Abstract
The filamentous cyanobacterium Nostoc punctiforme differentiates from vegetative cells into three distinct cell types, heterocysts, hormogonia, and akinetes, in response to different stimuli. Cultures growing with ammonium can be induced to form hormogonia or heterocysts upon removal of the combined nitrogen. A DNA microarray consisting of 94% of the open reading frames predicted from the 9.059-Mb N. punctiforme genome was used to generate a global transcription data set consisting of seven time points over a 24-h period of nitrogen deprivation, which results in heterocyst formation. This data set was compared to a similarly generated data set of nitrogen-starved N. punctiforme resulting in hormogonium formation that had previously been published (E. L. Campbell, H. Christman, and J. C. Meeks, J. Bacteriol. 190:7382-7391, 2008). The transition from vegetative cells to either heterocysts or hormogonia resulted in rapid and sustained expression of genes required for utilization of alternate nitrogen sources. Overall, 1,036 and 1,762 genes were found to be differentially transcribed during the heterocyst and hormogonium time courses, respectively, as analyzed with the Bayesian user-friendly software for analyzing time series microarray experiments (BATS). Successive transcription of heterocyst regulatory, structural, and functional genes occurred over the 24 h required to form a functional heterocyst. During hormogonium differentiation, some heterocyst structural and functional genes were upregulated, while the heterocyst master regulator hetR was downregulated. There are commonalities in differential expression between cells bound for differentiation into heterocysts or hormogonia, yet the two paths are distinguished by their developmentally specific transcription profiles.
INTRODUCTION
Nostoc punctiforme is a highly versatile cyanobacterium in its physiological and developmental capabilities. It grows as long chains of vegetative cells when nutrients are replete and is capable of photoautotrophy, photoheterotrophy, or respiratory heterotrophy in the dark. When specific nutrients are limited or other environmental signals are sensed, N. punctiforme is capable of differentiating into three distinct cell types: akinetes, hormogonia, and heterocysts (41). In response to combined nitrogen (N) starvation, either heterocysts or hormogonia will differentiate, but a single filament cannot undergo both developmental paths (43). Depletion of combined N is the only known stimulant of heterocyst formation. These specialized cells form singly within intervals of 10 to 15 vegetative cells and provide a microoxic compartment to house the oxygen-labile nitrogenase enzyme complex that catalyzes conversion of dinitrogen to ammonium. Oxygen partial pressure is reduced in these cells by the following: development of additional cell envelope material composed of glycolipids and polysaccharides; dismantlement of oxygen-evolving components of photosystem II; increased respiration through heterocyst-specific cytochrome oxidase enzymes (54). In various filamentous cyanobacteria, hormogonia may be induced by many other signals besides N starvation, including excess phosphate (17) or a change in osmolarity (38), ionic strength (5), or age (31) of the medium, a shift from white or green light to red light (15), or a yet-to-be-identified signal exuded from symbiotic plant partners (11).
Hormogonia form transiently as part of a conditional developmental life cycle. After the environmental cue is received, all cells in the filament divide, DNA replication ceases (31), and net protein accumulation (11) and photosynthetic pigment production (15) are arrested. This results in a filament with cells much smaller than the original vegetative cells, and the shapes of the filament end cells become tapered. Hormogonia can also be distinguished by their accumulation of gas vacuoles and storage granules (1). About 12 h after induction, filaments exhibit gliding motility, which requires contact with a surface. The hormogonia are motile until about 48 h after induction and, after another 24 h, the tapered end cells differentiate into heterocysts when cultured in the absence of combined N. The remaining cells then resume growth as normal vegetative cells, ultimately as filaments with regularly spaced heterocysts. Hormogonium formation is required for infection of a symbiotic plant partner. Following infection, filaments differentiate heterocysts at a much higher frequency per vegetative cell and with a different pattern (43). This is the only known naturally occurring event that substantially changes the pattern and frequency of heterocysts in the filament.
Heterocyst differentiation, as well as nitrogenase expression, is but one avenue for cyanobacteria to utilize an alternate N source, albeit the most complex and energy-intensive. Much of the work toward understanding heterocyst differentiation in the free-living state draws from study of a closely related cyanobacterium, Anabaena (Nostoc) sp. strain PCC 7120 (here, Anabaena 7120) (23, 58, 60). Neither hormogonium nor akinete differentiation has been documented in Anabaena 7120, but the mechanisms governing heterocyst differentiation largely appear to be conserved among the heterocyst-forming cyanobacteria. Many cyanobacteria, including N. punctiforme, can use several different compounds as exogenous N sources, including ammonium, nitrate, urea, certain amino acids, and dinitrogen (25, 32). Excluding dinitrogen, these compounds are all sufficient to suppress heterocyst differentiation; ammonium requires the least amount of energy to assimilate, and it represses expression of genes required for utilization of other N sources (25, 32). However, whether amino acids, such as arginine and glutamine, effectively repress heterocyst differentiation in all strains is equivocal (see the discussion in reference 25). When ammonium is removed from a culture actively growing with it as the N source, the global nitrogen regulator NtcA is activated and positively regulates genes required for nitrate-nitrite transport and assimilation (24) and for urea transport (52). In the absence of these other sources of combined N, heterocyst formation is initiated. The earliest-known regulator required for timely heterocyst formation to be differentially transcribed was nrrA, encoding a response regulator. nrrA transcription is directly activated by NtcA (45), and NrrA in turn enhances expression of hetR (18). HetR activates transcription of several heterocyst regulatory genes, including itself, ntcA, patS, and hetN, and is essential to heterocyst formation. PatS and HetN are negative regulators required for establishment and maintenance of the heterocyst spacing pattern, respectively (10, 58). PatS inhibits HetR binding to DNA, thus decreasing expression of hetR and other genes dependent on HetR (36). These regulatory interactions culminate in the expression of the genes whose products are required for synthesis of the heterocyst-specific glycolipids, polysaccharides, and cytochrome oxidases that are instrumental in maintaining low O2 concentrations (54). There is also a decrease in synthesis of photosystem II components and carbon dioxide fixation in the heterocyst (54). Unlike hormogonium differentiation, heterocyst formation is a terminal event, as the ability to replicate is sacrificed in order to provide surrounding vegetative cells with combined N.
Since N. punctiforme will differentiate nitrogen stress-induced heterocysts (NSI-Het) or hormogonia (NSI-Hrm), this cyanobacterium provides a unique experimental system for the identification of common and branched signal transduction pathways leading to each of these developmental outcomes. Previous studies have examined global gene expression patterns in cultures of N. punctiforme growing diazotrophically and hormogonia 24 h after induction by N starvation (13); 495 and 1,827 genes, respectively, were differentially transcribed in comparison to ammonium-grown cells, thus showing a much greater change in transcription in hormogonia- than heterocyst-containing filaments. Time course analyses have also been performed, comparing hormogonia induced by N starvation or by the presence of a hormogonium-inducing factor (HIF) from the symbiotic plant partner Anthoceros punctatus. This comparison revealed a massive change in gene expression and a number of temporal patterns driven by each stimulus (12). The HIF induction data set indicated that the plant factor strongly polarizes the cells toward hormogonium formation, and NSI leads to a vacillating response, where a transcriptional checkpoint is realized before ultimately reaching the necessary outcome for hormogonium differentiation. Within each of these time series experiments, a common set of genes showing elevated or repressed transcription was defined.
To further define the N starvation, or stress, response, we generated a global transcription data set consisting of seven time points from 0.5 to 24 h after the N step-down that led to heterocyst differentiation in N. punctiforme. This data set was compared to the above-described NSI hormogonium data set in an effort to distinguish the N stress response from each developmental response. Differential transcription between the 24-h time point after N step-down and steady-state diazotrophic growth was also examined and revealed a large number of genes differentially transcribed at 24 h that returned to expression levels prior to N step-down.
MATERIALS AND METHODS
Strains and culture conditions.
