Abstract
The TonB system couples cytoplasmic membrane proton motive force to TonB-gated outer membrane transporters for active transport of nutrients into the periplasm. In Escherichia coli, cytoplasmic membrane proteins ExbB and ExbD promote conformational changes in TonB, which transmits this energy to the transporters. The only known energy-dependent interaction occurs between the periplasmic domains of TonB and ExbD. This study identified sites of in vivo homodimeric interactions within ExbD periplasmic domain residues 92 to 121. ExbD was active as a homodimer (ExbD2) but not through all Cys substitution sites, suggesting the existence of conformationally dynamic regions in the ExbD periplasmic domain. A subset of homodimeric interactions could not be modeled on the nuclear magnetic resonance (NMR) structure without significant distortion. Most importantly, the majority of ExbD Cys substitutions that mediated homodimer formation also mediated ExbD-TonB heterodimer formation with TonB A150C. Consistent with the implied competition, ExbD homodimer formation increased in the absence of TonB. Although ExbD D25 was not required for their formation, ExbD dimers interacted in vivo with ExbB. ExbD-TonB interactions required ExbD transmembrane domain residue D25. These results suggested a model where ExbD2 assembled with ExbB undergoes a transmembrane domain-dependent transition and exchanges partners in localized homodimeric interfaces to form an ExbD2-TonB heterotrimer. The findings here were also consistent with our previous hypothesis that ExbD guides the conformation of the TonB periplasmic domain, which itself is conformationally dynamic.
INTRODUCTION
The TonB system of Gram-negative bacteria couples the proton motive force (PMF) of the cytoplasmic membrane (CM) to the active transport of a diverse range of large, scarce, or important nutrients across the unenergized outer membrane (1, 24, 25). Active transport across the outer membrane requires TonB-gated transporters, which are 22-stranded β-barrels with lumens occluded by an amino-terminal globular domain called the cork (37).
In addition to the TonB-gated transporters in the outer membrane, the TonB system consists of 3 integral CM proteins, TonB, ExbB, and ExbD. TonB and ExbD have identical topologies, each with a single transmembrane domain (TMD) and more than two-thirds of each protein in the periplasm (15, 20, 43). ExbB has 3 TMDs and significant cytoplasmic domains (21, 22). ExbB and ExbD appear to couple the energy of the CM PMF to conformational changes in the TonB carboxy terminus (33). The TonB carboxy terminus directly contacts the TonB-gated transporters and somehow transmits energy for active transport of substrates. Evidence suggests all three proteins form a complex in the CM, but the stoichiometry of this complex is unknown (2, 38). The cellular TonB/ExbD/ExbB ratio is 1:2:7 (16).
The only step in TonB energization currently known to require the PMF is characterized by a formaldehyde-cross-linkable interaction between the TonB and ExbD periplasmic domains. While TonB is conformationally responsive to changes in the PMF, the absence of protonatable residues in the TonB TMD, among other data, suggests that TonB responds to the PMF only by virtue of TonB-ExbD interactions through their periplasmic domains (5, 11, 29, 38, 45).
ExbD can also be trapped in formaldehyde-cross-linked homodimers in vivo. Unlike the formaldehyde-mediated TonB-ExbD complex, homodimer formation does not require PMF, TonB H20, or ExbD D25 (38). It is unknown if ExbD homodimers are functional. In vitro studies pinpointed a specific region of the ExbD periplasmic domain, residues 104 to 116, as a homomultimeric interface. Nuclear magnetic resonance (NMR) studies of the ExbD periplasmic domain (residues 43 to 141) demonstrated extensive aggregation of this isolated domain, with 4 to 7 copies of the domain forming a homomultimer with 1 mM protein at pH 7.0 (10). It is not known if this region mediates homodimerization in vivo.
