Abstract
Bacteriocins produced by Lactobacillus salivarius isolates derived from a gastrointestinal origin have previously demonstrated efficacy for in vivo protection against Listeria monocytogenes infection. In this study, comparative genomic analysis was employed to investigate the intraspecies diversity of seven L. salivarius isolates of human and porcine intestinal origin, based on the genome of the well-characterized bacteriocin-producing strain L. salivarius UCC118. This revealed a highly conserved megaplasmid-borne gene cluster in these strains involved in the regulation and secretion of two-component class IIb bacteriocins. However, considerable intraspecific variation was observed in the structural genes encoding the bacteriocin peptides. They ranged from close relatives of abp118, such as salivaricin P, which differs by 2 amino acids, to completely novel bacteriocins, such as salivaricin T, which is characterized in this study. Salivaricin T inhibits closely related lactobacilli and bears little homology to previously characterized salivaricins. Interestingly, the two peptides responsible for salivaricin T activity, SalTα and SalTβ, share considerable identity with the component peptides of thermophilin 13, a bacteriocin produced by Streptococcus thermophilus. Furthermore, the salivaricin locus of strain DPC6488 also encodes an additional novel one-component class IId anti-listerial bacteriocin, salivaricin L. These findings suggest a high level of redundancy in the bacteriocins that can be produced by intestinal L. salivarius isolates using the same enzymatic production and export machinery. Such diversity may contribute to their ability to dominate and compete within the complex microbiota of the mammalian gut.
INTRODUCTION
There is increasing evidence to suggest that bacteriocin production is a desirable probiotic trait that enables the establishment and persistence of the producing strains within the gastrointestinal tract (GIT) (2, 9, 15, 35). These antimicrobials may have a narrow or broad spectrum of inhibition (16). Although broad-spectrum bacteriocins, such as nisin, have been useful with respect to the control of spoilage and pathogenic organisms in food preservation (8), there has been increasing interest in narrow-spectrum bacteriocins, such as thuricin CD, due to their narrow specificity and minimal impact on nontarget beneficial GIT microbes (30). As such, these bacteriocins offer potential alternatives to traditional antibiotics with respect to controlling pathogens within the gut (29). In situ production of such narrow-spectrum bacteriocins by probiotics would overcome complications such as the proteolytic degradation of orally delivered antimicrobial peptides during gastric transit. Lactobacillus salivarius is a promising probiotic candidate frequently isolated from human, porcine, and avian GITs, many of which are producers of unmodified bacteriocins of class IIa (pediocin-like bacteriocins), class IIb (two-component bacteriocins), and class IId (linear non-pediocin-like bacteriocins) (4, 13, 31). Significantly, an in vivo demonstration of the anti-infective properties of the abp118-producing strain L. salivarius UCC118 has established the in vivo functionality of such bacteriocins (6). In addition, purified OR7, a class IIa bacteriocin produced by the chicken intestinal isolate L. salivarius NRRL B-30514, has also been successfully employed to reduce Campylobacter jejuni colonization in poultry (31).
Closely related variants of abp118 (a two-component class IIb bacteriocin) frequently occur in intestinally derived L. salivarius isolates from different hosts (2, 17, 26, 27), suggesting that this feature may be important for the successful establishment of L. salivarius within the GIT. Further evidence of the ecological advantage that this trait bestows upon the producing strains was provided by the salivaricin P-producing L. salivarius DPC6005, which prevailed over four components of a probiotic formulation within the porcine ileum (34). The production of analogous bacteriocins by genetically distinct strains and species is frequently ascribed to extensive horizontal gene transfer events occurring within the GIT (1, 2, 15, 22, 33). It is thus notable that abp118 is encoded on a megaplasmid in L. salivarius UCC118. Although the transfer-associated genes within the conjugative megaplasmid pMP118 appear nonfunctional in L. salivarius UCC118 (10), corresponding RepA-type megaplasmids are universally present in L. salivarius (17). Interestingly, Wescombe et al. demonstrated the in vivo conjugative transmission of bacteriocin-rich megaplasmids in Streptococcus salivarius species, which was associated with the numerical prominence of the species within the oral cavity (35).
The variable nature of L. salivarius with respect to the ability to produce bacteriocins and the nature of the bacteriocins produced is becoming increasingly apparent. It has been noted that many L. salivarius strains that harbor homologues of the abp118 structural genes do not display anti-Listeria activity (17). A recent comprehensive genomic analysis revealed that the three-component regulatory system responsible for the transcriptional regulation of abp118 production was not well conserved and was likely responsible for the bacteriocin-negative phenotype of these strains (28). Other strains produce closely related, yet distinct, bacteriocins, such as salivaricin P, which differs from abp118 by 2 amino acids (aa) in their respective β peptides (2). Interestingly, the nine associated polymorphisms (three in the α gene and six in the β gene) appear to be specific features associated with L. salivarius isolates derived from porcine intestinal origin (2). It would thus seem that gene acquisition, mutation, and/or decay, all of which are potential consequences of species adaptation to a specific ecological niche (3, 25), have impacted the adaptation of this widespread bacteriocin locus of L. salivarius, driving genome-wide specialization in response to a particular niche.
We conducted a genome-wide comparison of seven genetically distinct bacteriocin-producing intestinal L. salivarius strains isolated in our laboratory, which revealed that the salivaricin/abp118-associated bacteriocin locus is a site of high variability. In the case of the neonatal isolate L. salivarius DPC6488, two novel narrow-spectrum bacteriocins, salivaricin T, which displays considerable homology to a two-component class IIb bacteriocin associated with Streptococcus thermophilus, and salivaricin L, a one-component class IId bacteriocin exhibiting anti-Listeria activity, are characterized in this study.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
L. salivarius strains of both human and porcine intestinal origin, previously isolated and characterized in our laboratory, were included in this study (Table 1). Lactobacilli were routinely cultured under anaerobic conditions at 37°C in MRS medium (Difco Laboratories, Detroit, MI). Anaerobic conditions were maintained by the use of anaerobic jars and Anaerocult A gas packs (Merck, Darmstadt, Germany). Listeria innocua DPC3572 and Listeria monocytogenes NCTC 11994 were grown aerobically at 37°C in brain heart infusion (BHI) (Merck).
Table 1.
Bacterial strains used in this study
| Source | Strain | Relevant features | Reference |
|---|---|---|---|
| Lactobacillus delbrueckii subsp. bulgaricus LMG 6901 | Indicator strain | 2 | |
| Listeria innocua DPC3572 | Indicator strain | 2 | |
| Listeria monocytogenes NCTC11994 | Indicator strain | 24a | |
| Human | Lactobacillus salivarius UCC118 | Abp118 producer | 13 |
| L. salivarius DPC6196 | Harbors bacteriocin structural genes | 2 | |
| L. salivarius DPC6488 | Produces salivaricin T and salivaricin L | 23 | |
| Porcine | L. salivarius DPC6502 | Bacteriocin producer | 23 |
| L. salivarius 7.3 | Salivaricin P producer | 2 | |
| L. salivarius DPC6189 | Salivaricin P producer | 2 | |
| L. salivarius DPC6027 | Salivaricin P producer | 2 | |
| L. salivarius DPC6005 | Salivaricin P producer | 2 |
Pulsed-field gel electrophoresis analysis.
