Abstract
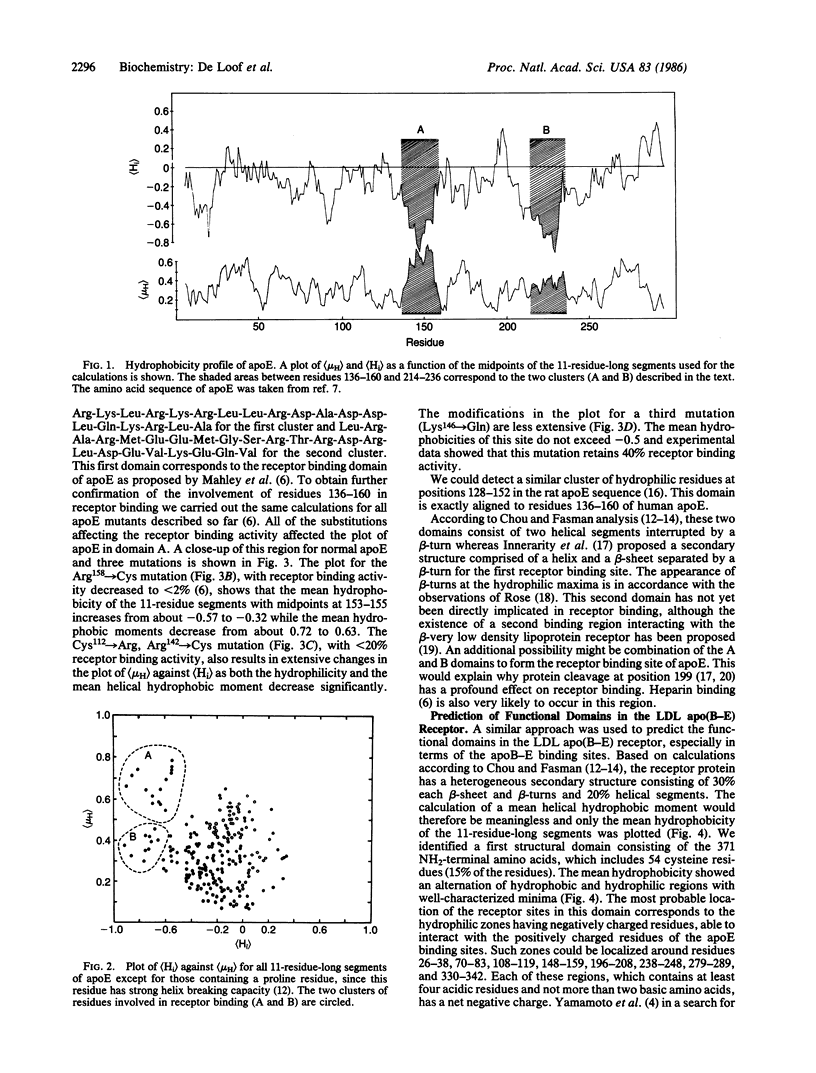
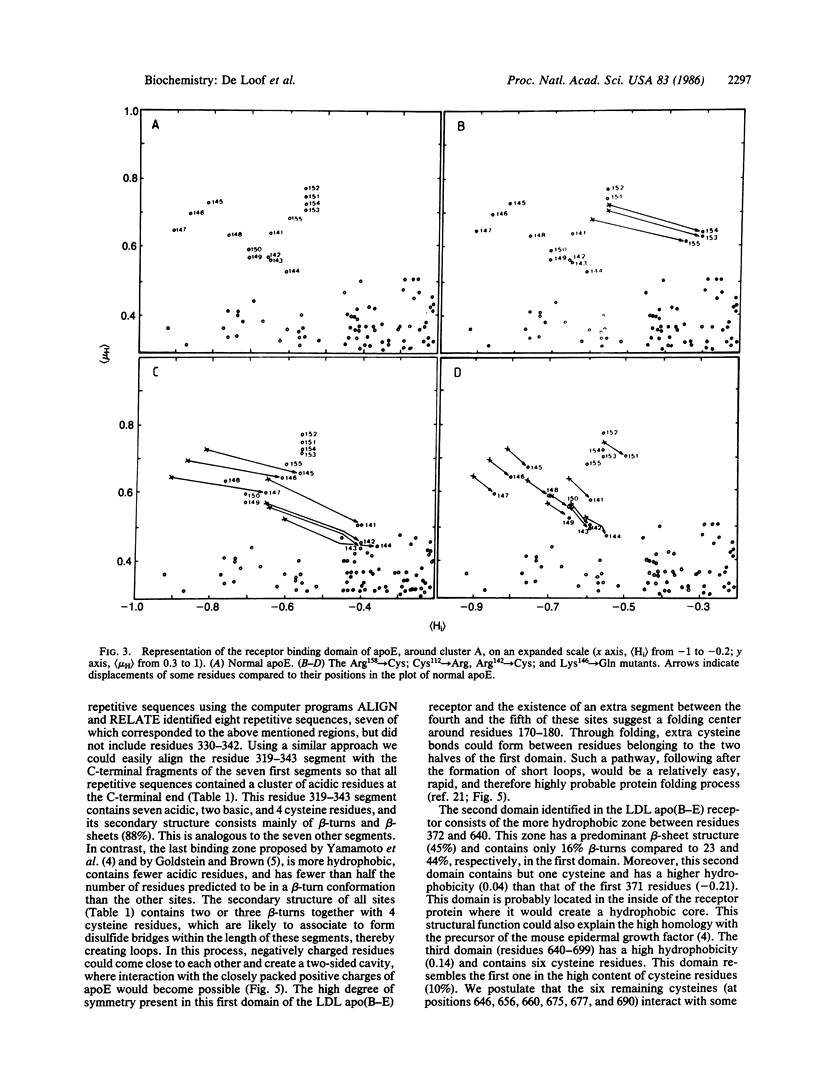
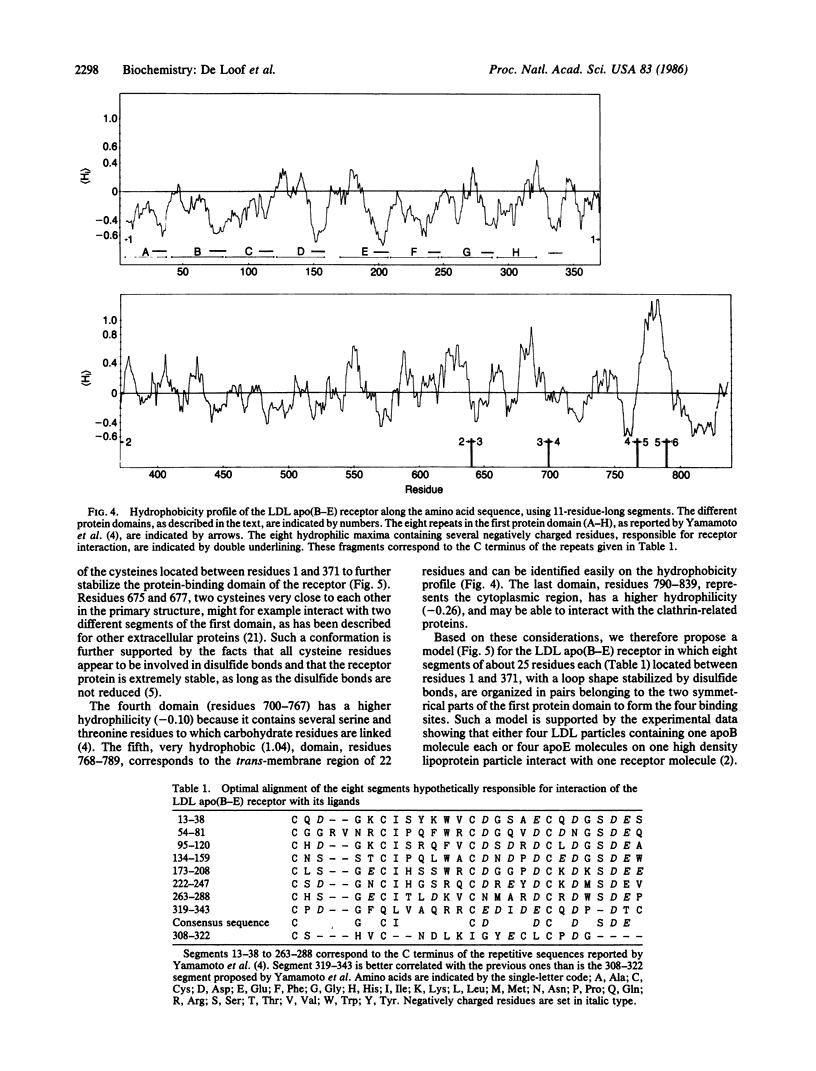
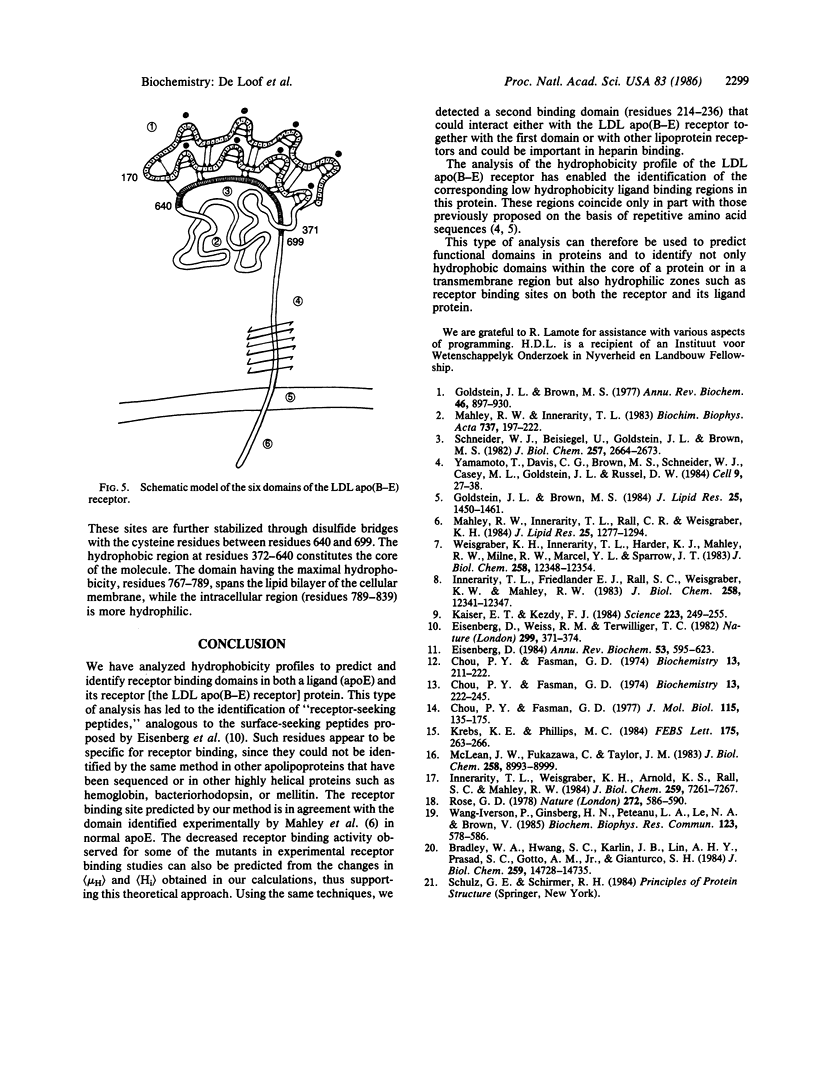
We have used mean hydrophobicity and hydrophobic moment calculations to predict the receptor binding domains in apolipoprotein E and in the low density lipoprotein apolipoprotein B-E receptor. In apolipoprotein E, two receptor binding domains, residues 136-160 and 214-236, having a high hydrophilicity and a high mean helical hydrophobic moment, were identified. The first domain has been located experimentally and mutations influencing the hydrophobicity parameters of the binding site have been shown to affect the receptor binding. The second domain is probably, either separately or in combination with the first domain, involved in receptor binding or in heparin binding. In the low density lipoprotein apolipoprotein B-E receptor, six protein domains were identified. In the first domain (residues 1-371), eight hydrophilic maxima, organized in pairs through disulfide bonds, form the four experimentally observed receptor binding sites. These sites consist of repeats of 26 amino acids but differ from those reported by others [Yamamoto, T., Davis, C. G., Brown, M. S., Schneider, W. J., Casey, M. L., Goldstein, J. W. & Russell, D. W. (1984) Cell 39, 27-38]. The second, more hydrophobic, domain (residues 372-640) forms the core of the receptor, explaining its homology with the precursor of mouse epidermal growth factor, while the cysteine residues in the third domain (residues 641-699), interacting with those of the first domain, further stabilize the molecule. Beyond the fourth hydrophilic domain (residues 700-767), to which carbohydrates are linked, a very hydrophobic membrane spanning region (residues 768-789) could be detected easily. The last domain (residues 790-839), situated in the cytoplasma, contains hydrophilic maxima, as this region might interact with clathrin-related proteins. These data suggest that hydrophobicity analysis can detect and predict protein domains: hydrophilic receptor sites as well as hydrophobic core-forming and membrane-spanning regions.
