Abstract
The control of Vibrio cholerae phoBR expression by PhoB involves its binding to Pho boxes at −35 (box 1), −60 (box 2), and −80 (box 3) from the putative phoB translation start site. These loci were located in the sense (box 1) and antisense (boxes 2 and 3) strands of the phoBR regulatory region, and PhoB binds to these individual boxes with distinct affinities. Fusions of sequences containing different combinations of these boxes upstream of the lacZ reporter in a plasmid demonstrated that only those carrying boxes 1, 2, and 3, or 1 alone, activated transcription under inorganic phosphate (Pi) limitation. When a fragment, including only boxes 1 and 2, was fused to lacZ, expression was no longer induced by low Pi, suggesting a repressive role for PhoB∼box2 (PhoB bound to box 2) over the transcriptional activity induced by PhoB∼box1. The similarity between lacZ expression levels from promoter fragments containing the three boxes or box 1 alone showed that PhoB∼box3 eliminated the repressive effect imposed by PhoB∼box2 on phoBR transcription. Complementation assays with a phoBR-containing plasmid demonstrated that the 234-bp promoter fragment carrying the three boxes is absolutely required for operon expression in Vibrio cholerae ΔphoBR cells. This was observed under Pi abundance, when phoBR was expressed at a basal level and, also in low Pi conditions, when Pho regulon genes were fully expressed. Thus, under Pi limitation, PhoB exerts dual regulatory functions by binding sequentially distinct Pho boxes to enable the fine-tuning and precise control of phoBR expression in V. cholerae cells.
INTRODUCTION
Vibrio cholerae is found most commonly in marine or estuarine habitats, free or in association with phytoplankton and zooplankton and other marine organisms (7). Certain strains, mainly of serotypes O1 and O139, can be pathogenic to humans and cause cholera, an acute diarrheal disease that is responsible for significant mortality and economical damage in several parts of the world (6).
To establish a successful infection, V. cholerae must survive the stomach acidic environment, reach the small intestine, and synthesize TCP, the colonization factor toxin-coregulated pilus, and then CT, the cholera toxin (4, 11, 19, 20, 32, 44), among other factors. The expression of TCP and CT is transcriptionally regulated by ToxT, a member of the ToxR-dependent cascade, in response to environmental stimuli (34).
To survive and multiply in environments as diverse as aquatic ecosystems and the host digestive tract, V. cholerae coordinately regulates gene expression to generate a suitable adaptive response. Aquatic and terrestrial ecosystems generally are poor in nutrients such as phosphorus (46), an essential element since it is a constituent of phospholipids, phosphoproteins, and nucleic acids, and it is involved in most metabolic pathways as ATP, inorganic phosphate (Pi), or coenzymes.
In many bacterial species, phosphorus-containing compound uptake and metabolism depend on the extracellular Pi concentration. Under Pi starvation conditions, these processes require a two-component regulatory system to couple stimulus-response mechanisms, favoring bacterial cell adaptation to the environment (12). In Escherichia coli, V. cholerae, and other species, the system is encoded by the phoBR operon, and in Bacillus subtilis and Streptomyces coelicolor it is encoded by its homologues phoPR and phoRP, respectively (18). PhoR is the sensor histidine kinase that monitors extracellular Pi availability, and PhoB (or PhoP) is the response regulator, which transcriptionally regulates a set of genes, the Pho regulon, that is involved in bacterial phosphate management (12, 13). Recent studies have shown that the PhoBR system has many additional roles in bacteria (12, 13, 18). In particular, roles for PhoBR have been demonstrated in V. cholerae motility, biofilm formation, intestinal colonization, and virulence (9, 10, 36, 37, 43, 47).
E. coli response to phosphate deficiency involves PhoR autophosphorylation and the transfer of Pi to its cognate response regulator, PhoB (26). Phosphorylated PhoB (PhoB∼Pi) binds with higher affinity than the nonphosphorylated protein to conserved 18-bp DNA sequences, called Pho boxes, in the regulatory regions of Pho regulon genes (28). The E. coli consensus Pho box, CT(GT)TCAT A(AT)A(AT) CTGTCA(TC), consists of two 7-bp direct repeats separated by a conserved 4-bp AT rich spacer, where PhoB binds as a head-to-tail dimer and interacts with the RNA polymerase σ70 subunit to control the transcription of downstream genes (3, 25). Most Pho regulon promoters contain a single Pho box that is sufficient to confer regulation. However, PhoB-binding sites can be found in two or more copies within some promoter regions (15–17), where the protein usually binds in a cooperative manner for appropriate gene expression (21, 35). The E. coli Pho regulon consists of about 40 genes and includes the phoBR operon (18, 50), which is transcriptionally autoregulated via the direct binding of PhoB to a single Pho box at −35, 10 bp upstream of the putative −10 region (28, 45).
In a previous work, we have shown that in V. cholerae phoR and phoB genes constitute an operon whose regulatory region contains a putative Pho box sequence located at −35, as in E. coli (47). Moreover, mutation in phoBR eliminated the expression of Pho regulon genes in strains of V. cholerae O1 under Pi limitation. Due to the involvement of the V. cholerae O1 phoBR operon in many physiological processes and in virulence (9, 36, 37, 43, 47), we decided to further investigate the mechanism underlying its expression.
In this work, we demonstrated that the V. cholerae phoBR operon is autoregulated by a novel and complex mechanism that involves PhoB binding to three Pho boxes, at −35 (box 1), −60 (box 2), and −80 (box 3) upstream of the putative phoB translation start site. These boxes are located in the sense (box 1) and antisense (boxes 2 and 3) strands, and PhoB interacts with them in a sequential and cooperative manner and with different affinities. PhoB bound to distinct boxes acts as a transcriptional activator or a repressor, fine-tuning the expression of the phoBR operon under Pi limitation. Furthermore, we provide evidence that phoBR is transcriptionally activated by proteins other than PhoB when cells are cultivated under Pi abundance.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and culture conditions.
Bacterial strains and plasmids used in this work are listed in Table 1. Cells were routinely propagated in LB agar or LB for liquid cultures (39). The minimal medium TG (5) was used for bacterial growth under high (6.5 mM; TGHP) or low (65 μM; TGLP) Pi concentration. When required, 100 μg/ml ampicillin, 100 μg/ml streptomycin, and/or 50 μg/ml kanamycin was added to the media. Cultures were grown at 37°C in a shaking water bath at 200 rpm or in an incubator.
Table 1.