Differentiation of the L. salivarius isolates was confirmed by pulsed-field gel electrophoresis (PFGE) as previously described (24). The megaplasmid content of the strains was also determined by PFGE following S1 nuclease treatment of high-molecular-weight DNA, also previously described (17).
DNA amplification, sequencing, and analysis.
Template DNA was extracted from L. salivarius DPC6005 and L. salivarius DPC6488 to amplify the genetic loci responsible for salivaricin production in the corresponding strains using primers designed to be specific for the locus of abp118 (accession number AF408405). A series of abp118-specific primers were used to amplify the salivaricin P locus of strain DPC6005 by routine PCR, generating a contiguous sequence of approximately 13 kb. The primer pair 5′ CCGCCGATATACTATTCGTGG 3′ and 5′ GAGAGTTAGACCTGATGAAG 3′ was also used to amplify the 13-kb salivaricin P gene cluster in its entirety using Extensor Hi-Fidelity PCR master mix (Abgene, Surrey, United Kingdom) under conditions recommended by the manufacturer.
A PCR amplicon previously generated using DPC6488 template DNA and primers specific for the amplification of the salivaricin P structural genes (24) generated approximately 900 bp of sequence data in the present study. Following sequence analysis, the abp118-specific primer pair described above was also employed to amplify the 12-kb salivaricin gene cluster of strain DPC6488. Oligonucleotide primers were synthesized by Sigma-Genosys (Poole, Dorset, United Kingdom), and purified amplicons were sequenced by Beckman Coulter Genomics (Essex, United Kingdom). Alignments and analyses of sequence data were performed using LASERGENE 6 software (DNAStar Inc., Madison, WI). Database searches were performed using the basic local alignment search tool (BLAST) on the National Center for Biotechnology Information (NCBI) server (http://www.ncbi.nlm.nih.gov). Open reading frames (ORFs) were identified using LASERGENE 6, ORF finder, and glimmer on the NCBI server and Genemark gene prediction software (http://exon.biology.gatech.edu).
Microarray-based comparative genomic hybridization.
Comparative genomic hybridizations were performed using a highly replicated custom microarray (Agilent Technologies, CA) designed based on the genome of L. salivarius UCC118, as previously described (5, 11, 28). The experimental procedures for genomic DNA extraction, fragmentation, and fluorescent labeling and cohybridization experiments with fluorescently labeled genomic DNA (gDNA) of the test and reference strains employed in this study were recently described (28). Two dye swap replicate hybridizations were performed for each strain tested, and self-hybridization of the reference strain was carried out as a control experiment. Analyses of the microarray data were also performed as described by Raftis et al. (28). The sequencing data obtained for our strains were in agreement with the empirically determined cutoff intervals used to discriminate between the presence, divergence, and absence of genes previously selected on the basis of a comparison of the output data of a BLASTn comparison of the UCC118-specific oligonucleotide probe set with the sequence of the draft genome of the type strain L. salivarius DSM20555 (accession number NZ_ACGT00000000), with the log2-transformed signal ratios determined for the hybridization reaction of the gDNA of the corresponding strain (28). Therefore, these cutoff intervals were also applied to the present data set. Cutoff intervals of ≥−1.5, < −1.5 and ≥−2.4, <−2.4 and ≥−4.5, <−4.5 and ≥−5.8, and <−5.8 corresponded to highly conserved, conserved, divergent, highly divergent, and absent features in the test strain relative to L. salivarius UCC118, respectively. Hierarchical clustering of strains was performed using the complete linkage clustering, and the results were visualized using Genesis software (32).
Purification of the antimicrobial peptides produced by L. salivarius DPC6488.
The antimicrobial peptides were purified from a 2-liter overnight culture of L. salivarius DPC6488 grown in MRS medium. The cells were harvested by centrifugation at 8,000 × g for 15 min, resuspended in 250 ml 70% (vol/vol) propan-2-ol containing 0.1% (vol/vol) trifluoroacetic acid (TFA), and stirred at room temperature for 3 h. Cells were removed by centrifugation and propan-2-ol by rotary evaporation. The resultant preparation was applied to a 5.0-g (20-ml volume) Strata C18 solid-phase extraction (SPE) column (Phenomenex, Cheshire, United Kingdom) preequilibrated with methanol and water. The column was washed with 35% (vol/vol) ethanol, and the antimicrobial peptides were subsequently eluted with 70% (vol/vol) propan-2-ol containing 0.1% (vol/vol) TFA. Following removal of the propan-2-ol from the preparation, 4-ml aliquots were applied to a Jupiter Proteo C12 reversed-phase–high-pressure liquid chromatography (RP-HPLC) column (250.0 by 10.0 mm; 4-μm particle size; 90-Å pore size; Phenomenex) preequilibrated with 25% (vol/vol) acetonitrile, 0.1% (vol/vol) TFA. The column was developed in a gradient of 25% to 55% (vol/vol) acetonitrile containing 0.1% (vol/vol) TFA from 5 to 50 min at a flow rate of 2.5 ml min−1. Individual fractions were assayed by well diffusion using the sensitive indicator strain Lactobacillus delbrueckii subsp. bulgaricus LMG 6901, and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) was performed to determine fractions containing the peptides of interest. Fraction 27, with a mass of 4,433 Da, corresponding to the salivaricin B peptide, was applied to a Luna analytical SCX cation-exchange HPLC column (250.0 by 4.6 mm; 5-μm particle size; 100-Å pore size; Phenomenex) for further purification, following removal of acetonitrile by rotary evaporation. Using buffer A (20 mM potassium phosphate containing 25% [vol/vol] acetonitrile, pH 2.5) and buffer B (20 mM potassium phosphate, 25% [vol/vol] acetonitrile, 1 M potassium chloride, pH 2.5), the column was preequilibrated with 10% buffer B and subsequently developed in a gradient of 10% to 65% buffer B from 5 to 45 min at a flow rate of 1.0 ml min−1. Individual fractions of interest were applied to a 200-mg (3-ml volume) Strata C18 SPE column (Phenomenex) preequilibrated with methanol and water. The column was washed with 30% (vol/vol) ethanol and 40% (vol/vol) propan-2-ol, followed by the elution of the antimicrobial component with 70% (vol/vol) propan-2-ol containing 0.1% (vol/vol) TFA. Similarly, fractions 38 to 40, with masses of 5,655 Da and 5,267 Da, corresponding to the respective SalTα and SalTβ peptides, were pooled, concentrated, and applied to the Luna analytical SCX cation-exchange HPLC column for separation and purification as described above, as were fractions 44 to 46, for which a mass of 4,117 Da was detected, corresponding to the mature salivaricin L peptide. Bacteriocin activity was monitored throughout purification by well diffusion assay using the sensitive indicator strain L. delbrueckii subsp. bulgaricus LMG 6901. MALDI-TOF MS was performed as previously described (7).