Full text
PDF
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradley W. A., Hwang S. L., Karlin J. B., Lin A. H., Prasad S. C., Gotto A. M., Jr, Gianturco S. H. Low-density lipoprotein receptor binding determinants switch from apolipoprotein E to apolipoprotein B during conversion of hypertriglyceridemic very-low-density lipoprotein to low-density lipoproteins. J Biol Chem. 1984 Dec 10;259(23):14728–14735. [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Beta-turns in proteins. J Mol Biol. 1977 Sep 15;115(2):135–175. doi: 10.1016/0022-2836(77)90094-8. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Conformational parameters for amino acids in helical, beta-sheet, and random coil regions calculated from proteins. Biochemistry. 1974 Jan 15;13(2):211–222. doi: 10.1021/bi00699a001. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of protein conformation. Biochemistry. 1974 Jan 15;13(2):222–245. doi: 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
- Eisenberg D. Three-dimensional structure of membrane and surface proteins. Annu Rev Biochem. 1984;53:595–623. doi: 10.1146/annurev.bi.53.070184.003115. [DOI] [PubMed] [Google Scholar]
- Eisenberg D., Weiss R. M., Terwilliger T. C. The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature. 1982 Sep 23;299(5881):371–374. doi: 10.1038/299371a0. [DOI] [PubMed] [Google Scholar]
- Goldstein J. L., Brown M. S. Progress in understanding the LDL receptor and HMG-CoA reductase, two membrane proteins that regulate the plasma cholesterol. J Lipid Res. 1984 Dec 15;25(13):1450–1461. [PubMed] [Google Scholar]
- Goldstein J. L., Brown M. S. The low-density lipoprotein pathway and its relation to atherosclerosis. Annu Rev Biochem. 1977;46:897–930. doi: 10.1146/annurev.bi.46.070177.004341. [DOI] [PubMed] [Google Scholar]
- Innerarity T. L., Friedlander E. J., Rall S. C., Jr, Weisgraber K. H., Mahley R. W. The receptor-binding domain of human apolipoprotein E. Binding of apolipoprotein E fragments. J Biol Chem. 1983 Oct 25;258(20):12341–12347. [PubMed] [Google Scholar]
- Innerarity T. L., Weisgraber K. H., Arnold K. S., Rall S. C., Jr, Mahley R. W. Normalization of receptor binding of apolipoprotein E2. Evidence for modulation of the binding site conformation. J Biol Chem. 1984 Jun 10;259(11):7261–7267. [PubMed] [Google Scholar]
- Kaiser E. T., Kézdy F. J. Amphiphilic secondary structure: design of peptide hormones. Science. 1984 Jan 20;223(4633):249–255. doi: 10.1126/science.6322295. [DOI] [PubMed] [Google Scholar]
- Krebs K. E., Phillips M. C. The contribution of alpha-helices to the surface activities of proteins. FEBS Lett. 1984 Oct 1;175(2):263–266. doi: 10.1016/0014-5793(84)80748-6. [DOI] [PubMed] [Google Scholar]
- Mahley R. W., Innerarity T. L. Lipoprotein receptors and cholesterol homeostasis. Biochim Biophys Acta. 1983 May 24;737(2):197–222. doi: 10.1016/0304-4157(83)90001-1. [DOI] [PubMed] [Google Scholar]
- Mahley R. W., Innerarity T. L., Rall S. C., Jr, Weisgraber K. H. Plasma lipoproteins: apolipoprotein structure and function. J Lipid Res. 1984 Dec 1;25(12):1277–1294. [PubMed] [Google Scholar]
- McLean J. W., Fukazawa C., Taylor J. M. Rat apolipoprotein E mRNA. Cloning and sequencing of double-stranded cDNA. J Biol Chem. 1983 Jul 25;258(14):8993–9000. [PubMed] [Google Scholar]
- Rose G. D. Prediction of chain turns in globular proteins on a hydrophobic basis. Nature. 1978 Apr 13;272(5654):586–590. doi: 10.1038/272586a0. [DOI] [PubMed] [Google Scholar]
- Schneider W. J., Beisiegel U., Goldstein J. L., Brown M. S. Purification of the low density lipoprotein receptor, an acidic glycoprotein of 164,000 molecular weight. J Biol Chem. 1982 Mar 10;257(5):2664–2673. [PubMed] [Google Scholar]
- Wang-Iverson P., Ginsberg H. N., Peteanu L. A., Le N. A., Brown W. V. Apo E-mediated uptake and degradation of normal very low density lipoproteins by human monocyte/macrophages: a saturable pathway distinct from the LDL receptor. Biochem Biophys Res Commun. 1985 Jan 16;126(1):578–586. doi: 10.1016/0006-291x(85)90645-x. [DOI] [PubMed] [Google Scholar]
- Weisgraber K. H., Innerarity T. L., Harder K. J., Mahley R. W., Milne R. W., Marcel Y. L., Sparrow J. T. The receptor-binding domain of human apolipoprotein E. Monoclonal antibody inhibition of binding. J Biol Chem. 1983 Oct 25;258(20):12348–12354. [PubMed] [Google Scholar]
- Yamamoto T., Davis C. G., Brown M. S., Schneider W. J., Casey M. L., Goldstein J. L., Russell D. W. The human LDL receptor: a cysteine-rich protein with multiple Alu sequences in its mRNA. Cell. 1984 Nov;39(1):27–38. doi: 10.1016/0092-8674(84)90188-0. [DOI] [PubMed] [Google Scholar]



