gallium ions in solution (see Fig. S1 in the supplemental material) and, consequently, can produce unreliable MICs. One semidefined and one defined medium were chosen. The semidefined medium, Iso-Sensitest, contains vitamins, amino acids, salts, hydrolyzed casein, and peptones. HHW was chosen as a defined medium, as it contains amino acids, vitamins, nucleotides, and glucose but no proteins (27). Of all the bacteria tested, P. aeruginosa was the most sensitive to gallium, and for all Pseudomonas strains, the IC90 of Ga-Cit was 2 to 8 times lower than that of Ga-DFOB (Table 1). The IC90 of Ga-Cit and Ga-DFOB were much lower for P. aeruginosa strains in HHW than in Iso-Sensitest. This is probably due to the larger trace amounts of iron in Iso-Sensitest or the presence of the undefined peptones and hydrolyzed casein that could potentially bind Ga. S. epidermidis strains were more sensitive to Ga-DFOB than Ga-Cit (Table 1). E. coli, Klebsiella pneumoniae, and Vibrio cholerae were slightly more sensitive to Ga citrate, while the sensitivity of S. aureus and Enterococcus faecalis to Ga-Cit and Ga-DFOB varied (Table 1). IC90 for S. aureus were comparable to reported MICs of Ga maltolate (∼4.4 mM) (4). Since the antibiofilm properties of Ga-Cit had not been investigated previously, we confirmed that Ga-Cit, like other Ga complexes, significantly inhibited biofilm production in P. aeruginosa at concentrations as low as 10 μM (Fig. 2).
Fig. 2.
Effect of subinhibitory concentrations of Ga-Cit on biofilm formation by P. aeruginosa PAO1. Black bars, biofilm formation; gray bars, optical density of the culture at 600 nm. Error bars represent standard deviations. Asterisks show values that are significantly different from the no-Ga control (P ≤ 0.005).
Uptake of Ga-Cit is more efficient than of Ga-DFOB.
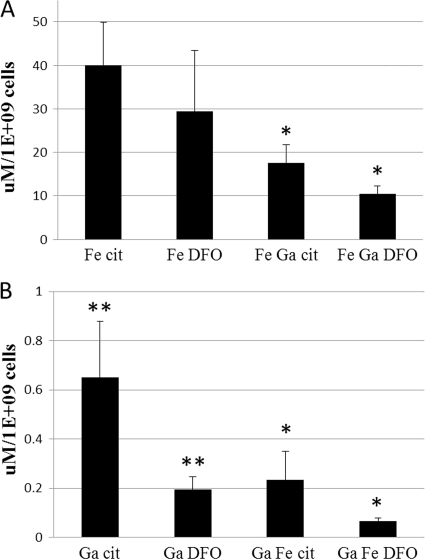
Levels of Ga in P. aeruginosa PAO1 exposed to Ga-Cit, Ga-DFOB, or equimolar mixtures of Ga and Fe citrate or of Ga- and Fe-DFOB (Fig. 3B) were analyzed using ICP-MS, and iron uptake was studied using a ferrozine assay (Fig. 3A). For both Ga and Fe there was a higher uptake for citrate complexes. The levels of Ga-Cit associated with the cells were three times higher than those of Ga-DFOB even in the presence of iron. In previous studies, Ga has been observed to associate with bacterial cells (30, 41), but as Ga tends to precipitate at neutral pH, it is not clear whether Ga was present inside the bacterium or trapped at the surface. To exclude the possibility of surface precipitation of gallium hydroxide, XPS analysis was performed to detect Ga at the bacterial surface. Since no Ga (or Fe) could be detected at the surface (data not shown), the concentrations measured by ICP-MS or the ferrozine assay were considered to be intracellular.
Fig. 3.
Ga and Fe uptake by P. aeruginosa. Cells of P. aeruginosa PAO1 were incubated in medium containing Ga or Fe or an equimolar mixture of Ga and Fe. (A) Uptake of Fe chelated with citrate and DFOB; (B) uptake of Ga chelated with citrate and DFOB. Significant differences were found in the uptake of Ga and/or Fe between the samples containing citrate and DFOB for Ga, and for Ga and Fe in combination, but not for samples containing only Fe (∗, P ≤ 0.05; ∗∗, P ≤ 0.005). Error bars represent standard deviations.
Neither Ga-Cit nor Ga-DFOB is toxic to mammalian cells.
Antibacterial compounds may display cytotoxic effects and trigger immune responses in humans and other animals (44). Increased levels of MCP-1, IL-8, and IL-6 in cells exposed to these compounds are indicative of inflammation (14). Monocyte-chemotactic protein 1 (MCP-1) is a factor specifically attracting monocytes, and IL-8 is a factor used to recruit neutrophils to the site of inflammation. To study the cytotoxic and inflammatory responses in mammalian cells exposed to Ga-Cit and Ga-DFOB, several relevant cell lines were chosen, such as bronchial epithelial cells, alveolar epithelial cells, murine fibroblasts, and murine macrophages (see Fig. S2A, S2B, and S3 in the supplemental material). For all but one cell line, toxicity was found to be very low and fluctuated from 0 to 10% in the presence or absence of serum. Slightly higher toxicity was found only for fibroblast cells exposed to Ga-DFOB when the assay was carried out without calf serum (23.2% ± 4%) (see Fig. S2A). This cytotoxicity was not concentration dependent and was comparable to the cytotoxicity of common antibiotics such as kanamycin and gentamicin in human and rat fibroblast cultures (13, 16).
Levels of MCP-1, IL-6, and IL-8 were similar to those in negative controls, indicating that no proinflammatory response was triggered in lung epithelial cells at concentrations up to 640 μM (highest concentration tested) for either gallium complex (see Fig S3 in the supplemental material).
Metabolomic analysis and identification of differentially expressed proteins.
As previous transcriptomic analysis only showed a few specific changes in gene expression connected to Ga-Cit exposure, we studied the P. aeruginosa metabolome in the presence of 20 μM Ga-Cit to learn more about the mechanism of Ga toxicity and the observed antibiofilm effects.