Peptide synthesis.
Synthetic analogues of mature salivaricin B and salivaricin L were synthesized according to the deduced amino acid sequences of slnT3 and ORF13 (slnL) using microwave-assisted solid-phase peptide synthesis (MW-SPPS) performed on a CEM Liberty microwave peptide synthesizer using an H-Ser-HMPB-ChemMatrix and an H-Cys(TRT)-HMPB-ChemMatrix resin (PCAS Biomatrix Inc., Quebec, Canada), respectively. The molecular masses of the synthetic analogues were confirmed using MALDI-TOF MS, and the antimicrobial activity of the crude peptides was assayed by well diffusion. Synthetic salivaricin L was purified by RP-HPLC using a Jupiter C5 column (250.0 by 10.0 mm; 10-μm particle size; 300-Å pore size; Phenomenex) developed in a gradient from 25% to 55% (vol/vol) acetonitrile containing 0.1% TFA from 10 to 50 min at a flow rate of 2.5 ml min−1. Fractions with the desired molecular mass, identified by MALDI-TOF MS, were pooled and lyophilized using a Genevac HT 4X lyophilizer (Genevac Ltd., Ipswitch, United Kingdom). The peptide was dissolved in 50% (vol/vol) acetonitrile at a concentration of 5 mg/ml and stored at −20°C under nitrogen. Appropriate dilutions of the peptide in 50 mM potassium phosphate buffer, pH 6.8, were used for bacteriocin assays.
Specific activity analysis of salivaricin L.
A microtiter plate assay system was used to determine the minimum concentration of the synthetic salivaricin L analogue required to inhibit growth of the indicators, L. innocua DPC3572 and L. monocytogenes NCTC 11994, by 50% (MIC50). The microtiter plate was first treated with bovine serum albumin (BSA) to prevent adherence of the peptide to the sides of the wells, as described previously (12). Each plate included triplicate assays at each concentration examined. Each well contained a total volume of 200 μl, comprised of purified synthetic salivaricin L and 150 μl of a 1-in-10 dilution of the indicator culture (A590 = 0.5) in BHI broth. Control wells contained medium only (blanks) or untreated indicator culture. The microtiter plate cultures were then incubated at 37°C, and the optical density at 590 nm (OD590) was recorded at hourly intervals for 6 h (Genios Plus; Tecan, Switzerland). Triplicate readings were averaged, and the blanks were subtracted from these readings. The amount of bacteriocin that inhibited the indicator strain by 50% was defined as 50% of the final OD590 ± 0.05 of the untreated control culture.
RESULTS
The seven L. salivarius test strains included in this study were of either human or porcine intestinal origin (Table 1) and were specifically selected for their ability to produce bacteriocins, which we propose constitutes an important probiotic trait (6, 31, 34). For comparative reasons, one isolate of human origin with a bacteriocin-negative phenotype, DPC6196, was also included in the genotypic analysis, as it harbors homologues of the abp118 bacteriocin structural genes (2). Due to the localization of the abp118 bacteriocin gene cluster on the megaplasmid of strain UCC118 (17), the megaplasmid content of the test strains was first compared with that of UCC118.
Analysis of the megaplasmid content of bacteriocin-producing L. salivarius isolates.
PFGE of total genomic DNA confirmed that the seven test strains were genetically distinct (data not shown), and the megaplasmid content of each, as determined by S1 PFGE, is outlined in Table 2. The human-derived isolates DPC6196 and DPC6488 were found to harbor megaplasmids of 180 and 195 kb, respectively. Smaller plasmids were also identified in these strains, two in DPC6488 of approximately 20 kb and 30 kb and one in DPC6196 of approximately 48 kb. The porcine-derived isolates, DPC6005, DPC6502, and 7.3, each contained megaplasmids of approximately 242 kb, similar to pMP118 (5), while DPC6027 and DPC6189 contained larger plasmids of 320 kb and 360 kb, respectively. Multiple megaplasmids were observed in L. salivarius DPC6189 and L. salivarius 7.3, which both appear to harbor an additional linear megaplasmid of approximately 195 kb. The linear nature of these megaplasmids was determined when, under different switching conditions, PFGE analyses revealed that the corresponding bands migrated to the same locations on the gel regardless of whether DNA from these isolates was digested with S1 nuclease (data not shown).
Table 2.
Megaplasmid content of intestinal L. salivarius strains
| Plasmid | Approx plasmid size (kb) for strain: |
|||||||
|---|---|---|---|---|---|---|---|---|
| UCC118 | DPC6196 | DPC6488 | DPC6502 | DPC6027 | DPC6189 | 7.3 | DPC6005 | |
| Circular megaplasmids | 242 | 180 | 195 | 242 | 320 | 360 | 195 | 242 |
| Linear megaplasmids | 195 | 195 | ||||||
| Smaller plasmids | 44, 20 | 48 | 30, 20 | |||||
Genomic diversity of L. salivarius strains.
Array comparative genomic hybridization (CGH) analyses revealed considerable genomic diversity between strain UCC118, the five Bac+ (bacteriocin-producing) porcine isolates, and the human-derived Bac+ strain DPC6488 and Bac− (bacteriocin negative, despite harboring homologues of the abp118 structural genes) strain DPC6196, consistent with results reported by Raftis et al. (28). We particularly noted a high level of plasticity within the UCC118 megaplasmid-associated bacteriocin abp118-encoding locus.
Genetic diversity of the salivaricin bacteriocin locus in L. salivarius.
The genetic determinants responsible for the production of abp118 are comprised of the abp118 α and β structural genes, accompanied by the genes encoding a cognate immunity protein, a three-component regulatory system, and an ABC transporter and transport accessory protein that are responsible for cleavage and secretion of the mature active bacteriocin (13) (Fig. 1). Six additional ORFs are also present on the 10.7-kb abp118 locus; three of them encode putative bacteriocin-like precursor peptides, two of which have no homologues while one was identified as a pre-salivaricin B homologue (4, 13). Each of the L. salivarius test isolates included in this study is bacteriocinogenic, with the exception of DPC6196, and sequence analysis revealed that the abp118 gene cluster shares greater than 90% sequence similarity with that of salivaricin P (data not shown). Despite this, CGH revealed considerable genetic diversity with respect to the bacteriocin locus across the test strains (Fig. 2). The porcine intestinal strain DPC6502 was particularly notable by virtue of lacking abp118-related homologues, despite having an antimicrobial-producing phenotype (Fig. 2) (24). This observation eliminates the possibility that the strain produces abp118, salivaricin P, and/or salivaricin B. Although CGH data indicated that the bacteriocin structural genes, the genes encoding the response regulator and the transport system of abp118, were conserved in each of the remaining porcine-derived isolates, DPC6005, DPC6027, DPC6189, and 7.3, the genes encoding the immunity and induction peptides and the histidine kinase component of the salivaricin regulatory systems of these isolates were divergent from their respective abp118 counterparts, as illustrated in Fig. 2. Indeed, sequence analysis of the salivaricin P locus of the prototype producing strain DPC6005 (data not shown) confirmed this.