Bacterial strains and plasmids used in this studya
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F−endA1 hsdR17 (rK− mK+) supE44 thi-1 recA1 gyrA (Nalr) relA1 Δ(lacZYA argF)U169 φ80dlacZΔM15 | 38 |
| BL21(DE3) | hsdS gal (λcIts857 ind1 Sam7 nin5 lacUV5-T7 gene 1) | 42 |
| V. cholerae strains | ||
| 569BSR | Classical Inaba, Smr, wild-type V. cholerae | 47 |
| WK3 | ΔphoBR mutant of 569BSR; Smr Kmr | 47 |
| Plasmids | ||
| pGEX4T-3 | High-copy vector containing an MCS preceded by GST coding region; Apr | GE Healthcare |
| pGEXphoWT | 0.7-kbp 569BSR phoB ORF inserted into BamHI/XhoI pGEX4T-3 restriction sites, fused 5′ to GST coding region; Apr | This work |
| pIC552 | Low-copy vector containing lacZ preceded by an MCS to insertion of a promoter region; Apr | 23 |
| pIC234 | pIC552 containing a region of 234 bp upstream of phoBR putative transcription start site regulating lacZ; Apr | 10 |
| pIC86 | pIC552 containing a region of 86 bp upstream of phoBR putative transcription start site regulating lacZ; Apr | This work |
| pIC62 | pIC552 containing a region of 62 bp upstream of phoBR putative transcription start site regulating lacZ; Apr | This work |
| pIC234mut | pIC552 containing the mutated region of 234 bp upstream of phoBR putative transcription start site regulating lacZ; Apr | This work |
| pIC86mut | pIC552 containing the mutated region of 86 bp upstream of phoBR putative transcription start site regulating lacZ; Apr | This work |
| pWKS30 | Low-copy vector; Apr | 49 |
| pGEM-T Easy | High-copy easy cloning vector; Apr | Promega |
| pWK5 | High-copy plasmid; contains phoBR operon, including entire regulatory region; Apr | 47 |
| pWK20 | 3 kbp pWK5 PstI fragment inserted into PstI pWKS30 restriction site; contains phoBR coding region plus 234-bp promoter; Apr | This work |
| pCG7 | pWK5 containing a SacII restriction site in the center of phoBR regulatory region; Apr | This work |
| pCG8 | 3-kbp pCG7 PstI fragment inserted into PstI pWKS30 restriction site; contains phoBR coding region plus 234-bp mutated promoter; Apr | This work |
| pLC1 | pWK5 reverse amplified, digested with BglII, and linked; contains phoBR plus 62-bp promoter; Apr | This work |
| pLC2 | pWK5 reverse amplified, digested with BglII, and linked; contains phoBR plus 86-bp promoter; Apr | This work |
| pLC3 | 2.3-kbp pLC1 BglII/PstI fragment, inserted into BamHI/PstI pWKS30 restriction sites; contains phoBR plus 62-bp promoter; Apr | This work |
| pLC4 | 2.3-kbp pLC2 BglII/PstI fragment, inserted into BamHI/PstI pWKS30 restriction sites; contains phoBR plus 86-bp promoter; Apr | This work |
| pLC5 | 2.3-kbp fragment of pCG7 amplified and cloned into vector pGEM-T Easy; contains phoBR plus 86-bp mutated promoter; Apr | This work |
| pLC6 | 2.3-kbp pLC5 NotI fragment, inserted into NotI pWKS30 restriction site; contains phoBR plus 86-bp mutated promoter; Apr | This work |
Sm, streptomycin; Km, kanamycin; Ap, ampicillin; Cm, chloramphenicol; MCS, multiple-cloning site; ORF, open reading frame.
Genetic methods and cloning procedures.
All genetic manipulations were carried out in E. coli DH5α (38). V. cholerae chromosomal DNA was purified as described previously (1) from overnight cells cultured in LB. Standard methods were used for minipreparations of plasmids, restriction enzyme digestions, and ligations (39). DNA fragments, unless specified, were purified from agarose gels with Illustra GFX PCR DNA (GE Healthcare) in accordance with the supplier's recommendations. The preparation of E. coli competent cells and electroporation were carried out as described previously (39), whereas V. cholerae competent cells were prepared and electroporated as described previously (29, 47). PCR amplifications were performed in an automatic thermal cycler (Ominigene) using standard conditions (14).
Different fragments of the putative phoBR regulatory region were cloned into pIC552 to generate lacZ fusions. To this end, pWK5 (47), a plasmid containing the phoBR operon and an ∼400-bp-long sequence upstream of phoB, was used as a template for PCR. Plasmid pIC234 containing the 234-bp phoBR full-length regulatory region upstream of lacZ was obtained in a previous work (10). Fragments 86 and 62 bp long of the phoBR regulatory region were obtained by PCR amplification with oligonucleotides pIC170F (5′GCGAGATCTGCCTGAACCTTA3′) and PhoBXhoI (5′TTCTTCTCGAGTGTTTTTCACCAACGCCA3′) as well as Met2E (5′CGGGCCATGGTTTGCAAAGGTCACAAAAG3′) and PhoBXhoI, respectively. Purified PCR products were digested with NcoI and XhoI and cloned upstream of the lacZ gene into pIC552 that had been opened with the same endonucleases (23), generating pIC86 and pIC62, respectively (Table 1).
To introduce a mutation at approximately −60 of the putative translation start site of phoB within Pho box 2, pWK5 was used as a template in a reverse PCR along with oligonucleotides CPMut1 (5′TCACCCGCGGATGAAAATTTGTTGC3′) and CPMut2 (5′ATTACCGCGGTTCAGGCCGAAATTC3′), containing sites for the restriction enzyme SacII at their 5′ ends. Pfx Triple Master (Eppendorf) was used according to the supplier's instructions, and the PCR product was digested with SacII and ligated. In this process, the AT-rich motif TTAT in the wild-type sequence was replaced by GCGG. The recombinant plasmid, namely, pCG7, was used as the template for subsequent amplifications. Different segments of the phoBR regulatory region containing the mutated sequence were cloned in pIC552 upstream of lacZ. To this end, mutated fragments of 234 and 86 bp were PCR amplified using the primers pairs PhoBNcoI (5′CCCCCCATGGTTTAAACCACATTGTTG3′) and PhoBXhoI as well as P170Fmut (5′GCGAGATCTGCCTGAACCGCG3′) and PhoBXhoI, respectively. Purified fragments were digested with NcoI and XhoI, cloned into pIC552, and opened with these enzymes, generating pIC234mut and pIC86mut (Table 1).
For the complementation analysis of the phoBR mutant strain WK3 (ΔphoBR) (Table 1), low-copy-number plasmids containing the phoBR operon under the control of different fragments of its own regulatory region were obtained as described below. Initially, pWK5 was digested with PstI to remove a 3-kbp fragment containing phoBR and the full-length regulatory region. The low-copy-number plasmid pWKS30 (49) was opened with the same enzyme and ligated to the phoBR PstI fragment, generating plasmid pWK20. To obtain plasmids containing the phoBR operon under the control of shorter fragments of its wild-type regulatory region, pWK5 was subjected to reverse PCR using Pfx Triple Master. P50F (5′GCGAGATCTGGTCACAAAAGT3′) and V50RA (5′GCGAGATCTAAAGTGGTCACCAAG3′) or P170F and V50RA were used to amplify fragments containing 62 or 86 bp, respectively. PCR products were digested with BglII and ligated, generating pLC1 and pLC2, respectively. Both plasmids then were double digested with PstI and BglII, and fragments of approximately 2.3 kbp were cloned into pWKS30 digested with the same enzymes, creating pLC3 (62 bp upstream of phoBR) and pLC4 (86 bp upstream of phoBR). To generate a plasmid with the full-length regulatory region mutated within Pho box 2, pCG7 was digested with PstI to remove a 3-kbp fragment that subsequently was cloned into the PstI site of pWKS30, generating plasmid pCG8. Plasmid with a version of the promoter containing Pho box 1 and mutated box 2 (2mut) was obtained using pCG7 as the template for PCR with Pfx Triple Master and the oligonucleotides LIV1 (5′AATAATTTCGCCTGAACCGC3′) and P50R (5′TCTAGAGGATCCCCGGGTA3′). The 2.3-kbp product (86 bp upstream of phoBR) was purified and cloned into pGEM-T Easy (Promega), resulting in plasmid pLC5. This in turn was digested with NotI to release the 2.3-kbp fragment that was inserted into the NotI site of pWKS30, generating plasmid pLC6.
For the expression of the 569BSR phoB gene in E. coli, its coding sequence was amplified using oligonucleotide primers PhoBini (5′GGCGCGGATCCAGAAGGATTCTGGTTGTTGAAG3′) and CterXhoI (5′GCGCCGCTCGAGTTAGGCTTTGG3′), with 569BSR genomic DNA as the template. The 0.7-kbp fragment was digested with BamHI and XhoI and cloned into pGEX4T-3 (GE Healthcare) digested with the same enzymes, generating pGEXphoWT, which contains the glutathione S-transferase (GST) coding sequence 5′ of phoB. All constructions were screened and confirmed by PCR and sequencing (ABI PRISM BigDye Terminator cycle sequencing; Applied Biosystems).
Overexpression of Vibrio cholerae O1 phoB and PhoB purification.