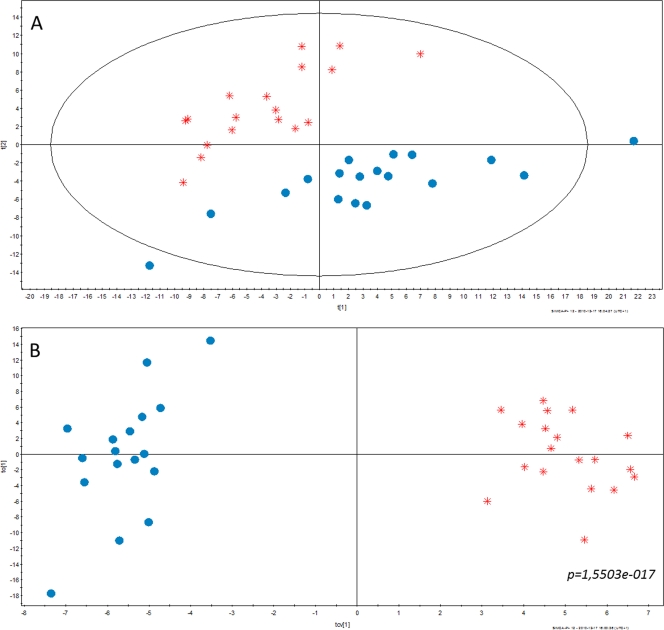
The optical densities of the cultures containing Ga-Cit (mean OD600, 0.92 ± 0.04) were only slightly lower than those of cultures grown in the presence of Na citrate (mean OD600, 1.07 ± 0.06), and the cultures were in late log phase. In the multivariate analysis of the metabolomic data, a clear separation of samples treated with gallium citrate and sodium citrate (reference) was observed. PCA was used to study the main features of the data and also to detect possible outliers (Fig. 4A). A separation between the two sample classes (Ga-Cit and Na citrate) could be detected in the first two principal components of the PCA, which suggest a clear alteration of the metabolome between the classes. This was also confirmed by the OPLS-DA analysis (P = 1.5503e-017) (Fig. 4B), which is a method more suitable than PCA for modeling and interpreting multivariate differences between sample classes. Altogether, 64 metabolites were significantly changed (P < 0.05), and of those, 19 were present at higher concentrations in the presence of gallium and 45 were present at lower concentrations (Table 2). A total of 26 metabolites were identified with high probability (scores from 900 to 1,000 using NIST MS Search). The rest of the metabolites were identified (scores from 650 to 900) only as belonging to a certain class of chemicals. Pyroglutamic and glutamic acid contributed to most of the separation between Ga-containing samples and controls, and they were decreased in the presence of Ga. In a recent metabolomic study, glutamate was claimed to be a metabolite of P. aeruginosa PAO1 critical for maintaining the balance between its versatile metabolic pathways (21). The concentrations of some fatty acids (FA) were increased (Table 2), which is noteworthy since lipid metabolism was previously shown to play a role in the mitigation of Al disturbance of Fe homeostasis in P. fluorescens (23). Benzoic acid derivatives were increased and ornithine was decreased, which correlates with the increase of pyochelin and decrease of pyoverdine biosynthesis pathways previously shown by transcriptomics analysis of P. aeruginosa grown in the presence of Ga (30).
Fig. 4.
Multivariate analysis of metabolomic data. (A) PCA scores plot, PC2 (t[2]) versus PC1 (t[1]) from the unsupervised PCA showing the maximum variation in the metabolomic data based on all detected metabolites. The plot shows a clear separation between the two sample classes, Ga citrate (red) and Na citrate (blue). Each symbol represents one sample described by all detected metabolites. (B) Cross-validated OPLS-DA scores plot, orthogonal component 1 (to[1]) versus predictive component 1(tcv[1]) from the supervised OPLS-DA showing the separation between the two sample classes. The separation was statistically significant (P = 1.5503e-017) as determined by analysis of variance of the cross-validated model.
Table 2.
Metabolites with increased or decreased concentrations in the presence of 20 μM Ga-Cit
| Metabolitea | P value | Changeb |
|---|---|---|
| UPSC_20292_GCTOF_Populus_RI_1873* | 0.03444619 | + |
| Isoleucine (2TMS) | 0.03050156 | + |
| Phosphoric acid (3TMS) | 0.0081851 | + |
| Mannose-6-phosphate methoxyamine (6TMS)* | 0.00245481 | + |
| Octadecanoic acid, n- (1TMS)* | 0.00088635 | + |
| Benzoic acid (1TMS)* | 0.00043407 | + |
| Glyceric acid-3-phosphate, d- (4TMS)* | 0.00020383 | + |
| l-Lysine (4TMS) | 0.00019975 | + |
| Uridine (4TMS) | 6.1046E-05 | + |
| Dodecamethylpentasiloxane | 2.5124E-05 | + |
| UPSC_20086_GCTOF_Populus_RI_2297* | 2.6964E-05 | + |
| Benzeneacetic acid, 2,5-bis[(trimethylsilyl)oxy]-, trimethylsilyl ester* | 1.1736E-05 | + |
| Octadecenoic acid (1TMS) | 1.9231E-06 | + |
| Mannose methoxyamine (5TMS)* | 4.4574E-07 | + |
| Hexadecenoic acid (1TMS) | 7.4169E-07 | + |
| Ethanolamine, N,N,O-TMS | 6.1749E-07 | + |
| UPSC_20080_GCTOF_Populus_RI_2185* | 3.4477E-07 | + |
| Rhamnose, dl- (1MEOX) (4TMS)* | 1.1798E-08 | + |
| 1,14-Tetradecanedioic acid-2TMS* | 1.0816E-10 | + |
| Pyroglutamic acid, dl- (2TMS) | 2.0597E-18 | − |
| Glutamic acid, dl- (3TMS) | 2.6057E-13 | − |
| Ornithine (4TMS) | 1.3566E-12 | − |
| Oleic acid* | 1.0368E-13 | − |
| Daphnetin* | 7.0079E-07 | − |
| Succinic acid (2TMS) | 1.5987E-07 | − |
| Nicotinamide (2TMS)* | 1.4841E-06 | − |
| Serine, dl- (3TMS) | 2.0835E-05 | − |