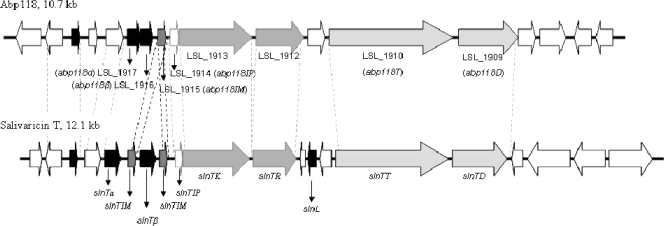
Fig. 1.
Comparative representation of the abp118 and salivaricin T gene clusters. The bacteriocin structural and predicted immunity and regulation and transport genes are indicated by black, charcoal, and gray arrows, respectively. ORFs encoding unknown proteins are represented by white arrows. The homologues of the putative protein products encoded by the salivaricin T gene cluster are listed in Table 3. The most notable regions of similarity between the respective structural and immunity genes are highlighted by the black dashed lines.
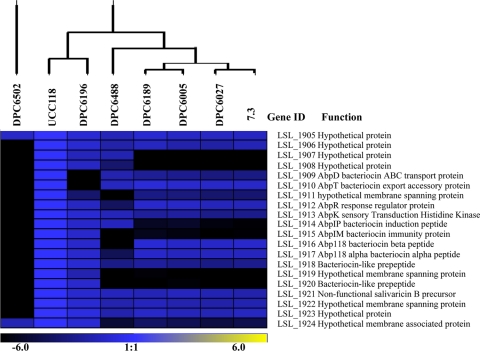
Fig. 2.
Graphical representation of the genetic diversity of the bacteriocin locus within L. salivarius test strains as determined by aCGH. The black, blue, and yellow regions represent absence, conservation, and overrepresentation of CDS, respectively, corresponding to the color legend.
Overall, the abp118 locus was most highly conserved in the human isolate DPC6196 (Fig. 2). Although displaying a Bac− phenotype, this isolate harbors homologues of abp118α and abp118β, differing by just 1 and 2 nucleotides, respectively (2). CGH analysis revealed divergence with respect to the abpT (LSL_1910) and abpD (LSL_1909) homologues, required for secretion of the mature active bacteriocin, in DPC6196, which is likely responsible for the Bac− phenotype of the strain. Previously, PCR analysis using primers specific for the salivaricin P structural genes indicated the presence of related genes in L. salivarius DPC6488 (24). However, the unique genetic variability pattern of this neonatal isolate instead indicated that the genes corresponding to abp118α (LSL_1917) and abp118β (LSL_1916) were divergent and absent, respectively (Fig. 2). In contrast, the genes encoding the abp118 regulatory and transport systems were perfectly conserved in this strain relative to UCC118, as was the gene encoding pre-salivaricin B (LSL_1921). As this strain is of human origin, an important consideration in the selection of commercial probiotics, the antimicrobial activity and corresponding genetic determinants of the isolate were further investigated.
Analysis of the gene products encoded by the salivaricin locus of L. salivarius DPC6488.
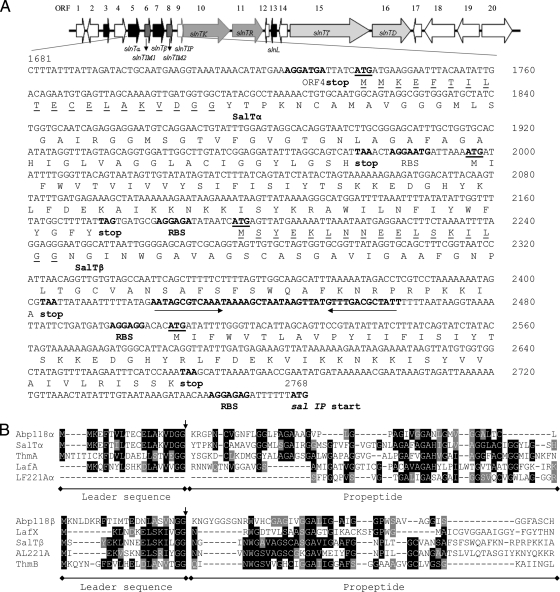
A single primer pair successfully amplified the individual salivaricin loci of L. salivarius UCC118, DPC6005, and DPC6488 in their entirety. Sequencing of the corresponding amplicon of L. salivarius DPC6488 allowed the elucidation of the most divergent region of the bacteriocin locus of the strain, which on the basis of CGH data corresponds to the sequence between LSL_1913 and LSL_1924 of strain UCC118 (Fig. 2), and confirmed the conservation of the regulatory and transport systems of abp118 in the strain. In silico analysis of the sequence data resulted in the identification of 20 ORFs (Fig. 1 and Table 3), five of which, ORF3 to -5, -7, and -13, encoded bacteriocin-like prepeptides. The putative products of ORF3 and ORF4 (correspondingly designated slnT3 and slnT4) were 98% and 97% identical to the salivaricin B precursor peptide (LSL_1921) and a bacteriocin-like prepeptide (LSL_1918) of UCC118, respectively. The deduced products of ORF5 and ORF7 also resembled bacteriocin precursor peptides with double-glycine leader sequences and were designated slnTα and slnTβ (Fig. 3A). The first 57 nucleotides of slnTα closely resemble those encoding the leader peptide of Abp118α and Sln1, differing by 2 nucleotides (and, in turn, 1 aa) (Fig. 3B). However, no significant homology was observed between the corresponding propeptide (61 aa) and Abp118α (45 aa). Indeed a BLAST search revealed that this peptide instead shared 47% identity with ThmA, one component of the two-peptide bacteriocin thermophilin 13 produced by S. thermophilus (20) (Fig. 3B). The bacteriocin-like precursor peptide encoded by slnTβ displayed similarity to acidocin LF221 A, one peptide of a putative two-component bacteriocin produced by Lactobacillus gasseri (18) (Fig. 3B). Only partial sequence of the complementary peptide of LF221 A (37 C-terminal amino acids) was available for comparison with the propeptide encoded by slnTα, with which it shared 30% similarity (Fig. 3B). While the leader sequence of SalTβ also shares 42% similarity with that of Abp118α, no significant similarity was observed between the leader sequence of SalTβ and that of Abp118β (Fig. 3B). Indeed, the SalTβ leader sequence was most similar (59%) to that of the LafX peptide of lacticin F, produced by Lactobacillus johnsonii (Fig. 3B) (14), while, perhaps most notably, the propeptide encoded by slnTβ shares 43% similarity with ThmB, the complementary peptide of ThmA (20).
Table 3.