Stock cultures of strain UCD 102 (an N. punctiforme strain ATCC 29133 spontaneous hormogonium-deficient mutant) were grown in 50 ml of Allan and Arnon minimal salts medium (3) diluted 4-fold (AA/4). Flasks containing 500 ml AA/4 supplemented with 2.5 mM NH4Cl and 5.0 mM morpholinepropanesulfonic acid (MOPS; pH 7.8) were inoculated from stock cultures and grown to a chlorophyll a (Chl a) concentration of 2 to 3 μg/ml for 7 days at 22°C with shaking at 200 rpm before being subjected to N step-down. Chl a was monitored spectrophotometrically in methanolic extracts (42). N step-downs were carried out by centrifuging cells at 1,000 × g for 5 min, washing three times with AA/4, and then suspending in combined N-free AA/4 medium. A 50-ml volume of cells was harvested at 0.5, 1, 3, 6, 12, 18, and 24 h after N step-down, frozen quickly in liquid nitrogen, and stored at −80°C prior to RNA extraction. Growth was maintained in constant light at about 14 μmol photons/m2/s from cool white fluorescent light bulbs. For growth rate studies, ammonium-grown cultures with a Chl a concentration of 1 μg/ml were subjected to N step-down. Changes in dry weight and Chl a concentration over time were monitored to generate growth curves. Dry weight was determined on samples collected on a tared 0.22-μm membrane filter, and filters were air dried.
RNA extraction and hybridization.
RNA extraction, cDNA synthesis, cDNA labeling, and hybridizations were performed as previously described (13). Briefly, 100 ml of culture containing a Chl a concentration of 2 to 3 μg/ml was concentrated to 500 μl, and RNA was isolated by phenol-chloroform extraction and lithium chloride precipitation and then further purified with Qiagen RNeasy columns. Fifteen micrograms of total RNA was used for cDNA synthesis, employing the Invitrogen SuperScript Plus indirect cDNA synthesis system, and labeled with Alexa Fluor dyes 555 and 647. Microarray slides were hybridized using a Tecan HS4800 hybridization station at 50°C. cDNA from each time point was mixed with the control time zero (ammonium-grown N. punctiforme) cDNA and hybridized. Each time point consisted of three biological replicates that were also dye swapped with the control time zero sample; since the slides were printed in duplicate, this resulted in 12 technical replicates to assess differential expression. Microarray slides were printed by the Microarray Core on the UC Davis campus and were composed of PCR products of internal gene fragments representing 6,893 computationally determined open reading frames (ORF) of the N. punctiforme genome (44).
Data analysis.
Slides were scanned with a GenePix Pro 4000B scanner (Molecular Devices). A grid of the spots was created, and spots were sized manually using GenePix Pro 3.0 software to create GenePix result files (.gpr). Data contained in .gpr files were normalized, and statistical analyses were performed using the R package LIMMA GUI (49) as described previously (13). The resultant top tables (tab-delimited text files containing normalized expression values for each ORF with statistical information, such as P, adjusted P, and B values) were used to define statistically significant differential expression for individual time point comparisons. The statistical term B is defined as the log odds that a gene is differentially expressed (eB), and it takes into account multiple testing, such as t and P values. The higher a positive B value, the greater the probability that the experimental value differs from the reference; a B of 0 reflects a 50% probability.
For time series analyses, normalized M values [log2(experimental) − log2(reference)] were computed from the same .gpr files as above using the R package LIMMA GUI. These tab-delimited text files contained normalized M values for each of the three biological replicates. The M values for duplicate spots were averaged and, with dye swaps, this resulted in 6 M values for each gene. These data were analyzed with the Bayesian user-friendly software for analyzing time series microarray experiments (BATS) (4), using the default settings. The NSI-Hrm time course data from Campbell et al. (12) were compared to the NSI-Het time course results obtained in this study. This comparison is biologically significant, because cultures having undergone the same environmental stress differentiate into two mutually exclusive developmental states. After significant genes were determined using BATS, K-means clustering was performed with Cluster 3.0 (16), and heat maps were generated with Java TreeView (48). Microsoft Excel was used to construct graphs. The raw data for the experiments in this paper, .gpr files, are accessible at http://microbiology.ucdavis.edu/Meeks/, under the heading microarray data and the paper title.
RESULTS AND DISCUSSION
Time course analyses using BATS.
For rigorous statistical analysis of time series data, BATS (4) was chosen. The BATS algorithm fits the time points of each gene to a polynomial function. A vector of coefficients from these polynomials is constructed and used to perform Bayesian statistical analysis to determine the time series that are significantly different than zero. This approach retains the temporal nature of the data to perform the analysis, rather than testing each point independently. In a study of genome-wide expression in human breast cancer cells, BATS had the greatest success in determining statistically significant genes with known biological relevance for time series data over the other three methods compared (46). Genes determined to be statistically relevant via BATS were clustered to determine similar patterns of expression within each time course. The number of K-means clusters chosen for each time course was determined by performing variance analysis, as described previously (12) (see also Fig. S1 in the supplemental material). In both time courses, variance leveled off at 10 clusters, but only 6 clusters were chosen for the NSI-Het time course because there was very little difference in transcriptional patterns with the additional 4 clusters. For comparison of the single time points, genes with a B value of ≥0 were deemed significant, as applied previously (13).
Use of BATS to determine differentially transcribed genes in the heterocyst time series analysis indicated 1,036 genes were significantly up- or downregulated (see Table S1 in the supplemental material). Compared to the 495 genes differentially transcribed during steady-state diazotrophic growth reported previously (13), and the approximately 431 genes obtained via deep RNA sequencing of Anabaena 7120 at 21 h after N step-down (23), the differences in up- or downregulation were quite large, but they were consistent with the 1,030 predicted significant genes at the 24-h time point as determined in R with B at ≥0 (see Fig. 8, below). These findings are also consistent with an estimate of 600 to 1,000 genes transcribed during heterocyst formation 48 h after N step-down in Anabaena variabilis, based on RNA-DNA hybridization (40). In the NSI hormogonium time course, 1,762 genes were detected through BATS (see Table S2 in the supplemental material), while a similar number of genes, 1,847, was found by R analysis to be differentially transcribed at 24 h in NSI hormogonia (12). These numbers show statistically significant changes from time zero values, irrespective of the extent of change. One commonly sees arbitrary selection of greater-than-2-fold changes in differential transcription, which are assumed to reflect biologically significant changes. Although the timing and direction of change may be suggestive, there is no method for predicting the biological relevance of any extent of changes in transcription levels in lieu of a biological assay.
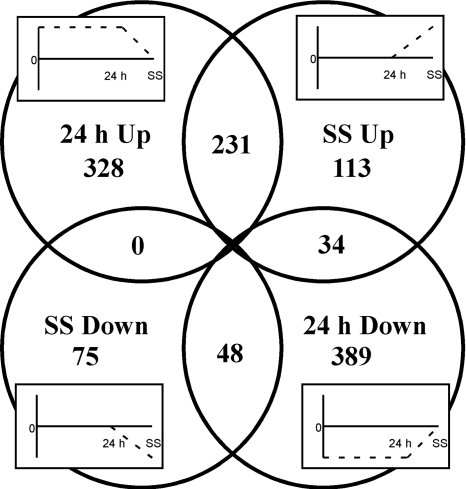
Fig. 8.
Venn diagram summarizing the comparison of 24-h post-N step-down to steady-state diazotrophic growth. Genes that were significantly up- or downregulated as determined by B values of ≥0 at 24 h and steady state (SS) were compared. The four plot insets depict the general patterns of change in expression for each set of genes indicated. For example, there are 75 genes downregulated at SS that show no change at 24 h.
Functional categorization of differentially transcribed genes revealed that the percentage of genes in each category for both time courses resembled the percentage represented in the genome (Table 1). While approximately 40% of the genes do not yet have a metabolic function assigned to them (unassigned category), their differential transcription supports their identification as encoding a physiologically relevant protein. These ORFs encoding unassigned proteins represent a largely unexplored portion of the N. punctiforme genome, the characterization of which may begin with systems-level studies such as these by defining their transcription in the context of known genes. Core metabolism represents the category of genes required for metabolic processes, such as energy conservation and biosynthesis, required for growth. The NSI-Hrm time course had a slightly higher number of genes in the core metabolic category than heterocysts, and most of these genes were downregulated, as all cells in the filament enter a nongrowth state. Conversely, the NSI-Het time course had a larger portion of genes in the adaptive metabolism category than NSI-Hrm, reflecting the extensive de novo structural and metabolic changes in the differentiation of a highly specialized heterocyst from a vegetative cell. The number of differentially transcribed genes assigned to transport function was similarly small in both time courses. Differential expression of putative computationally defined pseudogenes and selfish metabolism genes, such as those encoding transposases and phage remnants, was very minimal.
Table 1.