| Uric acid (4TMS) | 1.6225E-05 | − |
| Aspartic acid, dl- (3TMS) | 3.2943E-05 | − |
| Propanoic acid, 2-methyl-, 1-(1,1-dimethylethyl)-2-methyl-1,3-propanediyl | 6.275E-06 | − |
| Spermidine (5TMS) | 0.00010096 | − |
| UPSC_10004_GCTOF_Ath_RI_1145* | 3.7482E-05 | − |
| Malonic acid (2TMS) | 0.00023005 | − |
| Tryptamine, 5-hydroxy- (4TMS)* | 5.3545E-05 | − |
| β-d-Galactopyranoside, methyl 2,3-bis-O-(trimethylsilyl)-, cyclic methylboronate* | 5.9687E-05 | − |
| 3-Trimethylsiloxycapric acid, trimethylsilyl ester | 0.00025522 | − |
| Heptanoic acid, n- (1TMS)* | 0.00010886 | − |
| 1,3-Diaminopropane, N,N,N,N-TMS | 0.000231 | − |
| Dodecanoic acid, trimethylsilyl ester | 0.00109794 | − |
| Cyclohexene, 3-ethyl-1-trimethylsilyloxyl* | 0.00065972 | − |
| Pyrophosphoric acid (4TMS) | 0.0010718 | − |
| Threonic acid (4TMS)* | 0.00059617 | − |
| l-Glutamine (3TMS)* | 0.00134619 | − |
| 1,11-Bis(trimethylsiloxy)undecane* | 0.00193666 | − |
| Alanine, beta- (3TMS) | 0.00219231 | − |
| Asparagine, DL- (3TMS)* | 0.00165867 | − |
| UPSC_10154_GCTOF_Ath_RI_1762* | 0.00231544 | − |
| 4,6-Dimethyl-1-oxa-4,6-diazacyclooctane-5-thione* | 0.00352622 | − |
| β-d-Glucofuranose, 6-O-(trimethylsilyl)-, cyclic 1,2:3,5-bis(butylboronate)* | 0.00311849 | − |
| β-d-Galactopyranoside, methyl 2,3-bis-O-(trimethylsilyl)-, cyclic butylboronate* | 0.00318001 | − |
| 4-Aminobutyric acid (3TMS) | 0.00527924 | − |
| Tyramine (3TMS)* | 0.00373642 | − |
| Heptanoic acid, n- (1TMS) | 0.00790108 | − |
| M000000_A308001-101_MST_3084.1_EITTMS_* | 0.01138945 | − |
| Adenosine-5-monophosphate (5TMS) | 0.00861185 | − |
| UPSC_20470_GCTOF_Populus_RI_1253* | 0.00976594 | − |
| Glycine (2TMS) | 0.01312471 | − |
| Glucose, d- (1MEOX) (5TMS) | 0.017127 | − |
| Galactose methoxyamine (5TMS) | 0.01216915 | − |
| EITTMS_N12C_ATHR_1701.7_1135EC44_* | 0.01337805 | − |
| Piperidin-2-one, 3-amino- (2TMS) | 0.01678599 | − |
| Butanoic acid, 4-[bis(trimethylsilyl)amino]-, trimethylsilyl ester* | 0.03408965 | − |
| Ribose, 2-deoxy-, d- (1MEOX) (3TMS)* | 0.04348563 | − |
| Arginine, dl-, -NH3 (3TMS) | 0.04914455 | − |
Asterisks indicate metabolites identified as belonging to a certain class of chemicals (with a score from 650 to 900). Metabolites identified with a high score (from 900 to 1,000) in an NIST MS search have no asterisk. Metabolites with UPSC numbers are unknown compounds that belong to the Umea Plant Science Center database. TMS, trimethylsilyl.
+, increase; −, decrease.
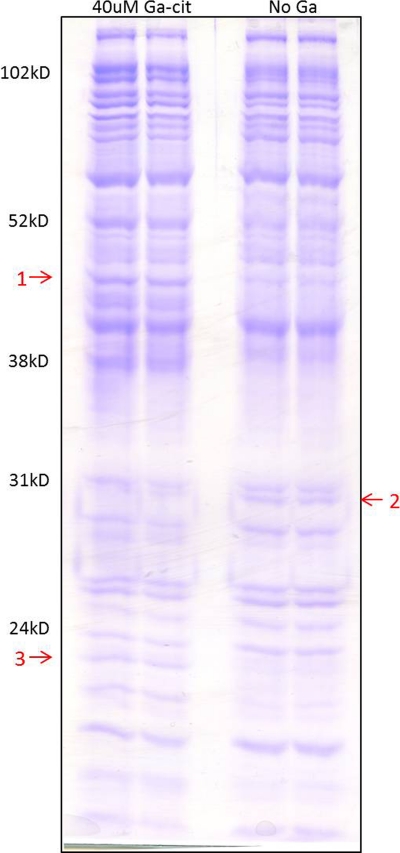
Protein profiles of cell lysates from samples with 20 or 40 μM Ga-Cit showed patterns that differed from those of control samples (Fig. 5). The differences were more pronounced at 40 μM. Three bands (Fig. 5) distinguished Ga-Cit-treated samples from control samples. Band 1 contained one protein identified as dihydrolipoamide succinyltransferase E2 subunit (GI 254234687; Mascot score, 143). The other two bands contained two proteins each: band 2 contained 50S ribosomal protein L2 (GI15599456; Mascot score, 198) and F0F1 ATP synthase subunit gamma (GI15600748; Mascot score, 85); band 3 contained riboflavin synthase subunit alpha (GI15599250; Mascot score, 185) and 3-ketoacyl-(acyl-carrier-protein) reductase (GI15598163; Mascot score, 87). Bands 1 and 3 were upregulated in the presence of Ga-Cit, and band 2 was upregulated in the control experiment. A decrease in expression of the genes coding for ribosomal proteins in the presence of Ga was shown previously, and our result is consistent with this finding (30). However, increased production of the other identified proteins has not been described before. It has been shown that reduction of Fe (III) by ferric siderophores occurs through nonenzymatic reaction with riboflavin, FADH, and FMNH2 (22). Furthermore, it has been shown that the expression of genes for riboflavin synthesis and secretion of riboflavin into the medium in a siderophore mutant of E. coli enabled it to survive under iron restriction (52). This could explain the upregulation of riboflavin synthase for bacteria exposed to Ga. Dihydrolipoamide succinyltransferase together with two other subunits constitutes the α-ketoglutarate dehydrogenase complex of the TCA cycle. Its upregulation in the presence of Ga-Cit suggests that enzymes in the TCA cycle are directly or indirectly affected by Ga.