Homologues of the deduced proteins encoded by the salivaricin T bacteriocin locus of L. salivarius DPC6488
| ORF (gene) | Size (aa) | Function | Homologue | % Identitya (no./total) | Reference |
|---|---|---|---|---|---|
| 1 (slnT1) | 65 | Conserved hypothetical protein | Conserved hypothetical protein of L. salivarius DSM20555 | 95 (62/65) | EEJ73426b |
| 2 (slnT2) | 87 | Conserved hypothetical protein | Conserved hypothetical protein of L. salivarius DSM20555 | 100 (87/87) | EEJ73427b |
| 3 (slnT3) | 57 | Bacteriocin-like prepeptide | Salivaricin B prepeptide | 98 (56/57) | 4 |
| 4 (slnT4) | 89 | Bacteriocin-like prepeptide | LSL_1918 of L. salivarius UCC118 | 98 (83/85) | 5 |
| 5 (slnTα) | 80 | Salivaricin T prepeptide SalT alpha | ThmA, amphipathic pore-forming peptide precursor of S. thermophilus | 45 (35/77) | 20 |
| 6 (slnTIM1) | 59 | Putative bacteriocin immunity protein | Abp118 IM (LSL_1915) of L. salivarius UCC118 | 82 (45/55) | 13 |
| 7 (slnTβ) | 75 | Salivaricin T prepeptide SalT beta | Acidocin LF221A produced by L. gasseri | 46 (31/67) | 18 |
| 8 (slnTIM2) | 54 | Putative bacteriocin immunity protein | Abp118 IM (LSL_1915) of L. salivarius UCC118 | 76 (38/50) | 13 |
| 9 (slnTIP) | 38 | Putative induction peptide | Abp118 IP (LSL_1914) of L. salivarius UCC118 | 100 (38/38) | 13 |
| 10 (slnTK) | 429 | Sensory transduction histidine kinase | AbpK of L. salivarius UCC118 | 99 (424/429) | 13 |
| 11 (slnTR) | 265 | Response regulator | Salivaricin response regulator of strain CECT5713/(AbpR 94%) | 95 (249/263) | ADJ79880b |
| 12 | 41 | Hypothetical protein | None | ||
| 13 (SlnL) | 59 | Bacteriocin-like prepeptide | Putative bacteriocin of Streptococcus sp. strain C105 | 60 (35/60) | EFX55741b |
| 14 | 74 | Hypothetical protein | None | ||
| 15 (slnTT) | 719 | Salivaricin T ABC transporter protein | AbpT (LSL_1910) of L. salivarius UCC118 | 99 (709/719) | 13 |
| 16 (slnTD) | 384 | Salivaricin T export accessory protein | AbpD (LSL_1909) of L. salivarius UCC118 | 97 (370/384) | 13 |
| 17 | 63 | Truncated transposase | Transposase ISLasa2b, IS1223 family (LSL_1957) | 97 (59/61) | 5 |
| 18 | 246 | Truncated transposase | Transposase ISLasa2b, IS1223 family (LSL_1957) | 94 (231/246) | 5 |
| 19 | 172 | Transposase | IS1223 family transposase (LSL_1958, LSL_0049) of UCC118 | 98 (169/172) | 5 |
| 20 | 273 | Hypothetical protein | Conserved hypothetical protein of L. salivarius CECT5713 | 95 (257/270) | ADJ79877b |
Percent identity was determined using BLAST.
Accession number of the sequence directly submitted to EMBL database.
Fig. 3.
Nucleotide sequence and deduced peptide sequence of the salivaricin T structural genes and the immunity genes of the salivaricin locus of L. salivarius DPC6488. (A) The 1,087-bp DNA sequence shown encodes the component peptides of salivaricin T produced by L. salivarius 6488 (SalTα and SalTβ), together with their leader sequences (underlined). The GG-processing sites are indicated by solid triangles. Two putative immunity genes are present downstream of slnTα and slnTβ. Putative ribosome binding sites (RBS) and start and stop codons are indicated in boldface, and start codons are also underlined. An inverted-repeat sequence (indicated by arrows), typical of a Rho-independent terminator sequence, was identified downstream of slnTβ, also indicated in boldface. (B) Alignment of salivaricin T component precursor peptides with those of abp118, acidocin LF221 A, lactacin F, and thermophilin 13. The arrows and black and gray shading indicate the cleavage sites of the double glycine leader sequences and identical and similar conserved residues of the alignment, respectively. The leader sequences of the Abp118α and SalTα prepeptides and SalTβ and LafX prepeptides share 95% and 59% identity, respectively. The SalTα propeptide displays the greatest identity (46%) with ThmA, and their respective complementary peptides, SalTβ and ThmB, share 33% identity. The SalTβ propeptide shares the greatest identity (41%) with acidocin LF221 A, and the partial available sequence of the LF221 A complementary peptide shares 24% identity with the propeptide of SalTα.
Further in silico analysis of this cluster identified two ORFs encoding putative immunity proteins downstream of the potential bacteriocin structural genes slnTα and slnTβ (Fig. 3A). The deduced protein sequences corresponding to slnT IM1 (ORF6) and slnT IM2 (ORF8) displayed 81% and 76% identity with Abp118 IM. Despite being present in two copies, these LSL_1915 homologues were not detected by CGH due to the divergence in the nucleotide sequences of the genes in DPC6488 (72% and 69% identity, respectively, with abpIM). Three ORFs downstream of the putative bacteriocin immunity gene, ORF9, -10, and -11, encode a putative induction peptide, a sensory transduction histidine kinase, and a response regulator, which shared 100%, 99%, and 94% identity with their respective abp118 counterparts, thus confirming conservation of the regulatory system of abp118 in strain DPC6488. The deduced products of ORF12 and -14 did not share homology with any previously known proteins. The 59-aa putative bacteriocin prepeptide encoded by ORF13 (slnL) consists of a double-glycine leader sequence of 18 aa and a mature peptide of 41 aa designated salivaricin L and shared the greatest homology (59% identity) with a putative bacteriocin precursor of the Streptococcus sp. strain C150 genome (accession no. EFX55741). This putative bacteriocin precursor also shared very weak homology (38% and 37%) with the precursor peptides of cerein 7B, produced by Bacillus cereus Bc7 (23), and sakacin Q, produced by Lactobacillus sakei (21), respectively. Interestingly, both cerein 7B and sakacin Q are single-peptide class IId bacteriocins that are produced simultaneously with cerein 7A and sakacin P by their respective producing strains (21, 23). The deduced protein products of ORF15 and -16 share 99% and 97% identity with those involved in the transport of abp118, AbpT, and AbpD, respectively, thus confirming conservation of the abp118 transport system in strain DPC6488. Three ORFs downstream of the putative bacteriocin transport genes encoded proteins homologous to the IS1223 family transposases. The protein products of overlapping ORF17 and -18 shared 97% and 94% identity with the 61 C-terminal amino acids and 246 N-terminal amino acids of transposase ISLasa2b encoded by LSL_1957 approximately 30 kb downstream of the abp118 bacteriocin locus on pMP118. In addition, the deduced product of ORF19 shares 98% identity with transposase ISLasa1b encoded by LSL_1958 of L. salivarius UCC118, indicative of a possible recombination event. The final ORF identified on the salivaricin locus of DPC6488 encoded a hypothetical protein of 274 aa, the C and N termini of which shared 97% and 87% identity with the smaller hypothetical proteins encoded by LSL_1907 (174 aa) and LSL_1908 (76 aa) of the abp118 locus, respectively.