Functional categorizations of BATS-determined significant genes
| Functional category | No. (%) of genes in category among: |
|||
|---|---|---|---|---|
| Genome | NSI-Het (n = 1,036) | NSI-Hrm (n = 1,762) | Common (n = 619) | |
| Adaptive | 1,017 (14) | 191 (18) | 259 (15) | 96 (16) |
| Core | 1,584 (22) | 333 (32) | 620 (35) | 231 (37) |
| Transport | 393 (5) | 70 (6.8) | 105 (6) | 40 (6.5) |
| Selfish | 238 (3) | 2 (0.2) | 9 (0.6) | 2 (0.3) |
| Unassigned | 3,548 (48) | 433 (42) | 759 (43) | 249 (40) |
| Pseudogenes | 555 (8) | 7 (0.7) | 10 (0.6) | 1 (0.2) |
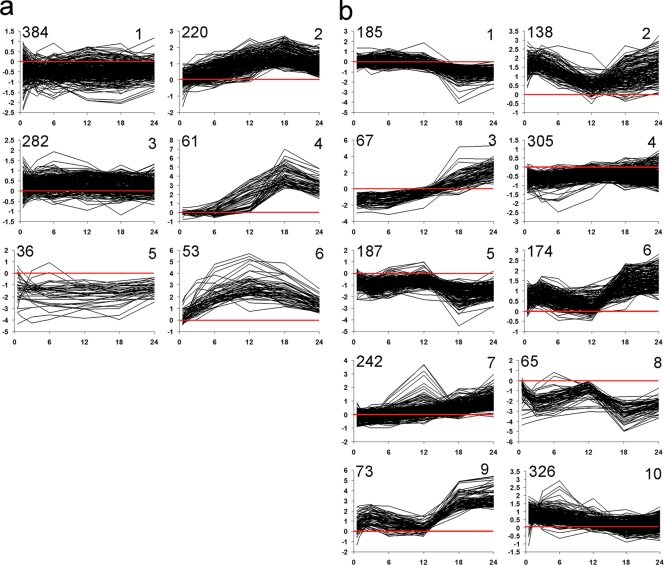
Each time course group was divided into clusters of genes with similar temporal patterns of expression by using the K-means clustering algorithm. The six clusters representing the NSI-Het time course (Fig. 1a) showed relatively simple patterns of temporal expression compared to the NSI-Hrm time course. The majority of differentially transcribed genes grouped into clusters 1 and 3, showing 384 downregulated and 282 upregulated, respectively, by less than 2-fold; rejection of the less-than-2-fold changes would yield 370 differentially transcribed genes. There are only 36 genes that showed strong downregulation (cluster 5); the remainder were divided among the upregulated genes, with cluster 2 representing those with late expression at moderate levels and clusters 6 and 4 showing high expression early and late, respectively. The temporal patterns in clusters 6, 4, and 2 were consistent with the sequential expression of genes during heterocyst development found in other heterocyst-forming cyanobacteria (9, 23), and their members will be further discussed below. The 10 NSI-Hrm clusters showed multiple distinct patterns of expression (Fig. 1b) that resembled the patterns determined previously using the complete data set (12), including the presence of a 12-h transcriptional checkpoint visible in clusters 2, 6, 7, and 8. Again, except for a few outliers, the clusters with the highest number of genes showed a less-than-2-fold change in expression (clusters 4 and 10). One reason to examine NSI hormogonia was to use a comparative approach in identifying aspects of the N stress response. Thus, processes relevant to the N stress response were analyzed here, while BATS analysis of those relevant to hormogonium biology will be addressed in a another publication prepared by our group (E. L. Campbell, H. D. Christman, and J. C. Meeks, submitted for publication).
Fig. 1.
Graphical representation of K-means clusters of 1,037 BATS-determined NSI-Het genes (a) and 1,762 BATS-determined NSI-Hrm genes (b). The x axis shows time (in h), and the y axis, M, shows the log2(experimental) − log2(reference) values. Minimum and maximum M values on the y axis are different in each plot to allow better visualization of the extent of statistically significant differentially transcribed genes. The red lines highlight where 0 lies on the y axes. The number of genes within each cluster is indicated in the upper left corner of each plot.
Genes in common between the NSI hormogonium and heterocyst time series.
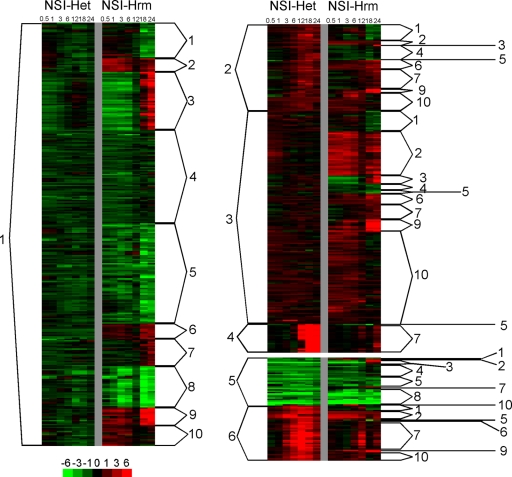
Of the total number of differentially transcribed genes in each time course, 619 were in common with the two time courses. Figure 2 is a heat map depiction of the expression values for the 619 common genes, organized by cluster number. About 55% (340) of the common genes were in NSI-Het-downregulated clusters 1 and 5. These represent the majority of all the genes downregulated during heterocyst differentiation: 305 of 384 and 35 of 36 for NSI-Het clusters 1 and 5, respectively (Fig. 2). Many of these genes showed an overall trend of downregulation in NSI-Hrm, but the pattern and magnitude of differential expression between many of them were different in these and other clusters. NSI-Hrm downregulated clusters 1, 4, 5, and 8, which are, in common with NSI-Het clusters 1 and 5, mostly genes encoding products for polymer synthesis and energy metabolism (see Table S2 in the supplemental material). These NSI-Hrm expression patterns are consistent with the documented entry into a nongrowth state by all cells of a hormogonium filament (11). The NSI-Het patterns implied that N starvation also results in a transient nongrowth state of vegetative cells that do not differentiate into heterocysts (elaborated below). The fact that the temporal patterns of change differed for the two developmental processes implies that either common regulatory mechanisms are activated in a different sequence following N step-down or that different mechanisms are involved.
Fig. 2.
Heat map representation of 619 genes in common between the NSI-Het and NSI-Hrm time courses. Color intensities of red and green areas indicate degrees of up- and downregulation, respectively, in reference to ammonium-grown cultures (color bar at bottom left). Numbers at the top are the time (in h) after N step-down. The numbers on the sides indicate K-means clusters for each time course.
Some of the common genes that are upregulated in NSI-Het also showed a degree of upregulation in NSI-Hrm. Of particular note, NSI-Het clusters 2, 4, and 6 contain some heterocyst regulatory, structural, and functional genes that have been shown to be under heterocyst developmental control in Anabaena 7120 (20). Most of these genes group into NSI-Hrm cluster 7 and are described in more detail below. NSI-Het cluster 3 represents genes with transient upregulation at different points in the time course; corresponding common genes in NSI-Hrm clusters 2, 6, and 9 are of interest because there is a transient decrease in expression at 12 h followed by a robust increase, characteristic of the transcriptional checkpoint described previously (12). These clusters have some genes that would more likely be involved in hormogonium function, such as pilin synthesis (NpR0117 and NpF0676) and chemotaxis signaling (NpF5962 and NpF2165).
The search for alternate N compounds for growth is realized in both developmental fates.