Fig. 5.
Identification of proteins induced or repressed by Ga-Cit. Samples of whole-cell lysates were prepared in duplicate and loaded on separate lanes of an SDS-PAGE gel for analysis. Arrows indicate proteins with altered expression in response to Ga-Cit. They were identified by MALDI TOF/TOF and partial peptide sequencing. Arrow 1, dihydrolipoamide succinyltransferase; arrow 2, 50S ribosomal protein L2 and FoF1 ATP synthase subunit gamma; arrow 3, riboflavin synthase subunit alpha and 3-ketoacyl-(acyl carrier protein) reductase.
Effect of the carbon source.
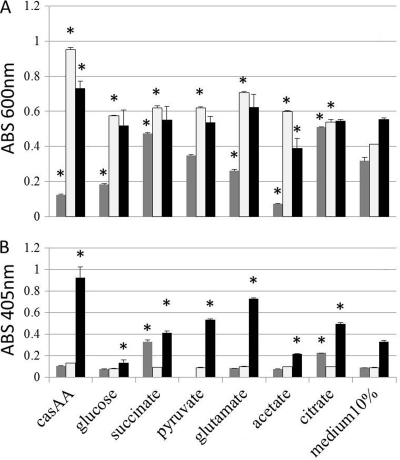
To test whether the shift in activity of the TCA cycle enzymes occurred in the presence of Ga-Cit, we tested growth of P. aeruginosa on different substrates of the TCA cycle. Indeed, in the presence of some salts (succinate and citrate), Ga-Cit displayed a lower inhibitory effect compared to that shown with the control (Fig. 6A), whereas salts such as acetate increased the inhibiting effect. The differences in Ga toxicity between P. aeruginosa grown on acetate and the same organism grown on citrate were large (Fig. 6), despite the fact that the growth curves on these substrates were similar without Ga-Cit (see Fig. S4 in the supplemental material). Casamino Acids and glucose, used as reference substrates, improved growth without Ga but increased the inhibitory effect in the presence of Ga-Cit. It should be noted that the substrates were added to 10% Iso-Sensitest medium, which could support P. aeruginosa growth. When Fe was present in excess, only base levels of pyoverdine were produced in the cultures (Fig. 6B). In cultures containing limited amounts of iron (10% Iso-Sensitest), production of pyoverdine differed depending on the substrates and was very low in cultures grown on glucose (Fig. 6B). Surprisingly, cultures containing Ga-Cit together with succinate or citrate produced higher levels of pyoverdine than those in medium with Ga-Cit alone. This indicates that pyoverdine synthesis was somewhat restored by succinate and citrate (Fig. 6B). Though no direct measurements of the activities of specific enzymes were performed, the growth experiments, together with the protein expression analysis, provide indirect evidence that certain metabolic pathways of P. aeruginosa are more affected by Ga than others.
Fig. 6.
Growth and pyoverdine production of P. aeruginosa PAO1 in the presence of Ga depends on the source of carbon. Bacteria were grown in 10% Iso-Sensitest complemented with 4 mg/ml of several substrates of the TCA cycle; Casamino Acids (casAA) and glucose were used as reference carbon sources. Optical density of the cultures (A) and pyoverdin production (B), both measured at the time point corresponding to the beginning of the stationary phase of growth in 10% Iso-Sensitest. Dark gray bars, presence of 20 μM Ga-Cit; light gray bars, presence of 20 μM FeCl3; black bars, 10% Iso-Sensitest. Error bars represent standard deviations; control samples contained 10% medium (without any additional carbon source) with or without Fe or Ga. Asterisks indicate values significantly different from the corresponding control value (P ≤ 0.005).
P. aeruginosa infection of macrophages.
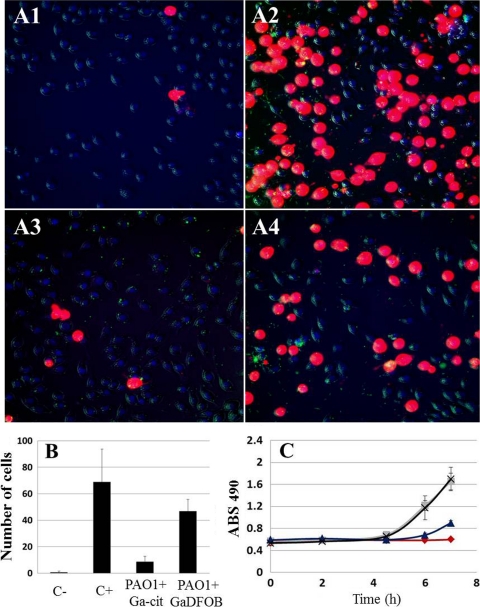
For any clinical application of antibacterial gallium complexes, it is important that the substances introduced are able to act in collaboration and synergy with the host immune system. To investigate this, an in vitro model for macrophage infection by P. aeruginosa was chosen. At 7 h postinfection, the macrophage toxicity conferred by infection (as assessed by measurements of LDH release in culture medium) was two times lower for samples containing 150 μM Ga-Cit than for samples with the same concentration of Ga-DFOB (Fig. 7). Under these experimental conditions, Ga-DFOB did not prevent induction of toxicity. Using live-cell microscopy, effects could be seen as early as 4.5 h postinfection, and the number of damaged macrophages in samples containing Ga-Cit was almost 3 times lower than in those containing Ga-DFOB. Interestingly, when a higher concentration of bacteria was used (MOI = 5), the differences between the two gallium compounds was negligible (data not shown), probably because the higher MOI was sufficient to induce macrophage toxicity without previous proliferation of bacteria. In human blood, most iron and Ga is bound to transferrin (4), which influences the concentrations of free Ga and Fe. However, the addition of apo-transferrin at different concentrations did not change the protective effect for the macrophages displayed by Ga citrate (see Fig. S5 in the supplemental material).