Isolation and characterization of the bacteriocin-like peptides produced by L. salivarius DPC6488.
The spectrum of activity of L. salivarius DPC6488, as determined by well diffusion assay of the neutralized cell-free supernatant (CFS), was previously found to be limited to 12 closely related species of lactic acid bacteria (including Enterococcus, Lactobacillus, and Streptococcus spp.) of a total of 62 indicator strains investigated. This activity was lost upon protease treatment with proteinase K, α-chymotrypsin, trypsin, or pepsin (24). Cross sensitivity assays using the CFS of DPC6488 and of the abp118 and salivaricin P producers (UCC118 and DPC6005, respectively) revealed that all 3 strains are immune to both abp118- and the DPC6488-associated antimicrobials. Interestingly, however, both human isolates were sensitive to the CFS of the salivaricin P producer DPC6005, despite the fact that their respective immunity peptides share greater than 76% sequence identity.
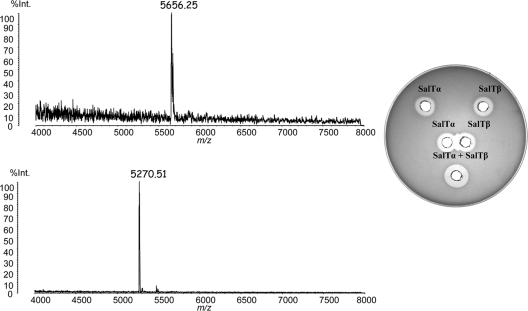
Further investigations were performed to establish the nature of the antimicrobials produced by DPC6488. Mass spectral analysis revealed that peptides with masses corresponding to those of the mature products of slnT3, slnTα, slnTβ, and slnL were present in the culture supernatant of DPC6488. To determine if these peptides were responsible for the antimicrobial activity of the strain, the peptides were separated and purified using cation-exchange chromatography. MS data confirmed the predicted molecular masses of the mature peptides encoded by slnTα, slnTβ, and slnL (5,655 Da, 5,269 Da, and 4,117 Da, respectively) in the individually eluted active fractions. The individual SalTα and SalTβ peptides exhibited antimicrobial activity against the sensitive indicator strain Lactobacillus delbrueckii subsp. bulgaricus LMG 6901, which was further enhanced when they were combined (Fig. 4). Thus, slnTα and slnTβ appear to encode a two-peptide bacteriocin that most closely resembles thermophilin 13, which we designated salivaricin T.
Fig. 4.
MALDI-TOF MS data for the salivaricin T component peptides SalTα (top left) and SlaTβ (bottom left) purified from a culture of strain L. salivarius DPC6488. Individual and combined antimicrobial activities of SalTα and SlaTβ against L. delbrueckii subsp. bulgaricus LMG 6901 (right).
MS analysis also established that the mature salivaricin B analogue encoded by salT3 is secreted by strain DPC6488 and identified the fraction containing a peptide of corresponding mass (4,433 Da). However, this peptide did not inhibit the L. delbrueckii subsp. bulgaricus indicator strain, nor did it display the anti-Listeria activity previously attributed to salivaricin B (reference 4 and data not shown). As noted above, a gene encoding a homologue of the salivaricin B precursor peptide is also located on the abp118 gene cluster (LSL_1921) but was designated nonfunctional in L. salivarius UCC118 (5, 13). Indeed, CGH demonstrates that this gene is conserved in six of the seven test strains employed in this study, and DNA sequence and MS data confirmed that the peptide encoded by slnT3 of DPC6488 shares 100% identity with salivaricin B. To investigate further, a synthetic analogue of mature salivaricin B was generated based on the deduced amino acid sequence of slnT3. However, we were again unable to detect antimicrobial activity from the synthetic peptide (data not shown). Furthermore, when either the synthetic analogue or the purified salivaricin B-containing fraction was combined with those containing SalTα and SalTβ or the individual purified component peptides of salivaricin P, Sln1 and Sln2, salivaricin B failed to enhance their antimicrobial activity against L. delbrueckii subsp. bulgaricus LMG 6901 or L. innocua (data not shown). These findings thus question the previously reported anti-Listeria potency of this bacteriocin (4).
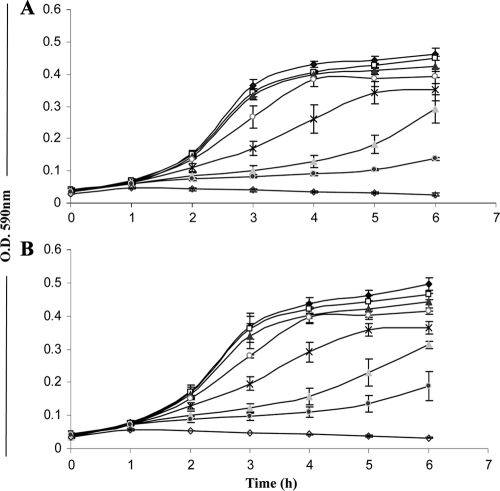
Interestingly, production of the fourth bacteriocin-like prepeptide encoded on the salivaricin gene cluster of strain DPC6488, with similarity to one-peptide class IId bacteriocins, was also confirmed by MALDI-TOF MS. While the peptide inhibited 3 of the 12 indicator strains sensitive to salivaricin T, L. delbrueckii subsp. bulgaricus, L. delbrueckii subsp. lactis, and L. ruminis, synergistic activity was not observed when it was combined with the salivaricin T component peptides SalTα and SalTβ. Moreover, the purified salivaricin L peptide also exhibited anti-Listeria activity. A synthetic analogue of the mature 41-aa peptide displaying activity similar to that of the natural peptide was employed to determine the specific activity of the bacteriocin, revealing an MIC50 of 20 μM for both L. innocua DPC3572 and L. monocytogenes NCTC 11994 (Fig. 5).
Fig. 5.
Inhibitory effects of synthetic salivaricin L on the growth of indicator strains L. innocua DPC3572 (A) and L. monocytogenes NCTC 11994 (B) at concentrations of 0 μM (♦), 1.0 μM (□), 5.0 μM (▲), 10.0 μM (○), 15.0 μM (×), 20.0 μM (▵), and 50.0 μM (♢). The error bars represent standard deviations based on triplicate data.