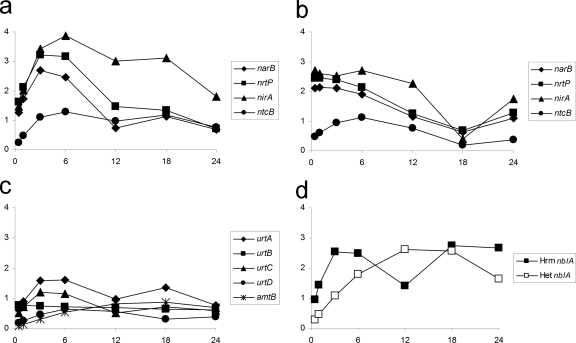
The earliest upregulated genes during the N stress response grouped into NSI-Het clusters 2 and 6 and NSI-Hrm clusters 2 and 6. Three genes, narB (NpR1526), nrtP (NpR1527), and nirA (NpR1528), encoding nitrate reductase, nitrate-nitrite permease, and nitrite reductase, were upregulated by 0.5 h in both time courses but showed a transient decrease in expression at 18 h in the NSI-Hrm data set (Fig. 3). The LysR-type transcriptional regulator encoded by ntcB (NpF1534) was also enhanced early in both states (Fig. 3). Transcription of the nitrate-nitrite metabolic genes is dependent on NtcB, as well as NtcA (27, 50). The presence of a nitrate-nitrite permease in terrestrial N. punctiforme is similar to that of the marine cyanobacterium Trichodesmium erythraeum (53), in contrast to the ABC-type nitrate-nitrite transporter, encoded by nrtABCD found in Anabaena 7120 and other freshwater cyanobacteria (26). N. punctiforme has putative homologs to nrtABCD that are downregulated in NSI-Het clusters 1 and 5 and NSI-Hrm clusters 1 and 5. However, it is most likely that the putative NrtABCD homologs in N. punctiforme function as something other than a transporter of nitrate and nitrite. A urea ABC transporter, encoded by urtABCD, was also upregulated early in the NSI-Het time course, but not in the NSI-Hrm time course, and was not transcribed at a high level as the nitrate-nitrite genes in NSI-Het (Fig. 3c).
Fig. 3.
Microarray data representing genes involved in alternate nitrogen source utilization. Plots consist of M values on the y axes and time (in h) on the x axes. (a) NSI-Het nitrate genes; (b) NSI-Hrm nitrate genes; (c) NSI-Het urea and ammonium transport genes; (d) NSI-Het/Hrm nblA.
A putative ammonium transporter (NpR3288) is marginally upregulated during heterocyst, but not hormogonium, differentiation (Fig. 3c). A gene (NpF0291) encoding the phycobiliprotein-degrading protein NblA is strongly upregulated early in both heterocyst and hormogonium differentiation (Fig. 3d). NblA has been shown to specifically degrade phycobiliproteins in the developing heterocyst but is not essential in the formation of functional heterocysts (7). NblA may play a more general role in N scavenging in developing hormogonia; there is an overall decrease in phycobiliprotein fluorescence in cells of hormogonium filaments, as visualized through epifluorescence microscopy, but there is no noticeable spatial pattern. It appears that under both developmental paths, the response to acquire alternate N sources is initiated very early. The gene encoding the nitrogen master regulator NtcA (NpF5511) was not significantly upregulated in hormogonia but did show the expected upregulation during heterocyst formation (Fig. 4b). It is possible that higher levels of activated NtcA are necessary for transcription of the urea genes, but not for those of nitrate-nitrite assimilation genes, or for nblA. There was no evidence for enhanced transcription of amino acid transporters in either of the developmental time courses, indicating that the presence of specific substrates may be required before transcription of these genes is enhanced.
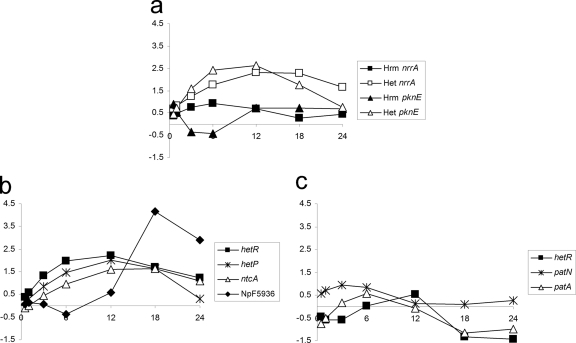
Fig. 4.
Microarray data representing heterocyst regulatory genes expressed in NSI-Het and NSI-Hrm. Plots consist of M values on the y axes and time (in h) on the x axes. (a) NSI-Het/Hrm nrrA and pknE genes; (b) NSI-Het regulatory genes; (c) NSI-Hrm regulatory genes.
The comparative transcriptomic approach for analysis of the N starvation response implies that this cyanobacterium immediately prepares to assimilate nitrate and nitrite, even in their absence, as well as recycle internal organic N in phycobiliproteins as initiated by NblA. Expression of the nitrate-nitrite metabolism genes could also be enhanced even more in the presence of these substrates. One secondary response, observed only in the NSI-Het time course, would include transport of urea, but apparently urea must be present and transported before the genes encoding urease are expressed. This sequence implies that transcriptional activation of the urease genes requires a substantial concentration of intracellular urea. Another secondary response that is marginally enhanced is ammonium transport, but again, only during NSI heterocyst differentiation. Apparently this cyanobacterium did not evolve to search for low concentrations of ammonium, as have other unicellular and filamentous cyanobacteria (25).
The transcriptional time course of heterocyst differentiation and of heterocyst-related genes differentially transcribed in NSI-hormogonia.
One of the earliest upregulated genes is nrrA in NSI-Het cluster 6 and NSI-Hrm cluster 10. The response regulator NrrA is required for timely completion of heterocyst differentiation, but it is not essential, as heterocysts eventually develop in the null mutant strain (18, 45). nrrA transcription was enhanced at 0.5 h in both time courses and remained elevated throughout, but the magnitude of expression was much less in hormogonia (Fig. 4a). These results imply that NrrA may be more involved in the general N stress response than specifically for heterocyst differentiation. The cognate histidine kinase for NrrA is unknown, and the only protein kinase with a similar expression pattern is a serine/threonine kinase, NpF6345, a pknE homolog (Fig. 4a). PknE in Anabaena 7120 is required for fully functional heterocysts (59), and overexpression represses heterocyst differentiation (47). The heterocyst master regulator hetR (NSI-Het cluster 6) is upregulated in a pattern similar to nrrA in NSI-Het (Fig. 4b), which is consistent with Northern blotting results (55), but its expression in NSI-Hrm (NSI-Hrm cluster 1) was downregulated more than 2-fold (Fig. 4c). The uncoupling of nrrA and hetR expression patterns during hormogonium differentiation could be due to modulation of the cognate histidine kinase activity for NrrA or to another factor influencing hetR transcription. Transcription of ntcA is upregulated by HetR and NtcA; therefore, low hetR transcription could explain why ntcA is not upregulated in NSI hormogonia.
It is difficult to glean any information on the involvement of the negative regulators PatS and HetN in the downregulation of hetR in hormogonia. In these experiments, RNA of less than 200 nucleotides was not isolated, so the kinetics of patS (42 bp) transcription is unknown. HetN in Anabaena 7120 contains the same RGSGR pentapeptide found in PatS, which is adequate for heterocyst suppression (56), but the putative hetN homolog in N. punctiforme (NpF3363) does not encode the RGSGR peptide, nor is NpF3363 upregulated during heterocyst differentiation. A conserved hypothetical protein, encoded by NpF5936, does contain this pentapeptide and was strongly upregulated by 18 h after N step-down (Fig. 4b); this could be the HetN-like negative regulator in N. punctiforme required for maintenance of the heterocyst pattern. However, NpF5936 is not differentially transcribed in NSI-Hrm, so it may not be responsible for the hormogonium-repressed level of hetR. Transcription of a gene required for proper heterocyst patterning, patN (41), was marginally enhanced at a low level early in the NSI-Hrm time course (Fig. 4c), but patN was not upregulated in the heterocyst time course. Mutation of patN results in a multiple singular heterocyst phenotype in which a higher concentration of heterocysts appears in the filament, but they remain evenly spaced with a shortened vegetative cell interval. Overexpression of patN does not repress heterocyst differentiation, as do patS and hetN (41), but this still does not rule out the possibility of an indirect role for PatN in attenuated hetR levels. Another heterocyst pattern gene, patA, was downregulated late in NSI-Hrm (Fig. 4c) but was not differentially transcribed during heterocyst development. The patA mutant in Anabaena 7120 results in heterocysts differentiating only at the filament termini (39) and suppresses the multiple contiguous heterocyst phenotype when hetR is overexpressed (8). Attenuation of patA expression in hormogonia may be a result of the same mechanism that depresses hetR. Expression of NpR1542, a hetP homolog, follows the same initial pattern as hetR in NSI-Het before declining by 24 h. However, NpR1542 is upregulated during steady-state diazotrophic growth (see Table S3 in the supplemental material). NpR1542 was not differentially transcribed during NSI-Hrm formation (Fig. 4b). HetP is required for heterocyst development beyond the proheterocyst stage (22). Overexpression of hetP was found to bypass the need for HetR during heterocyst differentiation in Anabaena 7120, and HetR likely activates hetP transcription (33).