Fig. 7.
Degree of cytotoxicity resulting from a P. aeruginosa PAO1 infection of J774 macrophages, as measured by LDH release and propidium iodide uptake. (A) Live-cell microscopy of J774 macrophages infected with P. aeruginosa PAO1 (green dots in panels A1 to A4) at an MOI of 0.5; at 4.5 h postinfection, more damaged cells were seen in the presence of Ga-DFOB than in Ga-Cit. (A1) No bacteria and no Ga; (A2) no Ga; (A3) bacteria and 150 μM Ga citrate; (A4) bacteria and 150 μM Ga-DFOB. (B) Propidium iodide toxicity test showing a higher number of damaged cells during macrophage infection by P. aeruginosa PAO1 with Ga-DFOB than with Ga-citrate 4.5 h postinfection (based on quantification of live-cell microscopy data). Macrophages were counted in seven fields for every condition; significant differences were found for all samples compared to the negative control (P ≤ 0.05). (C) LDH toxicity test showing a time-dependent increase in damaged cells during macrophage infection by P. aeruginosa PAO1 in samples with Ga-DFOB or Ga-Cit; in samples with Ga-DFOB, values are similar to positive controls, while in the presence of Ga citrate, the absorbance is half of that for Ga-DFOB. Squares, no Ga; crosses, bacteria and Ga-DFOB; diamonds, no bacteria and no Ga; triangles, bacteria and Ga-Cit.
DISCUSSION
In this study, we show that for several bacterial species, lower concentrations of Ga chelated with citrate than of Ga chelated with DFOB are needed to inhibit bacterial growth. The uptake of both Ga and Fe in P. aeruginosa was more efficient when the metal was present in the form of citrate complexes. The uptake of Ga in bacteria has been suggested to occur along the same routes as the uptake of Fe (7). Fe-DFOB can be taken up by P. aeruginosa through FoxA and FiuA receptors (5, 34), and Fe citrate can be taken up through the FecA receptor (36). There are more than 30 possible siderophore uptake systems in Pseudomonas (34), and among these, there are two well-defined uptake systems for pyoverdine and pyochelin. In fact, Ga has even been shown to be taken up by a P. aeruginosa mutant defective for both pyochelin and pyoverdine pathways as well as by a mutant with a mutation of the ferric citrate uptake system (30). This shows that the iron uptake network in this bacterium is dynamic and flexible. The metal affinity for siderophores can be characterized by a pM value at a specific pH. This value represents the negative logarithm of the metal ion concentration left in solution after complex formation and can differ with solution pH (8). For iron chelators, pM is designated pFe and is generally calculated for an Fe3+ concentration of 1 μM and a ligand concentration of 10 μM at the pH of serum (pH = 7.4). The higher the pM value, the higher the metal affinity for the ligand. DFOB, pyoverdine, and pyochelin all have higher pM values than citrate (pFeDFOB = 27; pFepyoverdine = 27; pFecitrate = 16). Consequently, iron bound to citrate can easily be taken up by, for example, vacant pyoverdine. In this way, both gallium and iron, initially bound to citrate, may be imported into the cell through several uptake systems. This ligand exchange was previously demonstrated to occur between ferrichrome siderophore and citrate and was reported to have a half-time of approximately 10 min (35). In contrast, the differences in affinity between DFOB and the other P. aeruginosa siderophores are very small, which means that Ga-DFOB and Fe-DFOB most likely enter P. aeruginosa only through dedicated FoxA and FiuA ports. On the other hand, enterobactin of E. coli has a pFe of 35 (8), which allows it to remove Ga even from DFOB, which might explain why IC90 of Ga-Cit and Ga-DFOB are similar for E. coli (Table 1). Increased sensitivity of S. epidermidis to Ga-DFOB is probably due to synergistic activity of Ga and DFOB, since DFOB alone is known to inhibit the growth of this species (3).
In this study, we found that the metabolic profile of P. aeruginosa was altered in the presence of gallium (Table 2). The metabolomic differences were observed while bacteria were still in the log phase and actively multiplying, which indicated that the effects were due to specific antibacterial effects of Ga-Cit. Concentrations of amino acids and many other metabolites were decreased, indicating that biosynthesis was affected by Ga-Cit. Glucose levels were also decreased, in agreement with the decreased transcription of genes for glucose transport and metabolism previously described by Kaneko et al. (30). Concentrations of pyroglutamic and glutamic acids were decreased the most, even though gene expression related to glutamate biogenesis or utilization has been reported to be unchanged in the presence of gallium (30). Glutamate is one of the central metabolites in P. aeruginosa, and its concentration has been shown to change in response to different carbon sources (21). Moreover, glutamate is a starting point for heme biosynthesis (KEGG database [http://www.genome.jp/kegg/kegg2.html]).