Given the inactivity of salivaricin B, and the antimicrobial activity of purified SalTα, SalTβ, and salivaricin L, it is apparent that the inhibition of L. delbrueckii subsp. bulgaricus strain LMG 6901 by L. salivarius DPC 6488 can be attributed to two novel bacteriocins encoded within a single gene cluster, which we designated salivaricin T, corresponding to the two-component peptide products encoded by slnTα and slnTβ, and salivaricin L, corresponding to the one-peptide class IId bacteriocin encoded by salL. Furthermore, the regulatory and export systems of the corresponding salivaricin locus are analogous to those of abp118 and salivaricin P, thereby highlighting the ability of these systems to recognize and produce various antimicrobial peptides.
DISCUSSION
CGH analysis revealed a high level of intraspecies diversity within the intestinal L. salivarius isolates employed compared to the genome of L. salivarius UCC118, largely consistent with a recent survey of the genomic diversity of 33 L. salivarius strains (28).
Bacteriocin production is a megaplasmid-encoded feature important for the probiotic functionality of L. salivarius UCC118 (5, 6), and CGH confirmed the presence of repA-type megaplasmids in each of the test strains investigated in this study. The megaplasmids of DPC6488 and DPC6196 were considerably smaller than pMP118; notably, a 67-kb nonfunctional conjugation transfer locus present in UCC118 (10) is absent in these strains. Indeed, this locus is also absent in DPC6502, despite the fact that it harbors a megaplasmid similar in size to pMP118. This indicates a considerable number of additional megaplasmid-encoded features in DPC6502, which is also the case for DPC6027 and DPC6189, which harbor megaplasmids considerably larger than pMP118. CGH analysis of the bacteriocin locus in the porcine salivaricin P-producing test strain DPC6005 produced results that were consistent with subsequent sequence analysis. This revealed that the salivaricin P gene cluster was organized in an arrangement similar to that of the genetic locus of abp118 (13, 27), with the most notable difference between the genes encoding the induction peptides (60% identity). It was not surprising that the N-terminal region of the corresponding sensory histidine kinase genes also varied slightly (93% identity). Similar variations were previously observed between the regulatory operons of the pln loci of Lactobacillus plantarum C11 and L. plantarum NC8, which are also otherwise highly homologous (19). Indeed, similar levels of variability were observed with respect to the bacteriocin loci of all of the salivaricin P-producing porcine isolates relative to UCC118, indicating that they may all be derived from a single ancestor. CGH also revealed that the bacteriocin-negative phenotype of the human isolate DPC6196 is likely due to a genetic defect in the bacteriocin transport system in the isolate.
Sequence analysis of the salivaricin locus of strain DPC6488 further validated the CGH data. Although the genes involved in regulation and transport of abp118 were highly conserved in DPC6488, the strain harbors structural genes for the production of two novel antimicrobials, a two-component class IIb bacteriocin, salivaricin T, and a one-peptide class IId bacteriocin, salivaricin L. It is understandable that strain DPC6488 was previously mistakenly characterized as a salivaricin P producer, as the primers designed to amplify the bacteriocin structural genes were complementary to the sequence encoding the double-glycine leader of Sln1 and the immunity genes of salivaricin P, which share considerable identity with the leader sequence of SalTα and the respective immunity genes of the corresponding salivaricin locus. Notably, the porcine isolate DPC6502, which was also previously characterized as a salivaricin P producer, lacked all abp118-related homologues, and so, the Bac+ phenotype of this strain will be the basis of further investigation.
The leader sequence of the SalTα precursor is almost identical to that of the Abp118α and Sln1 prepeptides (95% identity, with just 1 conservative amino acid difference [I/V] in 19), which may be a requirement for the efficient processing and export of the mature active peptide by the conserved ABC transporter. Indeed, while the mature salivaricin T peptides most closely resembled the component peptides of thermophilin 13 (20), the proteins involved in providing immunity to, regulating production of, and transporting these peptides are analogous to those of abp118. However, it should be noted that at present the thermophilin 13 gene cluster has not been reported. The presence of two putative immunity genes downstream of each of the salivaricin T structural genes is interesting, and this fact, combined with the individual activity of each of the component peptides, may indicate that salivaricin T evolved from two synergistically acting one-peptide bacteriocins. Interestingly, the one-peptide class IId bacteriocins cerein 7B and sakacin Q, with which salivaricin L shares weak homology, are also produced simultaneously with cerein 7A and sakacin P by their respective producing strains, B. cereus and L. sakei (21, 23). As observed with salivaricin T and salivaricin L, neither of these pairs of simultaneously produced bacteriocins exhibits synergistic activity. Moreover, salivaricin L exhibited activity antagonistic to the gastrointestinal pathogen L. monocytogenes, with an MIC50 of 20 μM.
L. salivarius DPC6488 was selectively isolated from the neonatal fecal population as a consequence of its antimicrobial phenotype. Therefore, it is tempting to suggest that salivaricin T and salivaricin L production may be an important mechanism for host colonization and prevalence of the producing strain, allowing it to outcompete closely related populations within the intestine. Notwithstanding this hypothesis, the apparent ability of L. salivarius to produce very differing bacteriocins using the same cellular machinery suggests a hitherto unknown versatility with respect to the production of this dominant probiotic trait.
ACKNOWLEDGMENTS
We thank Orla O'Sullivan and Rita Hickey for helpful discussions.
Eileen O'Shea is in receipt of a Teagasc Walsh Fellowship. This work was funded by the Food Institutional Research Measure of the Department of Agriculture, Fisheries and Food (grant no. 04R and DC) and the Alimentary Pharmabiotic Centre (which is funded by the Science Foundation of Ireland [SFI] through the Irish Government's National Development Plan). We and the work were supported by SFI (grant no. 07/CE/B1368).
Footnotes
Published ahead of print on 7 October 2011.