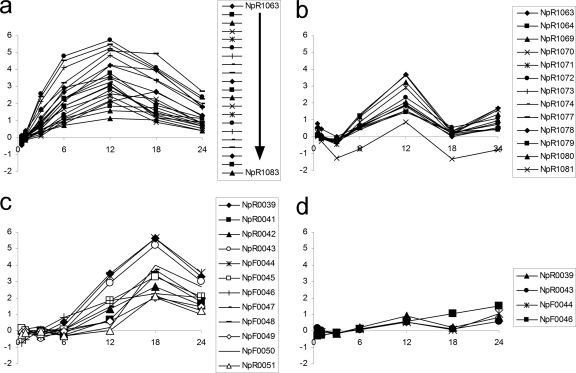
A contiguous group of 21 genes from NpR1063 to NpR1083 were upregulated early and extensively in NSI-Het (Fig. 5a). The majority of these genes are homologs of the Anabaena 7120 and A. variabilis proteins involved in synthesis of heterocyst-specific polysaccharides, including HepA and HepC (35). Thirteen of these genes, including hepA, were transiently upregulated in the NSI-Hrm data set (Fig. 5b), but their expression was much lower than NSI-Het and the maximal extent of upregulation coincided with the 12-h checkpoint. Enhanced transcription of genes in another chromosomal region, encoding homologs of proteins involved in heterocyst glycolipid synthesis and deposition, occurred later in NSI-Het (Fig. 5c). This region contains 12 genes composed of two divergently transcribed groups, NpR0039 to NpR0043 and NpF0044 to NpF0051, with similarity to a group of genes in Anabaena 7120 in a similar orientation (21). Included in these groups are hetM, hglC, hglG, hglE, devB, and devC. Four of these genes, including devB, hglE, and hetM, were also upregulated transiently in the NSI-Hrm time course, and all but one (NpF0046) displayed the pattern at the 12-h checkpoint followed by an increase at 24 h (Fig. 5d). Transcription of genes required for synthesis of heterocyst structural components was unexpected in hormogonia. It is possible that these polysaccharides and glycolipids are synthesized in hormogonia, but whether they are deposited to the wall is problematic, because growth and division eventually resumes, which is thought to be prevented by the glycolipid layer in heterocysts (54).
Fig. 5.
Microarray data representing heterocyst polysaccharide (a and b) and glycolipid synthesis (c and d) genes expressed in NSI-Het (a and c) and NSI-Hrm (b and d) time course studies. Plots consist of M values on the y axes and time (in h) on the x axes.
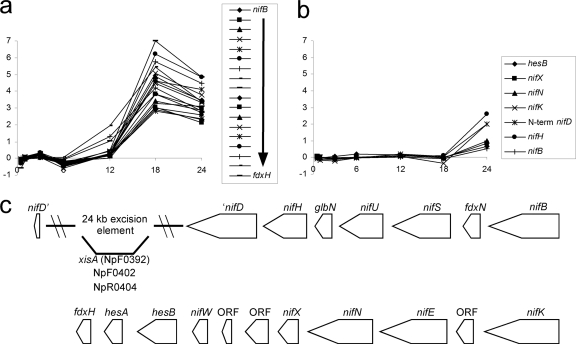
Also transcribed late are the heterocyst-specific genes encoding cytochrome oxidases and nitrogenase. In the NSI-Het time course, the nitrogenase genes (nif) were strongly upregulated by 18 h after N step-down (Fig. 6a). The core set of nif genes, nifB-fdxN-nifS-nifU-nifH-nifD-nifK-ORF-nifE-nifN-nifX-ORF-ORF-nifW-hesA-hesB-fdxH, encoding catalytic and accessory proteins, is conserved among the heterocyst-forming cyanobacteria (51). In Anabaena 7120 the fdxN and nifD genes are interrupted by 55-kb and 11-kb DNA insertion elements, respectively, that are excised during heterocyst differentiation (28). The N. punctiforme fdxN gene has no insertion element, and nifD contains a 24-kb element (44). Within the 24-kb nifD element, a gene encoding a hypothetical protein with a hemolysin-type calcium binding region (NpF0402) is upregulated by 3 h after step-down (NSI-Het cluster 6), and an unassigned gene with similarity to nuclear transport factors (NpR0404) is downregulated at 12 h (NSI-Het cluster 5). In Anabaena 7120, excisase (XisA) is required for excision of the 11-kb element from within nifD, but its expression pattern during heterocyst differentiation is not clear (19, 29). A xisA homolog (NpF0392) is present in the 24-kb nifD element of N. punctiforme, and its expression is not enhanced during heterocyst differentiation. Different from Anabaena 7120 is the presence of glbN located between nifH and nifU. GlbN is a hemoglobin-like protein believed to be involved in oxygen scavenging for cytochrome oxidase and localizes specifically to heterocysts (34). Transcription of glbN follows the same pattern as the surrounding nif genes in N. punctiforme.
Fig. 6.
(a and b) Microarray data representing nitrogenase genes expressed in NSI-Het (a) and NSI-Hrm (b) time course studies. Plots consist of M values on the y axes and time (in h) on the x axes. (c) Organization of the nif genes in N. punctiforme.
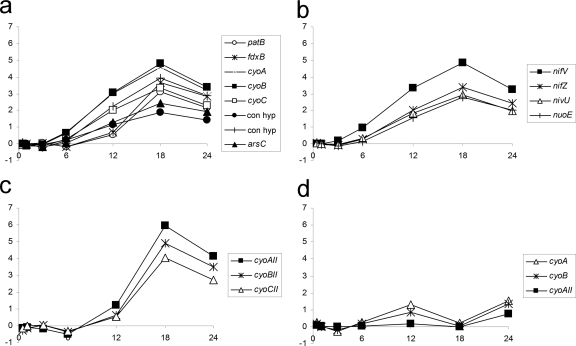
Present within the genomic vicinity of the conserved nif genes are other groups of similarly expressed genes. Genes encoding a regulator of the maintenance of the heterocyst spacing pattern, PatB (37), a ferredoxin, and one heterocyst-specific cytochrome oxidase (patB-fdxB-cyoABC) were upregulated by 12 h (Fig. 7a). Four genes near nifB similar to nifU, nifT, nifZ, and NADH:ubiquinone oxidoreductase (nuoE) were also upregulated by 12 h (Fig. 7b). A second set of cytochrome oxidase genes, cyoAII (NpR3537), cyoBII (NpR3536), and cyoCII (NpR3535), lies elsewhere in the chromosome, and their expression was enhanced by 18 h (Fig. 7c).
Fig. 7.
Microarray data representing cytochrome oxidase (a, c, and d) and putative nitrogenase-related (b) genes expressed in NSI-Het (a, b, and c) and NSI-Hrm (d) time course studies. Plots consist of M values on the y axes and time (in h) on the x axes.
A recent study employing RNA deep sequencing of Anabaena 7102 with 3 time points (6, 12, and 21 h after N deprivation) resulted in a broadly similar temporal sequence of gene expression, with apparent deviations in the absolute timing for a number of genes (23).
Most unusual, 6 genes were upregulated in hormogonia that are located in the group of conserved nif genes described above. These included hesB, nifX, nifE, nifK, the C-terminal nifD fragment, and nifH (Fig. 6b) (the sequences of the C- and N-terminal fragments of the insertion-interrupted nifD gene were printed in the array as distinct ORFs). The N-terminal fragment of nifD was not detected in the hormogonium but was found in the heterocyst time course. It is possible that the N-terminal nifD fragment was not detected because the 24-kb element was not removed during hormogonium differentiation, thus preventing accumulation of a stable N-terminal nifD message. If this were so, then nifK must be expressed from an unknown promoter in the absence of the excision event, possibly a promoter in the nifD element that transcribes the C-terminal nifD fragment and nifK. In Anabaena 7120, nifH and nifD were expressed in a strain unable to excise the 11-kb element but nifK was not expressed, indicating the need for excision before nifK expression. Transcription from nifH is not necessary for the excision event (30). In addition, two of the three cytochrome oxidase genes near patB, cyoA (NpF0336) and cyoB (NpF0337), were transiently expressed in the 12-h checkpoint pattern, as was cyoAII by 24 h (Fig. 7d).
A transition in gene expression after 24 h is required to establish steady-state diazotrophic growth.