Iron homeostasis in many bacteria, including P. aeruginosa, is regulated by Fur (ferric uptake regulator). Fur represses genes for Fe uptake when the level of this metal in the cell is sufficient and withdraws the repression when Fe is scarce (50). Additionally, Fur regulates expression of small RNAs, RyhB in E. coli and V. cholerae and PrrF1 and PrrF2 in P. aeruginosa (38, 51), responsible for translational inhibition of the synthesis of nonessential Fe proteins as a part of an iron-saving strategy when Fe is limited. Electron transport chain proteins involved in oxidative phosphorylation often depend on heme, and it is thought that when Fe is limited, heme biosynthesis becomes ineffective (38, 47). The presence of Al, which like Ga disturbs Fe metabolism, drives P. fluorescens to produce ATP through a modified TCA cycle instead of oxidative phosphorylation (47). This type of compensatory mechanism involving the TCA cycle has also been shown to appear in S. aureus during Fe starvation (20).
We observed that growth in the presence of different substrates of the TCA cycle and also on Casamino Acids modulated P. aeruginosa sensitivity to Ga-Cit (Fig. 6A). The effect could be explained by a catabolic repression phenomenon (45). Generally, carbon utilization in Pseudomonas is characterized by a strong hierarchy, with organic acids being a preferred source of carbon. Succinate, for example, is known to repress pathways for glucose metabolism, while glucose itself is a repressor for a number of other pathways, including mannitol and histidine utilization (45). Interestingly, the growth on glucose in the absence of Ga and Fe (Fig. 6A and B) was accompanied by lower production of pyoverdine than growth on other substrates. This indicates that pyoverdine production could also be regulated by different factors than Fe availability. Pyoverdine synthesis is induced by PvdS, a sigma factor that is known to be under the control of Fur (50). It was recently discovered that CysB, a master regulator of sulfur metabolism, also binds the pvdS promoter and positively regulates production of PvdS (28). The work of Kaneko et al. showed decreased transcription of genes for pyoverdine synthesis and pvdS in the presence of Ga (30). Consistent with this, we also observed very low levels of pyoverdine in 10% Iso-Sensitest medium in the presence of 20 μM Ga. On the other hand, at the same concentration of Ga but in the presence of succinate or citrate, more pyoverdine was produced (Fig. 6B). Consequently, nutritional signals in P. aeruginosa appear to interfere with Fe signaling. In Iso-Sensitest (or the similar tryptic soy broth medium), the presence of low concentrations of Ga might trigger bacteria to adjust their enzymatic reactions in a more favorable way with regard to energy acquisition and to avoid Fe-consuming pathways. Synthesis of pyoverdine could be too “expensive” under certain conditions, causing bacteria to instead rely on the alternative siderophore pyochelin (30). We hypothesize that some carbon sources induce expression of Fe-demanding pathways, and since Fe is not available, these pathways become costly for the cell and inhibit growth. Strong growth inhibition of P. aeruginosa by Ga-Cit was observed on Casamino Acids, and it implies that during infections when amino acids are the preferred carbon source, Ga could have a more potent effect. Activation and repression of catabolic pathways depend on concentration of the inducer (43). Concentrations of citrate in Ga-Cit solutions used in our study were lower than those used for bacterial growth on TCA substrates, and they most likely did not trigger citrate utilization.
Among other components that displayed decreased levels in the presence of Ga citrate, two are especially noteworthy, spermidine and 1,3-diaminopropane, a product of spermidine decomposition. Spermidine is one of the main polyamines in bacteria and has previously been shown to act as a translational regulator through several different mechanisms (26, 49). Successful formation of V. cholerae biofilms depends on the intact spermidine synthesis pathway (31), and inhibition of spermidine biosynthesis attenuates quorum sensing in Burkholderia pseudomallei (11). Consequently, it is likely that a decrease in spermidine levels upon exposure to Ga-Cit could contribute to the poor ability of P. aeruginosa cells to attach and form biofilms in the presence of Ga (30).
This work shows that Ga-Cit functions as an antibacterial agent against clinical isolates of several Gram-positive and Gram-negative pathogens. The broad-spectrum effects are important to take into consideration for possible future treatment of nosocomial and mixed infections of bacteria. At the concentrations effective against P. aeruginosa, Ga-Cit and Ga-DFOB caused neither proinflammatory responses in epithelial human cells nor cytotoxic effects. Detailed investigations of P. aeruginosa, the most susceptible species tested in our study, showed that complexation of Ga by different ligands not only is important in order to prevent precipitation but also influences the uptake of Ga. Moreover, it was observed that the effect of Ga-Cit differs depending on the carbon sources used by P. aeruginosa. This is important in terms of Ga-based drug development, since high concentrations of organic acid salts may enhance or mitigate the antibacterial effect of Ga. We suggest that Ga-Cit may cause activation of metabolic pathways that indicate Fe-saving behavior of the bacteria. The solution chemistry of Ga complexes and the interference of these complexes with the energy metabolism in bacteria is of great importance for understanding their biological activity. This study shows that the resulting antibacterial effect of Ga complexes is a combination of many factors, both chemical and biological, that should be taken into consideration if Ga compounds are to be used for battling infections.
Supplementary Material
ACKNOWLEDGMENTS
The Curth Nilsson Foundation for Scientific Research and Umeå University are acknowledged for funding.
We acknowledge Maria Fällman for kind permission to use the life cell microscope, Martin Welch for providing a part of the clinical isolates, and Erik Skaar for providing S. aureus Newman. PAO1 strain expressing the pJBA129 plasmid was kindly provided by Jens Bo Andersen and Michael Givskov. Igor Golovliov, Helena Lindgren, Johan Binesse, and Susanne Granholm are acknowledged for fruitful discussions.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 26 September 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Reference deleted.