Strain BL21(DE3) (Table 1) harboring pGEXphoWT was inoculated into 1 liter of LB-ampicillin and grown with aeration at 37°C until an optical density at 600 nm (OD600) of 0.7. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM, and the culture was further incubated under the same conditions for 4 h. Cells were collected by centrifugation and resuspended in phosphate-buffered saline (PBS) containing 1% (wt/vol) Triton X-100 at a proportion of 5 ml/g (wet weight) of cells. Aliquots of 1.25 mg lysozyme/g of cells were added to the suspension before sonication (Sonicator W-380; Heat Systems Ultrasonics, Inc.) until complete lysis. The lysate was centrifuged at 11,000 × g for 10 min at 4°C, and the soluble fraction, containing the major amount of fusion protein, was incubated with glutathione Sepharose resin (GE Healthcare) according to the manufacturer's instructions. The GST-PhoB fusion protein was purified and then cleaved by thrombin (GE Healthcare). The purity and concentration of the recombinant PhoB were determined by densitometric analysis of a Coomassie-stained SDS-PAGE 12.5% gel using the software Scion Image (IBM PC) and by measuring the OD280 (39). The soluble 26-kDa purified PhoB was trypsin digested (40) and analyzed by mass spectrometry on a Voyager DE PRO Biospectrometry workstation (Applied Biosystems). Samples containing purified PhoB were aliquoted and stored at −70°C.
Search for putative Pho box sequences upstream of phoBR.
A search for Pho box-like sequences within the 234-bp phoBR regulatory region was performed in both sense and antisense orientations using the GLAM2SCAN tool (8), part of the MEME suite (2), based on the consensus E. coli Pho box motif (24), which was constructed with the GLAM2 tool.
EMSA.
Electrophoretic mobility shift assay (EMSA) probes were duplex DNA oligonucleotides prepared either by annealing complementary single-stranded DNA oligonucleotides or by PCR using pWK5 or pCG7 (Table 1) as templates and specific primer pairs. For double-stranded probes, six oligonucleotides were used to generate three pairs: cx1fwd (5′CCAAGGTCACAAAAGTGTCATAAA3′) and cx1rev (5′TTTATGACACTTTTGTGACCTTGG3′), cx2fwd (5′TGAACCTTATATGAAAATTTGTTG3′) and cx2rev (5′CAACAAATTTTCATATAAGGTTCA3′), and cx3fwd (5′AGGTTTGGATATATGAATTTCGCC3′) and cx3rev (5′GGCGAAATTCATATATCCAAACCT3′). Each primer pair was incubated in annealing buffer (10 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM EDTA) (39) under the following conditions: 88°C for 2 min, 65°C for 10 min, 35°C for 10 min, and 25°C for 5 min. The PCR products and the annealed double-stranded oligonucleotides were purified from 15% polyacrylamide gels (39), quantified, and radiolabeled at the 5′ end with [γ-32P]ATP using T4 polynucleotide kinase (Invitrogen) (39). The binding of the purified PhoB to DNA probes and analysis of the products by gel electrophoresis were carried out as described previously (31). Reaction mixtures of PhoB with the 234-bp phoBR regulatory region contained 50 fmol of probe and increasing concentrations of PhoB (from 0 to 32 μM) in binding buffer (20 mM Tris-HCl, pH 7.0; 50 mM NaCl; 1 mM dithiothreitol [DTT]; 10 mM MgCl2; 100 μg/ml bovine serum albumin [BSA]; 0.5 μg/ml salmon sperm DNA) in a total volume of 10 μl. Competition assays were performed using 50 fmol of radiolabeled promoter fragment, PhoB at 32 μM, and 50, 250, or 500 fmol of unlabeled probe. Comparative EMSAs with 234-, 172-, and 62-bp fragments were performed using 50 fmol of labeled probes and increasing PhoB concentrations (0, 0.5, 2.5, 5, 10, 20, 30, and 37 μM). The specific ligation of PhoB to these individual sequences was tested using an excess of unlabeled probes (data not shown). EMSAs with sequences containing individual Pho box 1, 2, or 3 were carried out with 50 fmol of each probe and increasing concentrations of PhoB (from 0 to 37 μM). For competition assays, 50, 250, and 500 fmol of nonlabeled oligonucleotides was added to reaction mixtures containing 30 μM PhoB and 50 fmol of the corresponding labeled probe. To assess the relative binding affinity of PhoB for the individual Pho box 1, 2, or 3, reactions were carried out with 50 fmol of labeled Pho box 1 and PhoB at concentrations ranging from 0 to 30 μM (control). Another set of reaction mixtures contained PhoB at 30 μM, 50 fmol of labeled Pho box 1, and a total of 500 fmol of nonlabeled competitor probes as equimolar mixtures of Pho boxes 1, 2, and 3; Pho box 1; Pho box 2; and Pho box 3.
All reaction mixtures were incubated at room temperature for 30 min and then subjected to electrophoresis on a 7% nondenaturing polyacrylamide gel in 1× Tris-acetate-ETDA buffer (39). Gels were exposed to a PhosphoImager screen (Molecular Devices) cassette sealed against light for 24 h.
DNase I footprinting.
The oligonucleotide PhoBXhoI was radiolabeled at the 5′ end using [γ-32P]ATP and T4 polynucleotide kinase (39). After labeling, the oligonucleotide was precipitated and resuspended in diethyl pyrocarbonate (DEPC)-treated water. The 32P oligonucleotide PhoBXhoI and nonlabeled PhoBNcoI oligonucleotide were used as primers to amplify the 234-bp sequence upstream of phoBR using pWK5 (wild-type promoter) and pCG7 (mutant promoter) as templates. The 234-bp fragments were purified and quantified as described before. G+A sequencing (obtained by the chemical digestion of DNA) was carried out with 234-bp wild-type and mutated fragments. The reaction products were resuspended in 5 μl of sequencing buffer (T7 sequencing kit; GE Healthcare) and subjected to gel electrophoresis alongside DNase I digestions products (30).
Purified V. cholerae PhoB (0, 0.5, 2.5, and 10 μM) was incubated with 200 fmol of wild-type or mutated 234-bp DNA probe in binding buffer in a total volume of 49 μl. Reaction mixtures were incubated for 30 min at room temperature, and then 0.01 U of DNase I was added per tube for probe digestion. Reaction tubes were left at room temperature for 1 min, after which 140 μl of stop solution (192 mM sodium acetate, 32 mM EDTA, 0.14% SDS, 64 mg/ml herring sperm DNA) were added in each tube. DNA was extracted with phenol-chloroform and precipitated with sodium acetate and ethanol (39). The DNA pellet was resuspended in 5 μl of sequencing buffer. Products of DNase I digestion, as well as G+A sequencing, were subjected to electrophoresis on a 6% polyacrylamide gel under denaturing conditions. The gel was prerun at a constant voltage of 1,400 to 1,700 V and 40 W until the temperature reached 50°C, when samples were applied. After being run, the gel was dried at 80°C for 1 h and exposed to a PhosphoImager screen cassette sealed to light for 24 h.
Enzymatic assays.
For promoter activity assay, V. cholerae wild-type (569BSR) and ΔphoBR mutant (WK3) strains containing plasmid pIC552, pIC234, pIC86, pIC62, pIC234mut, or pIC86mut were grown in LB medium until an OD600 of 0.5. Cells were collected by centrifugation, resuspended in the same volume of TGHP (Pi abundance) or TGLP (Pi limitation), and cultivated for 16 h, at which time the promoter transcriptional activity was assessed by measuring β-galactosidase activity (9). PhoA (alkaline phosphatase) activity, the control of Pho regulon expression, was determined as previously described (47) in the same cells. Activities of both enzymes were expressed in U/μg of protein (22).
For complementation assays, V. cholerae ΔphoBR mutant (WK3) cells containing plasmid pWK20, pCG8, pLC3, pLC4, or pLC6 were cultivated as described above, and PhoA activity was determined as previously described (47).