Since fully formed heterocysts are visible with the characteristic presence of polar granules, loss of phycobilisome-dependent fluorescence, and expression of nitrogenase activity (based on acetylene reduction assays) at 24 h after N step-down in N. punctiforme (data not shown), we assumed that changes in transcriptional patterns would also be complete, or nearly so, by 24 h and be similar to steady-state N2 growth normalized to the ammonium-grown reference. However, even casual observation indicated differences between the 24-h (see Table S1 in the supplemental material) and steady-state N2-grown (13) data sets. Thus, we formally compared the two data sets. B values derived from the R (LIMMA GUI) statistical analysis were used for this comparison, with B values of ≥0 deemed statistically significant, and these genes also had P values of <0.0005.
A summary of this comparison, as depicted in a Venn diagram in Fig. 8, revealed that overall, a much larger set of genes is up- or downregulated at 24 h than during steady-state diazotrophic growth; 559 genes are upregulated 24 h after N step-down and 471 are downregulated, compared to the 378 and 123 that are up- or downregulated at steady state, respectively. A detailed list of these data is contained in Table S3 of the supplemental material. The largest number of genes in common between the two time points is upregulated. These 231 genes consist of a majority encoding known heterocyst regulatory and catalytic proteins, as well as those involved in nitrate-nitrite metabolism and urea transport that are part of the N stress response. The only other response regulator upregulated at both time points, in addition to nrrA, was NpF3659, a gene with homology to rpaA in Synechocystis sp. strain PCC 6803. RpaA was found to be involved in energy transfer between phycobilisomes and photosystems (6). Three protein kinases were also upregulated in common: NpR5250 and NpF5105 are related to tyrosine and serine/threonine kinases, respectively, with nothing known of their activities, while NpF6345 is the Anabaena 7120 pknE homolog and has been shown to be necessary for growth on N2 (59). Of the four transcriptional regulators upregulated in this group of 231 genes, NpF0307 is a homolog of sigE (alr4249) in Anabaena 7120. Based on visualization of a green fluorescent protein (GFP) transcriptional fusion, sigE appears to be specifically upregulated in heterocysts 16 h after N step-down (2). The upregulation of NpF0307 and the corresponding enhanced levels of nitrogenase and cytochrome oxidase transcription in N. punctiforme are similar to the pattern of sigE expression in Anabaena 7120. The other three putative transcriptional regulators are NpR1565, a PAS domain-containing CRP-like protein, NpF5754, a negative regulator of heat shock, and NpF1534, encoding NtcB, which regulates synthesis of the nitrate and nitrite reductases. There are also a number of core metabolism genes encoding proteins for energy metabolism, amino acid biosynthesis, and central metabolic oxidative pentose phosphate, glycolytic, and tricarboxylic acid pathway reactions among the upregulated genes. The latter are consistent with glucose catabolism in heterocysts for the supply of reductants to nitrogenase and respiration. Among the 48 downregulated genes in common are two histidine kinases that have very little similarity in Anabaena 7120, three putative motility genes, a response regulator receiver, sigma factor NpR1337 encoding SigJ, and some cell envelope and cofactor genes. Again, nearly 50% encode either conserved hypothetical or metabolically unassigned proteins.
The 389 genes that were downregulated at 24 h, but not differentially transcribed during steady-state N2 growth, were consistent with genes whose transcription pauses during a nongrowth period until steady-state growth with the new nitrogen source is initiated. These gene products are predominately related to core metabolism: energy generation, monomer biosynthesis, and protein synthesis, with some involved in sensing and signaling, as well as carbon storage. Such an extensive downregulation of core metabolic genes during heterocyst differentiation was not specifically identified in the deep RNA sequencing of Anabaena 7120 (23). The 328 genes upregulated at 24 h in N. punctiforme but not differentially transcribed during steady-state growth included 5 encoding transcriptional regulators, 14 for cofactors, 26 for protein polymerization, 35 for transport, and 21 genes involved in secondary metabolism (nostopeptolide production) that are organized as a genomic island of 70.3 kb (NpR3419 to NpR3452). In addition, genes encoding a putative TonB-ExbBD (NpR0781, NpR0782, and NpF0783) energizing complex are upregulated between 1 and 24 h after N step-down but not during steady-state N2-dependent growth. We speculate that these collective gene products could be involved in a general stress response that is no longer imposed once the cells acclimate to the new growth conditions and that a nostopeptolide secondary metabolite may function as a signal molecule that could be translocated by a TonB-dependent channel. Seventy-five genes were not differentially transcribed at 24 h but were downregulated during steady-state N2 growth. Expression of these genes may be important during growth with ammonium but not critical for diazotrophic growth. None is known as a heterocyst gene, and most are in the unassigned metabolic category. The 34 genes that were downregulated at 24 h and upregulated during steady-state growth included three stress-related genes, three response regulator genes, and a sigma factor, NpF4153, similar to Anabaena 7120 sigG. In Anabaena 7120, this sigma factor has been shown to be specifically upregulated in heterocysts via a promoter-GFP fusion at 9 h after N step-down (2).
It is highly probable that the products of genes common to 24-h and steady-state N2 growth that were upregulated from low levels of expression in the reference culture are localized in heterocysts, except for the 8 genes associated with nitrate assimilation and urea transport. This conclusion is supported by the common genes encoding heterocyst regulators, wall components, nitrogenase, and alternative cytochrome oxidases at steady state. The common transcriptional patterns of these well-known heterocyst genes give incentive to characterize other genes and gene products of known or unknown function that have similar transcription profiles. However, since heterocysts constitute less than 10% of the cells of a filament, it is difficult to detect downregulated genes specifically associated with heterocysts if they are also expressed in vegetative cells; an example of such a gene is that encoding ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco). Rubisco is absent in mature heterocysts (14); nevertheless, statistically significant differential expression of Rubisco was not detected during the time course study, at the 24-h time point, or during steady-state N2 growth in our studies. Similarly, phycobiliproteins are absent in heterocysts of laboratory cultures but, again, their differential expression was not significant.
We suggest that the genes downregulated at 24 h, but not during steady-state N2 growth, are most likely localized in vegetative cells, and their differential expression is a consequence of the cultures entering a transient nongrowth state in response to the shock of N starvation. Such a transition in growth and differential gene expression was previously suggested to occur in Anabaena 7120, but growth was not examined (18).
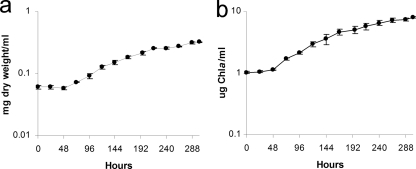
To test the above suggestion, growth of N. punctiforme was followed for up to 302 h after N step-down by monitoring the Chl a concentration and dry weight of cells over time (Fig. 9). There was a lag of about 48 h before exponential growth resumed, which was then followed by an extended period of light-limited linear growth. This lag is consistent with a period whereby the cells make the necessary changes to adapt to a new growth condition. By 24 h after N deprivation, everything necessary to form a fully functional heterocyst is expressed, but the vegetative cells and the unique physiological interdependence of heterocysts and vegetative cells in a filament require another 24 h of adaptation. The time period for adaptation is most likely variable, depending on cyanobacterial strain and growth conditions (light intensity, temperature, CO2 supplementation, etc.).
Fig. 9.
Growth curves based on mg (dry weight) per ml (a) and μg of chlorophyll a per ml (b) of N. punctiforme batch culture after N step-down.
Conclusions.
When cultures of N. punctiforme are grown in the presence of ammonium as the N source, transcription of genes encoding proteins for the assimilation of other N sources is repressed (time zero transcription data). This result verifies ammonium as the dominant N source in the hierarchy of environmental N compounds for growth, similar to other bacteria, including cyanobacteria (25, 32). BATS-enabled comparative global transcriptomic analyses of N stress-induced heterocyst and hormogonium differentiation have allowed us to predict the most likely immediately preferred alternative N sources, based on temporal gene expression profiles and the extent of differential expression, viz.: nitrate-nitrite > phycobilisome degradation via NblA ↔ urea > exogenous amino acids (such as glutamine and arginine, which repress heterocyst differentiation [data not shown]) > dinitrogen. In the absence of experiments with urea supplementation, the relationship between internal organic N and urea cannot be resolved.