- 2. Andrews J. M. 2001. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 48: 5–16 [DOI] [PubMed] [Google Scholar]
- 3. Antunes A. L. S., et al. 2008. Feasible identification of Staphylococcus epidermidis using desferrioxamine and fosfomycin disks. APMIS 116: 16–20 [DOI] [PubMed] [Google Scholar]
- 4. Baldoni D., Steinhuber A., Zimmerli W., Trampuz A. 2010. In vitro activity of gallium maltolate against staphylococci in logarithmic, stationary, and biofilm growth phases: comparison of conventional and calorimetric susceptibility testing methods. Antimicrob. Agents Chemother. 54: 157–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Banin E., et al. 2008. The potential of desferrioxamine-gallium as an anti-Pseudomonas therapeutic agent. Proc. Natl. Acad. Sci. U. S. A. 105: 16761–16766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beasley F. C., Heinrichs D. E. 2010. Siderophore-mediated iron acquisition in the staphylococci. J. Inorg. Biochem. 104: 282–288 [DOI] [PubMed] [Google Scholar]
- 7. Bernstein L. R. 1998. Mechanisms of therapeutic activity for gallium. Pharmacol. Rev. 50: 665–682 [PubMed] [Google Scholar]
- 8. Boukhalfa H., Reilly S. D., Michalczyk R., Iyer S., Neu M. P. 2006. Iron(III) coordination properties of a pyoverdin siderophore produced by Pseudomonas putida ATCC 33015. Inorg. Chem. 45: 5607–5616 [DOI] [PubMed] [Google Scholar]
- 9. Braud A., Hannauer M., Mislin G. L., Schalk I. J. 2009. The Pseudomonas aeruginosa pyochelin-iron uptake pathway and its metal specificity. J. Bacteriol. 191: 3517–3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cabrera G., Xiong A., Uebel M., Singh V. K., Jayaswal R. K. 2001. Molecular characterization of the iron-hydroxamate uptake system in Staphylococcus aureus. Appl. Environ. Microbiol. 67: 1001–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chan Y. Y., Chua K. L. 2010. Growth-related changes in intracellular spermidine and its effect on efflux pump expression and quorum sensing in Burkholderia pseudomallei. Microbiology 156: 1144–1154 [DOI] [PubMed] [Google Scholar]
- 12. Chenier D., et al. 2008. Involvement of fumarase C and NADH oxidase in metabolic adaptation of Pseudomonas fluorescens cells evoked by aluminum and gallium toxicity. Appl. Environ. Microbiol. 74: 3977–3984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chernikov V. G., et al. 2003. Comparison of cytotoxicity of aminoglycoside antibiotics using a panel cellular biotest system. Bull. Exp. Biol. Med. 135: 103–105 [DOI] [PubMed] [Google Scholar]
- 14. Chou S.-F., Chang S.-W., Chuang J.-L. 2007. Mitomycin C upregulates IL-8 and MCP-1 chemokine expression via mitogen-activated protein kinases in corneal fibroblasts. Invest. Ophthalmol. Vis. Sci. 48: 2009–2016 [DOI] [PubMed] [Google Scholar]
- 15. DeLeon K., et al. 2009. Gallium maltolate treatment eradicates Pseudomonas aeruginosa infection in thermally injured mice. Antimicrob. Agents Chemother. 53: 1331–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. El Mouedden M., Laurent G., Mingeot-Leclercq M. P., Tulkens P. M. 2000. Gentamicin-induced apoptosis in renal cell lines and embryonic rat fibroblasts. Toxicol. Sci. 56: 229–239 [DOI] [PubMed] [Google Scholar]
- 17. Emery T. 1986. Exchange of iron by gallium in siderophores. Biochemistry 25: 4629–4633 [DOI] [PubMed] [Google Scholar]
- 18. Enz S., Mahren S., Stroeher U. H., Braun V. 2000. Surface signaling in ferric citrate transport gene induction: interaction of the FecA, FecR, and FecI regulatory proteins. J. Bacteriol. 182: 637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Eriksson G. 1979. Algorithm for the computation of aqueous multicomponent, multiphase equilibria. Anal. Chim. Acta Computer Tech. Optimization 3: 375–383 [Google Scholar]
- 20. Friedman D. B., et al. 2006. Staphylococcus aureus redirects central metabolism to increase iron availability. PLoS Pathog. 2: e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Frimmersdorf E., Horatzek S., Pelnikevich A., Wiehlmann L., Schomburg D. 2010. How Pseudomonas aeruginosa adapts to various environments: a metabolomic approach. Environ. Microbiol. 12: 1734–1747 [DOI] [PubMed] [Google Scholar]
- 22. Halle F., Meyer J. M. 1992. Iron release from ferrisiderophores—a multistep mechanism involving a NADH FMN oxidoreductase and a chemical-reduction by FMNH2. Eur. J. Biochem. 209: 621–627 [DOI] [PubMed] [Google Scholar]
- 23. Hamel R., Appanna V. D. 2003. Aluminum detoxification in Pseudomonas fluorescens is mediated by oxalate and phosphatidylethanolamine. Biochim. Biophys. Acta Gen. Subj. 1619: 70–76 [DOI] [PubMed] [Google Scholar]
- 24. Harrison J. J., Ceri H., Stremick C. A., Turner R. J. 2004. Biofilm susceptibility to metal toxicity. Environ. Microbiol. 6: 1220–1227 [DOI] [PubMed] [Google Scholar]
- 25. Harrison J. J., et al. 2008. Copper and quaternary ammonium cations exert synergistic bactericidal and antibiofilm activity against Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 52: 2870–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Higashi K., et al. 2008. Selective structural change by spermidine in the bulged-out region of double-stranded RNA and its effect on RNA function. J. Biol. Chem. 283: 32989–32994 [DOI] [PubMed] [Google Scholar]
- 27. Hussain M., Hastings J. G., White P. J. 1991. A chemically defined medium for slime production by coagulase-negative staphylococci. J. Med. Microbiol. 34: 143–147 [DOI] [PubMed] [Google Scholar]
- 28. Imperi F., Tiburzi F., Fimia G. M., Visca P. 2010. Transcriptional control of the pvdS iron starvation sigma factor gene by the master regulator of sulfur metabolism CysB in Pseudomonas aeruginosa. Environ. Microbiol. 12: 1630–1642 [DOI] [PubMed] [Google Scholar]