RESULTS AND DISCUSSION
PhoB binds to multiple sites in the V. cholerae phoBR regulatory region in vitro.
In E. coli, the expression of the phoBR operon is autoregulated by PhoB binding to a single Pho box at −35 of the transcription initiation site of phoBR, as previously described (45).
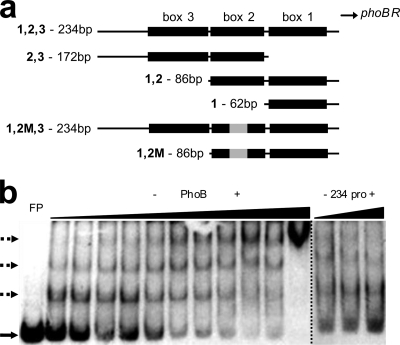
To investigate whether a similar regulatory mechanism occurs in V. cholerae cells, we performed EMSA using purified V. cholerae PhoB and a 32P-radiolabeled PCR fragment of 234 bp corresponding to the sequence immediately upstream of phoBR. The experiments were carried out with the unphosphorylated form of the protein, since it has been shown that E. coli PhoB does bind to the same Pho boxes in vitro but with lower affinity than its phosphorylated form (31). PhoB retarded the fragment mobility even at 1.5 μM, giving rise to two shifted bands in the EMSA gel (Fig. 1b). When PhoB was used at higher concentrations, a third complex was formed, which was represented in the gel by a very slow mobility band whose intensity increased with the protein concentration. EMSA with an unlabeled 234-bp fragment as the competitor DNA fragment showed the displacement of the labeled probe bound to PhoB, confirming the specificity of the protein-DNA interaction (Fig. 1b, right). The binding of purified PhoB to the phoBR regulatory region explains why in earlier EMSAs using protein extracts, only those from wild-type V. cholerae cells grown under Pi limitation shifted the phoBR 234-bp promoter fragment (data not shown).
Fig. 1.
(a) Scheme of phoBR regulatory region fragments used in this work. Pho boxes are represented by black rectangles; mutation in box 2 is represented by a gray rectangle. (b) EMSA pattern of the phoBR 234-bp regulatory region fragment radiolabeled with 32P by PhoB binding. In the left panel, 50 fmol of the fragment was incubated with increasing concentrations of purified PhoB (1.5, 2, 2.5, 3, 3.5, 4, 8, 12, 16, 20, and 32 μM). In the first lane, the FP (free probe) position is indicated by a solid arrow; dotted arrows point to three distinct shifted bands. In the right panel is the result of the competition assay using 50 fmol of radiolabeled 234-bp promoter fragment and PhoB at 32 μM against 50, 250, and 500 fmol of unlabeled probe.
Differently from E. coli, where the expression of phoBR is controlled by PhoB binding to a single Pho box (45), these results suggest that the V. cholerae phoBR regulatory region has multiple sites with different binding affinities for PhoB. In line with our results, it has been demonstrated that the expression of homologue operons, such as phoRP of Streptomyces coelicolor (41) and phoPR of B. subtilis (33), also requires the binding of the response regulator PhoP to multiple Pho boxes in their regulatory regions for efficient transcription.
Three putative Pho boxes were identified in the phoBR regulatory region.
A similarity search based on the E. coli Pho box consensus sequence (28) was carried out with the 234-bp regulatory region of V. cholerae phoBR using GLAM2 and GLAM2SCAN tools (2, 8). Three candidate sequences were found, at approximately −35 in the sense orientation and at −60 and −80 in the antisense orientation. Their sequences, positions, and orientations relative to the putative phoB translation initiation site are shown in Fig. 2a and b.
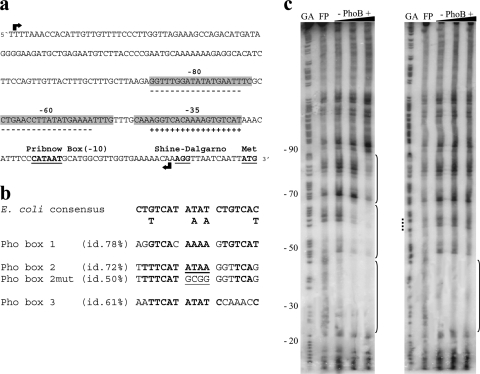
Fig. 2.
Mapping of Pho boxes in the phoBR upstream region of V. cholerae. (a) Diagram of the phoBR regulatory region, with the Pho boxes, identified by similarity search with GLAM2SCAN, and gaps. Sequences corresponding to footprint gaps are shown in gray; sequences identified as Pho boxes are marked with a plus (sense orientation) or minus (antisense orientation). Black arrows indicate the limits of the 234-bp sequence. (b) Pho box sequences in the phoBR regulatory region and their identities to the E. coli consensus Pho box and the box 2 mutated (box 2mut) version. (c) Results of DNase I protection assays of PhoB binding sites (brackets) in the phoBR regulatory region. Two hundred fmol of the radiolabeled wild-type 234-bp sequence (left) and 234-bp box 2 mutated version (right) were independently incubated with increasing concentrations of PhoB (0.5, 2.5, and 10 μM) and digested with DNase I. Digestion products were size fractionated by denaturing gel electrophoresis. GA is the sequencing ladder. The dotted line in panel c marks the positions of the mutated bases in Pho box 2mut. FP, free probe.
To confirm the location of PhoB binding sites in the phoBR regulatory region, a DNase I protection footprinting assay was performed (Fig. 2c). PhoB protected the DNA segment spanning nucleotides 20 to 90 from DNase I, which included three adjacent putative binding sites at −35, −60, and −80, with different binding affinities to PhoB (Fig. 2c, left). These sites correspond exactly to those identified by the PhoB binding motif search (Fig. 2a and b). It is interesting that relative binding affinities of PhoB to these sites seem to be in agreement with the identity level of each one to the E. coli consensus Pho box (Fig. 2b). The PhoB binding site at −35 corresponds to the sequence predicted in a previous work (47), with extensive identity (∼80%) to the Pho box in the regulatory region of the E. coli phoBR operon (28). These putative PhoB binding sites are referred to as boxes 1 (−35), 2 (−60), and 3 (−80).
Binding of PhoB to phoBR Pho boxes in vitro seems to be sequential and cooperative.
Since the three Pho boxes are in close proximity to each other in the phoBR regulatory region, the hypothesis of sequential and cooperative binding of PhoB to these sites was tested. To this end, a footprinting assay was performed as described earlier, using as a probe the 234-bp mutated fragment within the AT-rich core of Pho box 2. More precisely, the sequence TTAT was replaced by GCGG within Pho box 2 (Fig. 2b), because this AT-rich spacer between the two 7-bp direct repeats has a functional role in the intrinsic bending of the Pho box sequence upon PhoB binding in E. coli (24, 25). Thus, we expected to disturb the interaction of PhoB with box 2 and analyze its overall effect on PhoB binding to the phoBR regulatory region.
Interestingly, this mutation did not affect PhoB binding to box 1 but prevented the binding of the protein to boxes 2 and 3, even at higher protein concentrations (Fig. 2c, right). These findings suggested that the binding of PhoB to box 3 in vitro is dependent on the prior binding of the protein to box 2.
It has been shown that the expression of some E. coli Pho regulon genes, namely, psiE (16), ugpB (15), and pstS (17, 27), depends on the cooperative binding of PhoB to tandemly arranged Pho boxes in their regulatory regions. Such an arrangement seems to be required for the effective binding of PhoB to weakly conserved Pho box sequences, such as those in psiE and ugpB promoters, to enhance transcriptional activation (16). On the other hand, there are Pho regulon genes whose expression is repressed by the binding of PhoB to Pho boxes in their regulatory regions. This has been described, for instance, for the operons tagAB and tagDEF of B. subtilis, whose expression is both positively and negatively controlled by the two-component signal transduction system PhoPR, a homologue of E. coli PhoBR (13). In such cases, PhoP binding to Pho boxes is required for the transcriptional activation and/or repression of the operons under Pi starvation. Interestingly, some of these PhoP binding sites are located in the antisense strand of the promoters (21). Therefore, under Pi starvation, the activity of the Pho regulon in both E. coli and B. subtilis is modulated by the binding of their corresponding response regulators to Pho boxes to elicit specific responses to Pi limitation, which might involve the induction or repression of gene expression.