The seven-point time course of global differential gene expression leading to formation of a functional heterocyst provides much more detail than any other time course analysis presented to date. An advantage of DNA microarrays in defining global transcription profiles is the generation of sufficient data points with biological replicates for the rigorous statistical analyses we have presented here. A disadvantage is the limited dynamic range and narrow window leading to mechanistic studies on the regulation of transcription. The latter two limitations are largely negated by the deep RNA sequencing approaches that have recently been applied to Anabaena 7120 (23). However, RNA sequencing currently lacks the repetitions essential for confident identification of low-level differentially transcribed genes whose gene products are biologically significant, such as devH (NpR6193), which participates in regulation of heterocyst glycolipid synthesis (57). The overall sequence of differential gene expression in N. punctiforme is consistent with studies of Anabaena 7120 (19, 23, 54, 57). Genes encoding direct or indirect transcriptional regulators and signal transducers, such as NrrA, HetR, and PknE, are upregulated early, followed by gene products assembling, in sequence, the polysaccharide and glycolipid structural components of the heterocyst envelope, and finally those products for function of nitrogenase, including all of the nif proteins, cytochrome oxidases for oxygen protection, and carbon catabolic enzymes for reductant supply. Underlying this sequence are constitutively transcribed proteins that, unlike NtcA and HetR, are not statistically upregulated during the course of differentiation presented here but influence the presence and spacing pattern of heterocysts, such as PatA and PatN. Some unique features include differential expression of two genes within the nifD insertion element and lack of upregulation of xisA. A substantial number of genes encoding proteins of unassigned function were differentially transcribed, and these are targets for genetic disruption and phenotypic analyses. Specific emphasis should be placed on the 77 unassigned genes within the 231 genes upregulated that were in common between the 24-h and steady-state N2-grown data sets. Those 231 genes, minus the 8 genes encoding nitrate-nitrite assimilation and urea transport, plus the 48 downregulated genes in common reflect the minimum number of 271 genes differentially transcribed during formation of a functional heterocyst. The results presented here do not allow for a projection of the maximal number of genes that may be required.
Based on reports of developmental control over the transcription of especially nif genes of the heterocyst-localized nitrogenase (20, 54), their expression in hormogonia was unexpected. NSI hormogonia showed the same early N response as NSI heterocysts, but they lacked enhanced expression of heterocyst regulatory genes, which would be expected to elicit transcription of gene products for heterocyst structures and nitrogenase function. Expression of the heterocyst master regulator hetR was actually depressed, and ntcA transcripts remained at levels similar to those before N starvation. These data indicate that the general N stress response is coupled in cells destined to become hormogonia, and there is a link in the regulatory decision to become either a hormogonum or heterocyst.
Several observations indicate that the transition from growth on ammonium to N2 dependence is not particularly rapid, precise, or complete. First, there is an extensive growth lag phase of 24 h beyond the formation of a functional heterocyst, complete with N2 fixation. This lag correlates with the persistent downregulation of genes encoding proteins required for vegetative cell growth and division (core metabolism). Second, even during steady-state N2-dependent growth, genes encoding products for nitrate-nitrite assimilation and urea transport continue to be expressed, implying that the cultures retain the physiological status of N limitation. Perhaps the lack of trophic-level competitors has allowed this oxygenic photoautotrophic diazotroph to retain a lack of stringency in its transcriptional regulation.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the U.S Department of Energy (grant DE-FG02-08ER64693).
We thank Doug Risser, Daniella Ferreira, and Becky Parales for helpful comments during preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 14 October 2011.
REFERENCES
- 1. Adams D. G. 2000. Symbiotic interactions, p. 523–561 In Whitton B. A., Potts M. (ed.), The ecology of cyanobacteria, their diversity in time and space. Kluwer Academic Publishers, Dordrecht, Netherlands [Google Scholar]
- 2. Aldea M. R., Mella-Herrera R. A., Golden J. W. 2007. Sigma factor genes sigC, sigE, and sigG are upregulated in heterocysts of the cyanobacterium Anabaena sp. PCC7120. J. Bacteriol. 189: 8392–8396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Allan M. B., Arnon D. I. 1955. Studies on nitrogen-fixing blue-green algae. I. Growth and nitrogen fixation by Anabaena cylindrica Lemm. Plant Physiol. 30: 366–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Angelini C., Cutillo L., De Canditii D., Mutarelli M., Pensky M. 2008. BATS: a Bayesian user-friendly software for analyzing time series microarray experiments. BMC Bioinformatics 9: 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Armstrong R. E., Hayes P. K., Walsby A. E. 1983. Gas vacuole formation in hormogonia of Nostoc muscorum. J. Gen. Microbiol. 128: 263–270 [Google Scholar]
- 6. Ashby M. K., Mullineaux C. W. 1999. Cyanobacterial ycf27 gene products regulate energy transfer from phycobilisomes to photosystems I and II. FEMS Microbiol. Lett. 181: 253–260 [DOI] [PubMed] [Google Scholar]
- 7. Baier K., Lehm H., Stephan D. P., Lockau W. 2004. NblA is essential for phycobilisome degradation in Anabaena sp. strain PCC 7120 but not for development of functional heterocysts. Microbiology 150: 2739–2749 [DOI] [PubMed] [Google Scholar]
- 8. Buikema W. J., Haselkorn R. 2001. Expression of the Anabaena hetR gene from a copper-regulated promoter leads to heterocyst differentiation under repressing conditions. Proc. Natl. Acad. Sci. U. S. A. 98: 2729–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cai Y., Wolk C. P. 1997. Anabaena sp. strain PCC 7120 responds to nitrogen deprivation with a cascade-like sequence of transcriptional activations. J. Bacteriol. 179: 267–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Callahan S. M., Buikema W. J. 2001. The role of HetN in maintenance of the heterocyst pattern in Anabaena sp. PCC 7120. Mol. Microbiol. 40: 941–950 [DOI] [PubMed] [Google Scholar]
- 11. Campbell E. L., Meeks J. C. 1989. Characteristics of hormogonia formation by symbiotic Nostoc spp. in response to the presence of Anthoceros punctatus or its extracellular products. Appl. Environ. Microbiol. 55: 125–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Campbell E. L., Christman H., Meeks J. C. 2008. DNA microarray comparisons of plant factor- and nitrogen deprivation-induced hormogonia reveal decision-making transcriptional regulation patterns in Nostoc punctiforme. J. Bacteriol. 190: 7382–7391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Campbell E. L., Summers M. L., Christman H., Martin M. E., Meeks J. C. 2007. Global gene expression patterns of Nostoc punctiforme in steady-state dinitrogen-grown heterocyst-containing cultures and at single time points during the differentiation of akinetes and hormogonia. J. Bacteriol. 189: 5247–5256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cossar J. D., et al. 1985. Localization of ribulose 1, 5-bisphosphate carboxylase/oxygenase in the N2-fixing cyanobacterium Anabaena cylindrica. FEMS Microbiol. Lett. 28: 65–68 [Google Scholar]
- 15. Damerval T., Guglielmi G., Houmard J., de Marsac N. T. 1991. Hormogonium differentiation in the cyanobacterium Calothrix: a photoregulated developmental process. Plant Cell 3: 191–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. de Hoon M. J. L., Imoto S., Nolan J., Miyano S. 2004. Open source clustering software. Bioinformatics 20: 1453–1454 [DOI] [PubMed] [Google Scholar]
- 17. Douglas D., Peat A., Whitton B. A., Wood P. 1987. Influence of iron status on structure of the cyanobacterium (blue-green alga) Calothrix parientina. Cytobiosis 47: 155–165 [Google Scholar]
- 18. Ehira S., Ohmori M. 2006. NrrA, a nitrogen-responsive response regulator facilitates heterocyst development in the cyanobacterium Anabaena sp. strain PCC 7120. Mol. Microbiol. 59: 1692–1703 [DOI] [PubMed] [Google Scholar]
- 19. Ehira S., Ohmori M., Sato N. 2003. Genome-wide expression analysis of the response to nitrogen deprivation in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 10: 97–113 [DOI] [PubMed] [Google Scholar]