REFERENCES
- 1. Bacun-Druzina V., Mrvcic J., Butorac A., Stehlik-Tomas V., Gjuracic K. 2009. The influence of gene transfer on the lactic acid bacteria evolution. Mljekarstvo 59: 181–192 [Google Scholar]
- 2. Barrett E., et al. 2007. Salivaricin P: one of a family of two component anti-listerial bacteriocins produced by intestinal isolates of Lactobacillus salivarius. Appl. Environ. Microbiol. 73: 3719–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cai H., Rodriguez B. T., Zhang W., Broadbent J. R., Steele J. L. 2007. Genotypic and phenotypic characterization of Lactobacillus casei strains isolated from different ecological niches suggests frequent recombination and niche specificity. Microbiology 153: 2655–2665 [DOI] [PubMed] [Google Scholar]
- 4. Cataloluk O. 2001. Molecular characterization of the gene encoding for the salivaricin B activity and its flanking sequences. Turk. J. Biol. 25: 379–386 [Google Scholar]
- 5. Claesson M. J., et al. 2006. Multireplicon genome architecture of Lactobacillus salivarius. Proc. Natl. Acad. Sci. U. S. A. 103: 6718–6723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Corr S. C., et al. 2007. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc. Natl. Acad. Sci. U. S. A. 104: 7617–7621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cotter P. D., et al. 2006. Complete alanine scanning of the two-component lantibiotic lacticin 3147: generating a blueprint for rational drug design. Mol. Microbiol. 62: 735–747 [DOI] [PubMed] [Google Scholar]
- 8. Cotter P. D., Hill C., Ross R. P. 2005. Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3: 777–788 [DOI] [PubMed] [Google Scholar]
- 9. Dawid S., Roche A. M., Weiser J. N. 2007. The blp bacteriocins of Streptococcus pneumoniae mediate intraspecies competition both in vitro and in vivo. Infect. Immun. 75: 443–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fang F., et al. 2008. Characterization of endogenous plasmids from Lactobacillus salivarius UCC118. Appl. Environ. Microbiol. 74: 3216–3228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fang F., et al. 2009. Allelic variation of bile salt hydrolase genes in Lactobacillus salivarius does not determine bile resistance levels. J. Bacteriol. 191: 5743–5757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Field D., et al. 2010. Studies with bioengineered Nisin peptides highlight the broad-spectrum potency of Nisin V. Microb. Biotechnol. 3: 473–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Flynn S., et al. 2002. Characterization of the genetic locus responsible for the production of ABP-118, a novel bacteriocin produced by the probiotic bacterium Lactobacillus salivarius subsp. salivarius UCC118. Microbiology 148: 973–984 [DOI] [PubMed] [Google Scholar]
- 14. Fremaux C., Ahn C., Klaenhammer T. R. 1993. Molecular analysis of the lactacin F operon. Appl. Environ. Microbiol. 59: 3906–3915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hillman J. D. 2002. Genetically modified Streptococcus mutans for the prevention of dental caries. Antonie Van Leeuwenhoek 82: 361–366 [PubMed] [Google Scholar]
- 16. Le Blay G., Lacroix C., Zihler A., Fliss I. 2007. In vitro inhibition activity of nisin A, nisin Z, pediocin PA-1 and antibiotics against common intestinal bacteria. Lett. Appl. Microbiol. 45: 252–257 [DOI] [PubMed] [Google Scholar]
- 17. Li Y., et al. 2007. Distribution of megaplasmids in Lactobacillus salivarius and other lactobacilli. J. Bacteriol. 189: 6128–6139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Majhenic A. C., et al. 2004. DNA analysis of the genes encoding acidocin LF221 A and acidocin LF221 B, two bacteriocins produced by Lactobacillus gasseri LF221. Appl. Microbiol. Biotechnol. 63: 705–714 [DOI] [PubMed] [Google Scholar]
- 19. Maldonado A., Jimenez-Diaz R., Ruiz-Barba J. L. 2004. Induction of plantaricin production in Lactobacillus plantarum NC8 after coculture with specific Gram-positive bacteria is mediated by an autoinduction mechanism. J. Bacteriol. 186: 1556–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marciset O., Jeronimus-Stratingh M. C., Mollet B., Poolman B. 1997. Thermophilin 13, a nontypical antilisterial poration complex bacteriocin, that functions without a receptor. J. Biol. Chem. 272: 14277–14284 [DOI] [PubMed] [Google Scholar]
- 21. Mathiesen G., Huehne K., Kroeckel L., Axelsson L., Eijsink V. G. 2005. Characterization of a new bacteriocin operon in sakacin P-producing Lactobacillus sakei, showing strong translational coupling between the bacteriocin and immunity genes. Appl. Environ. Microbiol. 71: 3565–3574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nes I. F., Diep D. B., Holo H. 2007. Bacteriocin diversity in Streptococcus and Enterococcus. J. Bacteriol. 189: 1189–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Oscáriz J. C., et al. 2006. Purification and sequencing of cerein 7B, a novel bacteriocin produced by Bacillus cereus Bc7. FEMS Microbiol. Lett. 254: 108–115 [DOI] [PubMed] [Google Scholar]
- 24. O'Shea E. F., et al. 2009. Characterization of enterocin- and salivaricin-producing lactic acid bacteria from the mammalian gastrointestinal tract. FEMS Microbiol. Lett. 291: 24–34 [DOI] [PubMed] [Google Scholar]
- 24a. O'Shea E. F., O'Connor P. M., Cotter P. D., Ross R. P., Hill C. 2010. Synthesis of trypsin-resistant variants of the Listeria-active bacteriocin salivaricin P. Appl. Environ. Microbiol. 76: 5356–5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. O'Sullivan O., et al. 2009. Comparative genomics of lactic acid bacteria reveals a niche-specific gene set. BMC Microbiol. 9: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pilasombut K., et al. 2006. Purification and amino acid sequence of a bacteriocin produced by Lactobacillus salivarius K7 isolated from chicken intestine. Songklanakarin J. Sci. Technol. 28: 121–131 [Google Scholar]
- 27. Pingitore V. E., Hebert E. M., Nader-Macias M. E., Sesma F. 2009. Characterization of salivaricin CRL 1328, a two-peptide bacteriocin produced by Lactobacillus salivarius CRL 1328 isolated from the human vagina. Res. Microbiol. 160: 401–408 [DOI] [PubMed] [Google Scholar]
- 28. Raftis E. J., Salvetti E., Torriani S., Felis G. E., O'Toole P. W. 2011. Genomic diversity of Lactobacillus salivarius. Appl. Environ. Microbiol. 77: 954–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rea M. C., et al. 2011. Effect of broad and narrow spectrum antimicrobials on Clostridium difficile and microbial diversity in a model of the distal colon. Proc. Natl. Acad. Sci. U. S. A. 108: 4639–4644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rea M. C., et al. 2010. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc. Natl. Acad. Sci. U. S. A. 107: 9352–9357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stern N. J., et al. 2006. Isolation of a Lactobacillus salivarius strain and purification of its bacteriocin, which is inhibitory to Campylobacter jejuni in the chicken gastrointestinal system. Antimicrob. Agents Chemother. 50: 3111–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sturn A., Quackenbush J., Trajanoski Z. 2002. Genesis: cluster analysis of microarray data. Bioinformatics 18: 207–208 [DOI] [PubMed] [Google Scholar]
- 33. Van Reenen C. A., Chikindas M. L., Van Zyl W. H., Dicks L. M. 2003. Characterization and heterologous expression of a class IIa bacteriocin, plantaricin 423 from Lactobacillus plantarum 423, in Saccharomyces cerevisiae. Int. J. Food Microbiol. 81: 29–40 [DOI] [PubMed] [Google Scholar]
- 34. Walsh M. C., et al. 2008. Predominance of a bacteriocin-producing Lactobacillus salivarius component of a five-strain probiotic in the porcine ileum and effects on host immune phenotype. FEMS Microbiol. Ecol. 64: 317–327 [DOI] [PubMed] [Google Scholar]
- 35. Wescombe P. A., et al. 2006. Megaplasmids encode differing combinations of lantibiotics in Streptococcus salivarius. Antonie Van Leeuwenhoek 90: 269–280 [DOI] [PubMed] [Google Scholar]