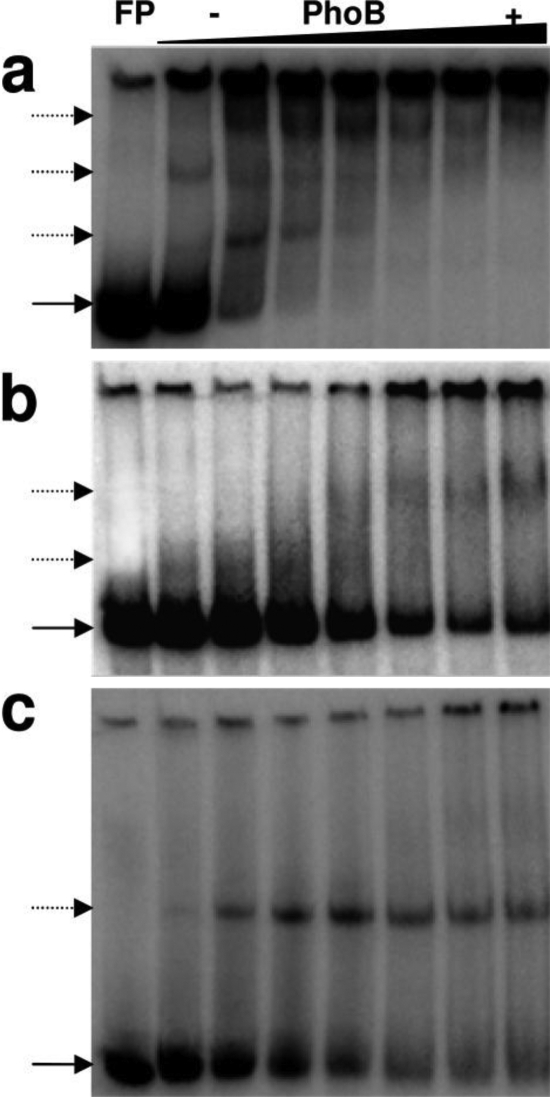
The putative PhoB binding sites identified in the V. cholerae phoBR regulatory region are located in the sense (box 1 at −35) and antisense strands (box 2 at −60 and box 3 at −80). Therefore, the binding of PhoB to these sequences under Pi limiting conditions might result in the activation and/or repression of phoBR to fine-tune the transcription of Pho regulon genes. To further explore the interaction between V. cholerae PhoB and Pho boxes, we performed EMSAs with purified protein and three distinct double-stranded oligonucleotide probes, corresponding to the sequences of the individual Pho boxes (Fig. 3). At the lowest concentration tested (2 μM), PhoB retarded the electrophoretic mobility of the box 1 (−35)-containing probe only and did not bind to Pho box 2 or box 3 probe, even at the highest concentration used (37 μM) (Fig. 3a, b, and c).
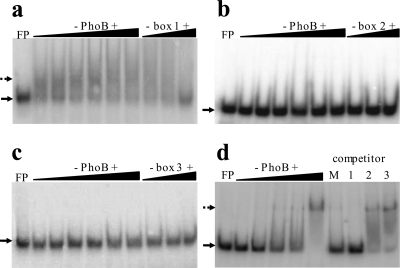
Fig. 3.
Binding of PhoB to individual Pho boxes in the phoBR regulatory region. Fifty fmol of radiolabeled double-stranded oligonucleotides corresponding to each one of the three Pho boxes was used in EMSAs with increasing PhoB concentrations. The binding patterns (dotted arrows) of Pho box 1 (a), Pho box 2 (b), and Pho box 3 (c) to PhoB at 2, 5, 10, 20, 30, and 37 μM are presented. Competition assays were carried out with 37 μM PhoB plus 50, 250, and 500 fmol of competing nonlabeled oligonucleotides. (d) The labeled oligonucleotide corresponding to Pho box 1 was incubated with increasing PhoB concentrations (0.5, 2, 5, 10, and 30 μM). Competition assay was performed with PhoB at 30 μM against 500 fmol of competing nonlabeled Pho boxes containing fragments. Lane M, equimolar mixture of Pho boxes 1, 2, and 3 (a total of 500 fmol); lane 1, 500 fmol of Pho box 1; lane 2, 500 fmol of Pho box 2; lane 3, 500 fmol of Pho box 3. FP, free probe (solid arrows).
The relative binding affinity of PhoB to the three Pho boxes was further analyzed by competition EMSA. To this end, the labeled complex of PhoB fused to box 1 (PhoB∼box1) was incubated with unlabeled oligonucleotides corresponding to box 1 itself or box 2 or 3 to test their ability to displace the bound protein from box 1. Only unlabeled Pho box 1 containing probe and the mixture of the three phoBR Pho boxes effectively displaced PhoB from the complex (Fig. 3d). These results confirmed the higher binding affinity of PhoB to box 1 relative to boxes 2 and 3. They also showed that box 1, at −35, was the only one within the phoBR regulatory region that functioned independently as a PhoB binding site in vitro.
Additional EMSAs (Fig. 4a, b, and c) were performed to compare the binding of PhoB to phoBR promoter regions containing distinct combinations of the three Pho boxes. The PhoB binding patterns to fragments with boxes 1, 2, and 3 (Fig. 4a) and to Pho box 1 only (Fig. 4c) confirm the results presented before (Fig. 1b and 3a, respectively).
Fig. 4.
Binding patterns of PhoB to three distinct phoBR promoter fragments. Fifty fmol of labeled double-stranded probes were used with increasing PhoB concentrations (0, 0.5, 2.5, 5, 10, 20, 30, and 37 μM). Binding patterns of probes containing boxes 1, 2, and 3 (a), boxes 2 and 3 (b), or box 1 alone (c) are shown. In the first lane, the free probe (FP) position is indicated by a solid arrow; dotted arrows point to shifted bands.
Although PhoB did not bind in vitro to probes containing Pho box 2 or 3 separately (Fig. 3b and c, respectively), it did bind to the promoter version containing boxes 2 and 3 (Fig. 4b). However, the mobility shift of this fragment was observed only at PhoB concentrations above 5 μM, about 10-fold higher than that required to retard fragments with boxes 1, 2, and 3 or box 1 (Fig. 4a and c), thus confirming the hypothesis that the binding of PhoB to boxes 2 and 3, in this order, is somehow assisted by the complex PhoB∼box1.
Taken together, these results suggest that (i) PhoB presents higher binding affinity in vitro to box 1 than to box 2 or 3, and (ii) the binding of PhoB to Pho boxes 1, 2, and 3 in vitro occurs in a sequential order and seems to be cooperative, as described for the PhoB-dependent regulation of the E. coli gene ugpB (15).
Binding of PhoB to Pho boxes in the sense and antisense strands of phoBR promoter region fine-tunes the expression of the operon.
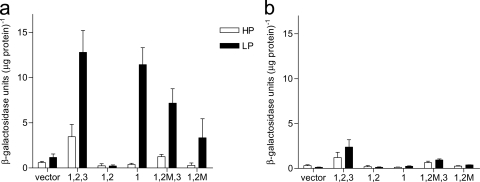
To test whether the in vitro binding pattern of PhoB to phoBR Pho boxes matches the in vivo binding of the protein to the phoBR regulatory region and correlates with promoter activity, wild-type 569BSR and its isogenic phoBR mutant, WK3, were transformed with plasmids containing the reporter gene lacZ under the control of distinct combinations of Pho boxes 1, 2, and 3 (Fig. 1a). Promoter activities were estimated by β-galactosidase levels in transformed cells grown under Pi limitation (TGLP medium) or Pi abundance (TGHP medium) for 16 h, when the expression of both the phoBR operon and Pho regulon (measured by alkaline phosphatase expression) still represents peak levels (data not shown).