- 20. Elhai J., Wolk C. P. 1990. Developmental regulation and spatial pattern of expression of the structural genes for nitrogenase in the cyanobacterium Anabaena. EMBO J. 9: 3379–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fan Q., et al. 2005. Clustered genes required for synthesis and deposition of envelope glycolipids in Anabaena sp. strain PCC 7120. Mol. Microbiol. 58: 227–243 [DOI] [PubMed] [Google Scholar]
- 22. Fernandez-Pinas F., Leganes F., Wolk C. P. 1994. A third genetic locus required for the formation of heterocysts in Anabaena sp. strain PCC 7120. J. Bacteriol. 176: 5277–5283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Flaherty B. L., Van Nieuwerburgh F., Head S. R., Golden J. W. 2011. Directional RNA deep sequencing sheds new light on the transcriptional response of Anabaena sp. strain PCC 7120 to combined-nitrogen deprivation. BMC Genomics 12: 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Flores E., Frias J. E., Rubio L. M., Herrero A. 2005. Photosynthetic nitrate assimilation in cyanobacteria. Photosynth. Res. 83: 117–133 [DOI] [PubMed] [Google Scholar]
- 25. Flores E., Herrero A. 1994. Assimilatory nitrogen metabolism and its regulation, p. 487–517 In Bryant D. A. (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Boston, MA [Google Scholar]
- 26. Frias J. E., Flores E., Herrero A. 1997. Nitrate assimilation gene cluster from the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 179: 477–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Frias J. E., Flores E., Herrero A. 2000. Activation of the Anabaena nir operon promoter requires both NtcA (CAP family) and NtcB (LysR family) transcription factors. Mol. Microbiol. 38: 613–625 [DOI] [PubMed] [Google Scholar]
- 28. Golden J. W., Carrasco C. D., Mulligan M. E., Schneider G. J., Haselkorn R. 1988. Deletion of the 55-kilobase-pair DNA element from the chromosome during heterocyst differentiation of Anabaena sp. strain PCC 7120. J. Bacteriol. 170: 5034–5041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Golden J. W., Weist D. R. 1988. Genome rearrangement and nitrogen fixation in Anabaena blocked by inactivation of xisA gene. Science 242: 1421–1423 [DOI] [PubMed] [Google Scholar]
- 30. Golden J. W., Whorff L. L., Wiest D. R. 1991. Independent regulation of nifHDK operon transcription and DNA rearrangement during heterocyst differentiation in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 173: 7098–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Herdman M., Rippka R. 1988. Cellular differentiation: hormogonia and baeocytes. Methods Enzymol. 167: 232–242 [Google Scholar]
- 32. Herrero A., Muro-Pastor A. M., Flores E. 2001. Nitrogen control in cyanobacteria. J. Bacteriol. 183: 411–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Higa K. C., Callahan S. M. 2010. Ectopic expression of hetP can partially bypass the need for hetR in heterocyst differentiation by Anabaena sp. strain PCC 7120. Mol. Microbiol. 77: 562–574 [DOI] [PubMed] [Google Scholar]
- 34. Hill D. R., et al. 1996. GlbN (cyanoglobin) is a peripheral membrane protein that is restricted to certain Nostoc spp. J. Bacteriol. 178: 6587–6598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huang G., et al. 2005. Clustered genes required for the synthesis of heterocyst envelope polysaccharide in Anabaena sp. strain PCC 7120. J. Bacteriol. 187: 1114–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huang X., Dong Y., Zhao J. 2004. HetR homodimer is a DNA-binding protein required for heterocyst differentiation, and the DNA-binding activity is inhibited by PatS. Proc. Natl. Acad. Sci. U. S. A. 101: 4848–4853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jones K. M., Haselkorn R. 2002. Newly identified cytochrome c oxidase operon in the nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120 specifically induced in heterocysts. J. Bacteriol. 184: 2491–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Knight C. D., Adams D. G. 1996. A method for studying chemotaxis in nitrogen fixing cyanobacterium-plant symbioses. Physiol. Mol. Plant Pathol. 49: 73–77 [Google Scholar]
- 39. Liang J., Scappino L., Haselkorn R. 1992. The patA gene product, which contains a region similar to CheY of Escherichia coli, controls heterocyst pattern formation in the cyanobacterium Anabaena 7120. Proc. Natl. Acad. Sci. U. S. A. 89: 5655–5659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lynn M. E., Bantle J. A., Ownby J. D. 1986. Estimation of gene expression in heterocysts of Anabaena variabilis by using DNA-RNA hybridizations. J. Bacteriol. 167: 940–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Meeks J. C., Campbell E. L., Summers M. L., Wong F. C. 2002. Cellular differentiation in the cyanobacterium Nostoc punctiforme. Arch. Microbiol. 178: 395–403 [DOI] [PubMed] [Google Scholar]
- 42. Meeks J. C., Castenholz R. W. 1971. Growth and photosynthesis in an extreme thermophile Synechococcus lividus (Cyanophyta). Arch. Mikrobiol. 78: 25–41 [DOI] [PubMed] [Google Scholar]
- 43. Meeks J. C., Elhai J. 2002. Regulation of cellular differentiation in filamentous cyanobacteria in free-living and plant-associated symbiotic growth states. Microbiol. Mol. Biol. Rev. 66: 94–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Meeks J. C., et al. 2001. An overview of the genome of Nostoc punctiforme, a multicellular, symbiotic cyanobacterium. Photosyn. Res. 70: 85–106 [DOI] [PubMed] [Google Scholar]
- 45. Muro-Pastor A. M., Olmedo-Verd E., Flores E. 2006. All4312, an NtcA-regulated two-component response regulator in Anabaena sp. strain PCC7120. FEMS Microbiol. Lett. 256: 171–177 [DOI] [PubMed] [Google Scholar]
- 46. Mutarelli M., et al. 2008. Time-course analysis of genome-wide gene expression data from hormone-responsive human breast cancer cells. BMC Bioinformatics 9: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Saha S. K., Golden J. W. 2011. Overexpression of pknE blocks heterocyst development in Anabaena sp. strain PCC 7120. J. Bacteriol. 193: 2619–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Saldanha A. J. 2004. Java Treeview: extensible visualization of microarray data. Bioinform. Appl. Notes 20: 3246–3248 [DOI] [PubMed] [Google Scholar]
- 49. Smyth G. K. 2005. Limma: linear models for microarray data, p. 397–420 In Gentleman R., Carry V., Dudoit S., Irizarry R., Huber W. (ed.), Bioinformatics and computational biology solutions using R and Bioconductor. Springer, New York, NY [Google Scholar]
- 50. Suzuki I., Horie N., Sugiyama T., Omata T. 1995. Identification and characterization of two nitrogen-regulated genes of the cyanobacterium Synechococcus sp. strain PCC 7942 required for maximum efficiency of nitrogen assimilation. J. Bacteriol. 177: 290–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Thiel T. 2004. Nitrogen fixation in heterocyst-forming cyanobacteria, p. 73–110 In Klipp W., Masepohl B., Gallon J. R., Newton W. E. (ed.), Genetics and regulation of nitrogen-fixing bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 52. Valladares A., Montesinos M. L., Herrero A., Flores E. 2002. An ABC-type, high-affinity urea permease identified in cyanobacteria. Mol. Microbiol. 43: 703–715 [DOI] [PubMed] [Google Scholar]
- 53. Wang Q., Li H., Post A. F. 2000. Nitrate assimilation genes of the marine diazotrophic, filamentous cyanobacterium Trichodesmium sp. strain WH9601. J. Bacteriol. 182: 1764–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wolk C. P., Ernst A., Elhai J. 1994. Heterocyst metabolism and development, p. 769–823 In Bryant D. A. (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Boston, MA [Google Scholar]
- 55. Wong F .C. Y., Meeks J. C. 2001. The hetF gene product is essential to heterocyst differentiation and affects HetR function in the cyanobacterium Nostoc punctiforme. J. Bacteriol. 183: 2654–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wu X., Liu D., Lee M. H., Golden J. W. 2004. patS minigenes inhibit heterocyst development of Anabaena sp. strain 7120. J. Bacteriol. 186: 6422–6429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xu X., Elhai J., Wolk C. P. 2007. Transcriptional and developmental responses by Anabaena to deprivation of fixed nitrogen, p. 383–422 In Herrero A., Flores E. (ed.), The Cyanobacteria: molecular biology, genomics and evolution. Horizon Scientific Press, Norwich, United Kingdom [Google Scholar]
- 58. Yoon H. S., Golden J. W. 1998. Heterocyst pattern formation controlled by a diffusible peptide. Science 282: 935–938 [DOI] [PubMed] [Google Scholar]
- 59. Zhang C.-C., Friry A., Peng L. 1998. Molecular and genetic analysis of two closely linked genes that encode, respectively, a protein phosphatase 1/2A/2B homolog and a protein kinase homolog in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 180: 2616–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang C.-C., Laurent S., Sakr S., Peng L., Bedu S. 2006. Heterocyst differentiation and pattern formation in cyanobacteria: a chorus of signals. Mol. Microbiol. 59: 367–375 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.