- 28a. Jiye A., et al. 2005. Extraction and GC/MS analysis of the human blood plasma metabolome. Anal. Chem. 77: 8086–8094 [DOI] [PubMed] [Google Scholar]
- 29. Jonsson P., et al. 2005. High-throughput data analysis for detecting and identifying differences between samples in GC/MS-based metabolomic analyses. Anal. Chem. 77: 5635–5642 [DOI] [PubMed] [Google Scholar]
- 30. Kaneko Y., Thoendel M., Olakanmi O., Britigan B. E., Singh P. K. 2007. The transition metal gallium disrupts Pseudomonas aeruginosa iron metabolism and has antimicrobial and antibiofilm activity. J. Clin. Invest. 117: 877–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee J., et al. 2009. An alternative polyamine biosynthetic pathway is widespread in bacteria and essential for biofilm formation in Vibrio cholerae. J. Biol. Chem. 284: 9899–9907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lemire J., Mailloux R., Auger C., Whalen D., Appanna V. D. 2010. Pseudomonas fluorescens orchestrates a fine metabolic-balancing act to counter aluminium toxicity. Environ. Microbiol. 12: 1384–1390 [DOI] [PubMed] [Google Scholar]
- 33. Lindgren H., et al. 2009. The 58-kilodalton major virulence factor of Francisella tularensis is required for efficient utilization of iron. Infect. Immun. 77: 4429–4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Llamas M. A., et al. 2006. The heterologous siderophores ferrioxamine B and ferrichrome activate signaling pathways in Pseudomonas aeruginosa. J. Bacteriol. 188: 1882–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lovenberg W., Rabinowitz J. C., Buchanan B. B. 1963. Studies on chemical nature of clostridial ferredoxin. J. Biol. Chem. 238: 3899–3913 [PubMed] [Google Scholar]
- 36. Marshall B., Stintzi A., Gilmour C., Meyer J. M., Poole K. 2009. Citrate-mediated iron uptake in Pseudomonas aeruginosa: involvement of the citrate-inducible FecA receptor and the FeoB ferrous iron transporter. Microbiology 155: 305–315 [DOI] [PubMed] [Google Scholar]
- 37. Menon S., Wagner H. N., Jr., Tsan M. F. 1978. Studies on gallium accumulation in inflammatory lesions: II. Uptake by Staphylococcus aureus: concise communication. J. Nucl. Med. 19: 44–47 [PubMed] [Google Scholar]
- 38. Mey A. R., Craig S. A., Payne S. M. 2005. Characterization of Vibrio cholerae RyhB: the RyhB regulon and role of ryhB in biofilm formation. Infect. Immun. 73: 5706–5719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miller M. J., et al. 2009. Utilization of microbial iron assimilation processes for the development of new antibiotics and inspiration for the design of new anticancer agents. Biometals 22: 61–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Monson R. E., Foulds I., Foweraker J., Welch M., Salmond G. P. 2011. The Pseudomonas aeruginosa generalized transducing phage, ϕ;PA3, is a new member of the ϕ;KZ-like group of ‘jumbo’ phages, and infects model laboratory strains and clinical isolates from cystic fibrosis patients. Microbiology 157: 859–867 [DOI] [PubMed] [Google Scholar]
- 41. Olakanmi O., et al. 2010. Gallium disrupts iron uptake by intracellular and extracellular Francisella strains and exhibits therapeutic efficacy in a murine pulmonary infection model. Antimicrob. Agents Chemother. 54: 244–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Österlund C., Lilliehook B., Ekstrand-Hammarström B., Sandström T., Bucht A. 2005. The nitrogen mustard melphalan activates mitogen-activated phosphorylated kinases (MAPK), nuclear factor-kappa B and inflammatory response in lung epithelial cells. J. Appl. Toxicol. 25: 328–337 [DOI] [PubMed] [Google Scholar]
- 43. Ozbudak E. M., Thattai M., Lim H. N., Shraiman B. I., van Oudenaarden A. 2004. Multistability in the lactose utilization network of Escherichia coli. Nature 427: 737–740 [DOI] [PubMed] [Google Scholar]
- 44. Ramstedt M., et al. 2009. Bacterial and mammalian cell response to poly(3-sulfopropyl methacrylate) brushes loaded with silver halide salts. Biomaterials 30: 1524–1531 [DOI] [PubMed] [Google Scholar]
- 45. Rojo F. 2010. Carbon catabolite repression in Pseudomonas: optimizing metabolic versatility and interactions with the environment. FEMS Microbiol. Rev. 34: 658–684 [DOI] [PubMed] [Google Scholar]
- 46. Schultz-Hauser G., Koster W., Schwarz H., Braun V. 1992. Iron(III) hydroxamate transport in Escherichia coli K-12: FhuB-mediated membrane association of the FhuC protein and negative complementation of fhuC mutants. J. Bacteriol. 174: 2305–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Singh R., et al. 2009. An ATP and oxalate generating variant tricarboxylic acid cycle counters aluminum toxicity in Pseudomonas fluorescens. PLoS One 4: e7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Smith R. M., Martell A. E. 1989. Critical stability constants, vol. 6, suppl. 2 Plenum Press, New York, NY [Google Scholar]
- 49. Tabor C. W., Tabor H. 1985. Polyamines in microorganisms. Microbiol. Rev. 49: 81–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vasil M. L. 2007. How we learnt about iron acquisition in Pseudomonas aeruginosa: a series of very fortunate events. Biometals 20: 587–601 [DOI] [PubMed] [Google Scholar]
- 51. Wilderman P. J., et al. 2004. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl. Acad. Sci. U. S. A. 101: 9792–9797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Worst D. J., Gerrits M. M., Vandenbroucke-Grauls C. M. J. E., Kusters J. G. 1998. Helicobacter pylori ribBA-mediated riboflavin production is involved in tron acquisition. J. Bacteriol. 180: 1473–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.