Similar β-galactosidase activity levels were observed in wild-type cells, grown under Pi limitation (LP), harboring either boxes 1, 2, and 3 fused to lacZ (box1,2,3∼lacZ) or box1∼lacZ (Fig. 5a). Under Pi abundance (HP), wild-type cells with box1,2,3∼lacZ showed low levels of β-galactosidase activity, while wild-type cells carrying box1∼lacZ responded similarly to those harboring promoterless lacZ (Fig. 5a). These data suggest that phoBR expression from Pho box 1 is strictly dependent on Pi limitation in vivo. However, the full-length phoBR regulatory region (with boxes 1, 2, and 3; 234 bp) apparently responds to other stimuli, which could explain its small but detectable in vivo activity in Pi rich medium, in agreement with previous findings (9, 10, 36, 47, 48).
Fig. 5.
β-Galactosidase activity levels in the wild-type (a) and ΔphoBR mutant cells (b) harboring vectors with different versions of the phoBR regulatory region fused to lacZ. Vector, promoterless lacZ; 1,2,3, boxes 1, 2, and 3 fused to lacZ; 1,2, boxes 1 and 2 fused to lacZ; 1, box 1 fused to lacZ; 1,2 M,3, box 1, mutated box 2, and box 3 fused to lacZ; 1,2 M, box 1 and mutated box 2 fused to lacZ. Strains were grown under high (HP) and low (LP) phosphate conditions for 16 h.
Surprisingly, wild-type cells carrying box1,2∼lacZ cultured either in Pi limitation (LP) or Pi abundance (HP) showed virtually no β-galactosidase activity, indicating the repression of lacZ transcription. In comparison, wild-type cells harboring box1,2mut∼lacZ presented low levels of β-galactosidase activity, but only under Pi limitation (LP), suggesting that transcription from the fragment with boxes 1 and 2 is a PhoB-dependent process and that the mutation in box 2 positively affected the transcription of phoBR (Fig. 5a).
The low level of β-galactosidase activity in wild-type cells carrying the construct of box 1 and mutated box 2 fused to lacZ (box1,2mut∼lacZ) relative to those with box1,2,3∼lacZ and box1∼lacZ under Pi limitation (LP) was a surprising result. It indicates that PhoB does interact with the mutant Pho box 2, although with a lower affinity than it does with the wild-type sequence, and decreases (but does not cancel) lacZ expression induced by PhoB∼box1.
In fact, the β-galactosidase activity in the wild-type strain transformed with the plasmid carrying box1,2mut∼lacZ was higher than that in cells harboring box1,2∼lacZ but did not reach the levels found in wild-type or box1∼lacZ cells.
We also investigated the effect of the mutated Pho box 2 on lacZ expression driven by the 234-bp full-length phoBR regulatory region in cells grown under low Pi (LP) conditions. Interestingly, in the wild-type strain carrying box1,2mut,3∼lacZ, the β-galactosidase activity level was slightly lower than that in cells carrying the construct box1,2,3∼lacZ (Fig. 5a), showing that PhoB interacted with the mutated box 2 in vivo. However, the fact that higher β-galactosidase activity was observed in wild-type cells carrying box1,2mut,3∼lacZ than in those harboring box1,2mut∼lacZ suggests that PhoB binds to box 3 and partially relieves the transcriptional repression imposed by PhoB∼box2mut (Fig. 5a). It is worthy of note that the interaction of PhoB with box 3 in this particular case seems to have been hindered by the abnormal associations of the PhoB∼box2mut construct, which is in agreement with in vitro results described before (Fig. 2c).
In contrast, purified PhoB did not bind to the mutant Pho box 2 in vitro, as observed by the footprint assay (Fig. 2c, right). This provides evidence for a distinct binding pattern of PhoB to the regulatory region of phoBR in vivo, which might depend on the assistance of unknown cellular factors. Taken together, these results suggest that in wild-type V. cholerae cells under Pi limitation, PhoB∼box2 imposes a strong repression on the transcriptional activity of phoBR promoter induced by PhoB∼box1, which is attenuated by a mutation within Pho box 2 or reversed by the binding of PhoB to box 3. A particularly noteworthy aspect of this regulatory mechanism is the strong positive effect of PhoB∼box3 on the transcription of phoBR in wild-type cells under Pi limitation. Accordingly, in wild-type cells carrying the construct box1,2∼lacZ the β-galactosidase activity was close to zero, but in those harboring the full-length wild-type phoBR promoter (wild-type and box1,2,3∼lacZ) it reached its highest level (Fig. 5a), highlighting the essential role of Pho box 3 in the positive control of phoBR expression in vivo under Pi limitation. The fact that similar β-galactosidase activity levels were found in wild-type cells transformed with plasmid carrying box1,2,3∼lacZ or box1∼lacZ under low Pi conditions strongly indicates that in vivo PhoB∼box3 overcomes the negative effect of PhoB∼box2 on the transcription of the phoBR operon.
Another important feature of the system is the location of boxes 2 and 3 in an antisense orientation with respect to the phoBR operon. The presence of PhoB binding sites in different strands of the regulatory region supports the hypothesis that PhoB exerts distinct functions in the control of phoBR expression in V. cholerae. As discussed previously, the binding of PhoP to the noncoding strand of tagAB and tagDEF in B. subtilis is critical for its negative role in the control of the expression of these genes (21). Thus, the antisense location of Pho box 2 could partially explain the repression of phoBR transcription mediated by PhoB∼box2 (Fig. 5a). However, Pho box 3 (at −80), immediately upstream of box 2 and also on the noncoding strand of phoBR, on the contrary, cancelled PhoB∼box2-mediated repression in vivo.
As expected, ΔphoBR mutant cells harboring plasmids with different fragments of the phoBR regulatory region upstream of lacZ presented very low β-galactosidase activities that were independent of Pi levels (Fig. 5b). Interestingly, in ΔphoBR cells carrying box1,2,3∼lacZ, independently of the extracellular Pi concentration the full-length promoter activity level was low and similar to that found in the wild type and box1,2,3∼lacZ under Pi abundance (Fig. 5b). These results strengthen the hypothesis that unknown activators with binding sites within the phoBR regulatory region play roles in its transcriptional control.
Pho regulon expression in V. cholerae requires a sequence containing Pho boxes 1, 2, and 3 immediately upstream of the translation start codon of the phoB gene.
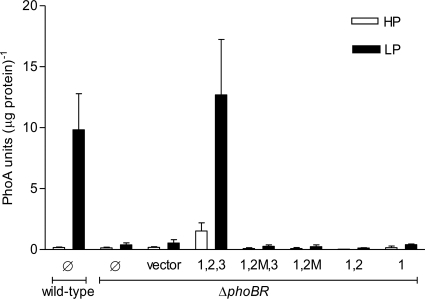
To better understand the regulatory role of each of the three phoBR Pho boxes in the expression of the operon in vivo, ΔphoBR mutant cells were complemented with low-copy pWKS30 derivatives containing phoBR genes under the control of different fragments of its regulatory region (Table 1). Cells were grown under high (HP) and low (LP) Pi levels for 16 h, and the expression of the Pho regulon genes was indirectly monitored by the activity of alkaline phosphatase (PhoA), the natural reporter of the regulon (Fig. 6). Only ΔphoBR cells complemented with box1,2,3∼phoBR expressed PhoA at a level similar to that of the wild-type strain under Pi limitation. This observation corroborates the hypothesis that the three Pho boxes are required simultaneously for the efficient expression of the Pho regulon (Fig. 5a and 6).
Fig. 6.
PhoA activity levels in the V. cholerae ΔphoBR mutant complemented with low-copy-number plasmids containing the phoBR operon under the control of different versions of its regulatory region. PhoA activity was measured in cells grown under high (HP) and low (LP) Pi conditions. ΔphoBR cells and ΔphoBR cells transformed with the empty vector were used as negative controls, and the wild-type strain was used as a positive control. Ø, no plasmid; vector, empty plasmid; 1,2,3, boxes 1, 2, and 3 fused to phoBR; 1,2 M,3, box 1, mutated box 2, and box 3 fused to phoBR; 1,2 M, box 1 and mutated box 2 fused to phoBR; 1,2, boxes 1 and 2 fused to phoBR; 1, box 1 fused to phoBR.
These findings, together with those of the lacZ fusion assays (Fig. 5), point to a complex mode of regulation of the V. cholerae phoBR operon. β-Galactosidase activity assays revealed that the phoBR promoter region containing only Pho box 1 was transcriptionally activated by PhoB under low Pi conditions in wild-type cells. However, this version of the phoBR promoter was not active in the phoB-deficient background, since no PhoA expression was detected under low Pi conditions. An obvious explanation is that ΔphoBR is a PhoB-deficient strain and a basal level of the protein is required to induce phoBR expression to a detectable level. On the other hand, ΔphoBR mutant cells complemented with the full phoBR regulatory region (box1,2,3∼phoBR) presented PhoA activity under low Pi, suggesting phoBR expression. The slightly higher PhoA activity in ΔphoBR and box1,2,3∼phoBR cells relative to the wild type might be due to the vector used for the complementation assays, which is present in 6 to 8 copies per cell (49). Promoter versions with box1,2mut,3 or box1,2mut did not drive the transcription of phoBR in ΔphoBR cells under Pi limitation, since no PhoA expression was observed. Nevertheless, they activated lacZ transcription in wild-type cells (Fig. 5a). These findings indicate that the expression of Pho regulon genes in vivo depends on a basal level of PhoB in the cells which, in turn, relies on the presence of a full-length regulatory region of phoBR.
Concluding remarks.
V. cholerae Pho regulon members have important roles, not only in the response to environmental phosphate limitation but also in pathogenesis (9, 10, 36, 37, 43, 47, 48). Therefore, it was not surprising to discover a tight control over the expression of phoBR and, consequently, of PhoB-dependent genes.
Data presented here indicate that the expression of the V. cholerae phoBR operon depends on a very peculiar binding pattern of PhoB to three Pho boxes. Moreover, the interaction of PhoB with distinct boxes either activates or represses phoBR expression. Based on these observations and on the set of results of the present study, we propose a hypothesis to explain the molecular events involved in V. cholerae phoBR autoregulation. Under Pi limiting conditions, basally expressed PhoB, activated by phosphorylation, binds with high affinity to Pho box 1, which is closest to the σ70 binding site on the phoBR regulatory region. PhoB∼box1 complex would cause a considerable bending in the DNA molecule, thus enabling interaction between PhoB and the RNA polymerase to activate transcription, as previously observed in E. coli (24). As the PhoB cellular level increases by positive autoregulation, the protein would bind to the lower affinity box 2, upstream of box 1, in the antisense promoter strand. Operon fusion assays showed virtually no lacZ transcription from the regulatory sequence carrying only boxes 1 and 2 in wild-type cells. Thus, we suggest that the binding of PhoB to box 2 would introduce changes in the spatial organization of the promoter sequence, reducing the interaction between PhoB∼box1 and the RNA polymerase σ70. More precisely, PhoB∼box 2 (in the antisense strand) might bend the DNA in the opposite orientation, affecting negatively the interaction of PhoB∼box1-RNA polymerase, thereby reducing the transcriptional rate of phoBR.
As shown above, the binding of PhoB to box 3 seems to require PhoB∼box2, which is not surprising, since box 3 contains more mismatches relative to the consensus sequence than box 1 and box 2 and, from the three loci, is the one with the lowest binding affinity for PhoB. Pho box 3 is in the antisense strand of the phoBR promoter, thus the complex PhoB∼box3 might further alter the structure of the regulatory sequence and the spatial arrangement of the bound molecules with respect to each other. This might bring PhoB∼box 1 into close proximity to the RNA polymerase, thus activating the transcription of phoBR. Rounds of the binding/unbinding of PhoB to these boxes and the resulting changes in structural features of the regulatory region ultimately would keep the transcription under tight control and the expression levels of the operon adjusted to the cell needs. Thus, PhoB exerts a dual regulatory function, as an activator and repressor, by binding to distinct Pho boxes in a sequential mode and with different affinities, enabling V. cholerae cells to fine-tune the expression of the phoBR operon under Pi limitation. Under Pi abundance, the V. cholerae phoBR operon does seem to be expressed, but at a lower level, apparently by the interaction of unknown activators to its regulatory region. This would ensure a basal level of PhoB, which is required not only for the Pi-independent roles that PhoBR plays in the cells but also for the expression of the Pho regulon when V. cholerae faces Pi-limiting conditions.
ACKNOWLEDGMENTS
We are grateful to Celso Pereira, Eduardo Camacho, and Lilian Ayres Sá for technical support.
We thank FINEP/Genoprot Project 0107054600, FAPERJ, CNPq/INBEB Project, and CAPES for financial support.
Footnotes
Published ahead of print on 7 October 2011.
REFERENCES
- 1. Ausubel F. M., et al. 1995. Short protocols in molecular biology. John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 2. Bailey T. L., et al. 2009. MEME suite: tools for motif discovery and searching. Nucleic Acids Res. 37:W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blanco A. G., Canals A., Bernues J., Sola M., Coll M. 2011. The structure of a transcription activation subcomplex reveals how sigma(70) is recruited to PhoB promoters. EMBO J. 30:3776–3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cash R. A., et al. 1974. Response of man to infection with Vibrio cholerae. I. Clinical, serologic, and bacteriologic responses to a known inoculum. J. Infect. Dis. 129:45–52 [DOI] [PubMed] [Google Scholar]
- 5. Echols H., Garen A., Garen S., Torriani A. 1961. Genetic control of repression of alkaline phosphatase in E. coli. J. Mol. Biol. 3:425–438 [DOI] [PubMed] [Google Scholar]
- 6. Faruque S. M., Albert M. J., Mekalanos J. J. 1998. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62:1301–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Faruque S. M., Nair G. B. 2002. Molecular ecology of toxigenic Vibrio cholerae. Microbiol. Immunol. 46:59–66 [DOI] [PubMed] [Google Scholar]
- 8. Frith M. C., Saunders N. F., Kobe B., Bailey T. L. 2008. Discovering sequence motifs with arbitrary insertions and deletions. PLoS Comput. Biol. 4:e1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goulart C. L., et al. 2009. Molecular analysis of VCA1008: a putative phosphoporin of Vibrio cholerae. FEMS Microbiol. Lett. 298:241–248 [DOI] [PubMed] [Google Scholar]
- 10. Goulart C. L., et al. 2010. A ToxR-dependent role for the putative phosphoporin VCA1008 in bile salt resistance in Vibrio cholerae El Tor N16961. Microbiology 156:3011–3020 [DOI] [PubMed] [Google Scholar]
- 11. Herrington D. A., et al. 1988. Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J. Exp. Med. 168:1487–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hsieh Y. J., Wanner B. L. 2010. Global regulation by the seven-component Pi signaling system. Curr. Opin. Microbiol. 13:198–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hulett F. M. 1996. The signal-transduction network for Pho regulation in Bacillus subtilis. Mol. Microbiol. 19:933–939 [DOI] [PubMed] [Google Scholar]
- 14. Innis M. A., Gelfand D. H. 1990. Optimization of PCRs, p. 3–12 In Innis M. A., Gelfand D. H., Sninsky J. J., White T. J. (ed.), PCR protocols. A guide to methods an applications. Academic Press, Inc., San Diego, CA [Google Scholar]
- 15. Kasahara M., Makino K., Amemura M., Nakata A., Shinagawa H. 1991. Dual regulation of the ugp operon by phosphate and carbon starvation at two interspaced promoters. J. Bacteriol. 173:549–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim S. K., et al. 2000. Dual transcriptional regulation of the Escherichia coli phosphate-starvation-inducible psiE gene of the phosphate regulon by PhoB and the cyclic AMP (cAMP)-cAMP receptor protein complex. J. Bacteriol. 182:5596–5599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kimura S., Makino K., Shinagawa H., Amemura M., Nakata A. 1989. Regulation of the phosphate regulon of Escherichia coli: characterization of the promoter of the pstS gene. Mol. Gen. Genet. 215:374–380 [DOI] [PubMed] [Google Scholar]
- 18. Lamarche M. G., Wanner B. L., Crepin S., Harel J. 2008. The phosphate regulon and bacterial virulence: a regulatory network connecting phosphate homeostasis and pathogenesis. FEMS Microbiol. Rev. 32:461–473 [DOI] [PubMed] [Google Scholar]
- 19. Lee S. H., Butler S. M., Camilli A. 2001. Selection for in vivo regulators of bacterial virulence. Proc. Natl. Acad. Sci. U. S. A. 98:6889–6894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee S. H., Hava D. L., Waldor M. K., Camilli A. 1999. Regulation and temporal expression patterns of Vibrio cholerae virulence genes during infection. Cell 99:625–634 [DOI] [PubMed] [Google Scholar]
- 21. Liu W., Eder S., Hulett F. M. 1998. Analysis of Bacillus subtilis tagAB and tagDEF expression during phosphate starvation identifies a repressor role for PhoP-P. J. Bacteriol. 180:753–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265–275 [PubMed] [Google Scholar]
- 23. Macián F., Perez-Roger I., Armengod M. E. 1994. An improved vector system for constructing transcriptional lacZ fusions: analysis of regulation of the dnaA, dnaN, recF and gyrB genes of Escherichia coli. Gene 145:17–24 [DOI] [PubMed] [Google Scholar]
- 24. Makino K., et al. 1996. DNA binding of PhoB and its interaction with RNA polymerase. J. Mol. Biol. 259:15–26 [DOI] [PubMed] [Google Scholar]
- 25. Makino K., Amemura M., Kim S. K., Nakata A., Shinagawa H. 1993. Role of the sigma 70 subunit of RNA polymerase in transcriptional activation by activator protein PhoB in Escherichia coli. Genes Dev. 7:149–160 [DOI] [PubMed] [Google Scholar]
- 26. Makino K., et al. 1989. Signal transduction in the phosphate regulon of Escherichia coli involves phosphotransfer between PhoR and PhoB proteins. J. Mol. Biol. 210:551–559 [DOI] [PubMed] [Google Scholar]
- 27. Makino K., et al. 1988. Regulation of the phosphate regulon of Escherichia coli. Activation of pstS transcription by PhoB protein in vitro. J. Mol. Biol. 203:85–95 [DOI] [PubMed] [Google Scholar]
- 28. Makino K., Shinagawa H., Amemura M., Nakata A. 1986. Nucleotide sequence of the phoB gene, the positive regulatory gene for the phosphate regulon of Escherichia coli K-12. J. Mol. Biol. 190:37–44 [DOI] [PubMed] [Google Scholar]
- 29. Marcus H., Ketley J. M., Kaper J. B., Holmes R. K. 1990. Effects of DNase production, plasmid size, and restriction barriers on transformation of Vibrio cholerae by electroporation and osmotic shock. FEMS Microbiol. Lett. 56:149–154 [DOI] [PubMed] [Google Scholar]
- 30. Maxam A. M., Gilbert W. 1992. A new method for sequencing DNA. Biotechnology 24:99–103 [PubMed] [Google Scholar]
- 31. McCleary W. R. 1996. The activation of PhoB by acetylphosphate. Mol. Microbiol. 20:1155–1163 [DOI] [PubMed] [Google Scholar]
- 32. Miller V. L., Mekalanos J. J. 1985. Genetic analysis of the cholera toxin-positive regulatory gene toxR. J. Bacteriol. 163:580–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Paul S., Birkey S., Liu W., Hulett F. M. 2004. Autoinduction of Bacillus subtilis phoPR operon transcription results from enhanced transcription from EsigmaA- and EsigmaE-responsive promoters by phosphorylated PhoP. J. Bacteriol. 186:4262–4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Peterson K. M., Mekalanos J. J. 1988. Characterization of the Vibrio cholerae ToxR regulon: identification of novel genes involved in intestinal colonization. Infect. Immun. 56:2822–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Prágai Z., et al. 2004. Transcriptional regulation of the phoPR operon in Bacillus subtilis. J. Bacteriol. 186:1182–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pratt J. T., Ismail A. M., Camilli A. 2010. PhoB regulates both environmental and virulence gene expression in Vibrio cholerae. Mol. Microbiol. 77:1595–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pratt J. T., McDonough E., Camilli A. 2009. PhoB regulates motility, biofilms, and cyclic di-GMP in Vibrio cholerae. J. Bacteriol. 191:6632–6642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Raleigh E. A., et al. 1988. McrA and McrB restriction phenotypes of some E. coli strains and implications for gene cloning. Nucleic Acids Res. 16:1563–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 40. Shevchenko A., et al. 1996. A strategy for identifying gel-separated proteins in sequence databases by MS alone. Biochem. Soc. Trans. 24:893–896 [DOI] [PubMed] [Google Scholar]
- 41. Sola-Landa A., Rodriguez-Garcia A., Franco-Dominguez E., Martin J. F. 2005. Binding of PhoP to promoters of phosphate-regulated genes in Streptomyces coelicolor: identification of Pho boxes. Mol. Microbiol. 56:1373–1385 [DOI] [PubMed] [Google Scholar]
- 42. Studier F. H., Moffat B. A. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113–130 [DOI] [PubMed] [Google Scholar]
- 43. Sultan S. Z., Silva A. J., Benitez J. A. 2010. The PhoB regulatory system modulates biofilm formation and stress response in El Tor biotype Vibrio cholerae. FEMS Microbiol. Lett. 302:22–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Taylor R. K., Miller V. L., Furlong D. B., Mekalanos J. J. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. U. S. A. 84:2833–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tommassen J., de Geus P., Lugtenberg B., Hackett J., Reeves P. 1982. Regulation of the Pho regulon of Escherichia coli K-12. Cloning of the regulatory genes phoB and phoR and identification of their gene products. J. Mol. Biol. 157:265–274 [DOI] [PubMed] [Google Scholar]
- 46. Vieira R. P., et al. 2008. Relationships between bacterial diversity and environmental variables in a tropical marine environment, Rio de Janeiro. Environ. Microbiol. 10:189–199 [DOI] [PubMed] [Google Scholar]
- 47. von Krüger W. M., Humphreys S., Ketley J. M. 1999. A role for the PhoBR regulatory system homologue in the Vibrio cholerae phosphate-limitation response and intestinal colonization. Microbiology 145:2463–2475 [DOI] [PubMed] [Google Scholar]
- 48. von Krüger W. M., et al. 2006. The phosphate-starvation response in Vibrio cholerae O1 and phoB mutant under proteomic analysis: disclosing functions involved in adaptation, survival and virulence. Proteomics 6:1495–1511 [DOI] [PubMed] [Google Scholar]
- 49. Wang R. F., Kushner S. R. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195–199 [PubMed] [Google Scholar]
- 50. Wanner B. L., Chang B. D. 1987. The phoBR operon in Escherichia coli K-12. J. Bacteriol. 169:5569–5574 [DOI] [PMC free article] [PubMed] [Google Scholar]