In this study, cysteine scanning of a 30-residue region of the ExbD periplasmic domain identified regions involved in homodimer formation in vivo, some of which mapped to the region identified in the NMR studies and some of which mapped to the opposite end of the solution structure, suggesting that the ExbD TMD contributes substantially to the conformation of a dynamic ExbD carboxy terminus. Most importantly, the same set of ExbD Cys substitutions that mediated spontaneous disulfide-linked homodimer formation also mediated spontaneous heterodimer formation with TonB A150C in vivo.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. Single-amino-acid substitutions were generated for all plasmids using 30-cycle extralong PCR. All ExbD Cys substitutions were constructed using this method, with pKP999 (exbD) as the template unless otherwise noted. pKP885, pKP899, pKP905, and pKP911 were derivatives of pKP660 (exbB exbD). All ExbD D25N Cys substitutions were derivatives of pKP1064 (exbD D25N), except for the following: pKP1050 was a derivative of pKP1005 (exbD K97C), pKP1051 was a derivative of pKP1011 (exbD F103C), pKP1052 was a derivative of pKP1017 (exbD T109C), pKP1053 was a derivative of pKP1023 (exbD L115C), pKP1082 was a derivative of pKP1029 (exbD T121C), and pKP1429 was a derivative of pKP1024 (exbD M116C). Forward and reverse primers were designed with the desired base change flanked on both sides by 12 to 15 homologous bases (primer sequences are available upon request). DpnI digestion was used to remove the template plasmid. The sequences of the exbB segment and the exbD gene were confirmed by DNA sequencing at the Penn State Genomics Core Facility, University Park, PA.
Table 1.
Strains and plasmids used in this study
| Strain or plasmida | Genotype or phenotype | Reference |
|---|---|---|
| Strains | ||
| W3110 | F− IN(rrnD-rrnE)1 | 18 |
| RA1017 | W3110 ΔexbBD::kan ΔtolQRA | 29 |
| RA1045 | W3110 ΔexbD ΔtolQR | 5 |
| KP1344 | W3110 tonB P14::blaM | 33 |
| KP1509 | W3110 ΔexbD ΔtolQR ΔtonB::kan | 38 |
| Plasmids | ||
| pKP381 | TonB(H20A) | 29 |
| pKP945 | TonB(C18G, A150C) | 38 |
| pKP660 | exbB exbD in pBAD24 | 38 |
| pKP885 | ExbB ExbD(K97C) | Present study |
| pKP899 | ExbB ExbD(F103C) | Present study |
| pKP905 | ExbB ExbD(T109C) | Present study |
| pKP911 | ExbB ExbD(L115C) | Present study |
| pKP1000 | ExbD(A92C) | 38 |
| pKP1049 | ExbD(D25N, A92C) | 38 |
| pKP999 | exbD in pPro24 | 38 |
| pKP1001 (L93C) | ||
| pKP1002 (T94C) | ||
| pKP1003 (E95C) | ||
| pKP1004 (G96C) | ||
| pKP1005 (K97C) | ||
| pKP1006 (K98C) | ||
| pKP1007 (D99C) | ||
| pKP1008 (T100C) | ||
| pKP1009 (T101C) | ||
| pKP1010 (I102C) | ||
| pKP1011 (F103C) | ||
| pKP1012 (F104C) | ||
| pKP1013 (R105C) | ||
| pKP1014 (A106C) | ||
| pKP1015 (D107C) | ||
| pKP1016 (K108C) | ||
| pKP1017 (T109C) | ||
| pKP1018 (V110C) | ||
| pKP1019 (D111C) | ||
| pKP1020 (Y112C) | ||
| pKP1021 (E113C) | ||
| pKP1022 (T114C) | ||
| pKP1023 (L115C) | ||
| pKP1024 (M116C) | ||
| pKP1025 (K117C) | ||
| pKP1026 (V118C) | ||
| pKP1027 (M119C) | ||
| pKP1028 (D120C) | ||
| pKP1029 (T121C) | ||
| pKP1064 | ExbD(D25N) | 38 |
| pKP1217 (D25N, L93C) | ||
| pKP1216 (D25N, T94C) | ||
| pKP1218 (D25N, E95C) | ||
| pKP1219 (D25N, G96C) | ||
| pKP1050 (D25N, K97C) | ||
| pKP1233 (D25N, K98C) | ||
| pKP1203 (D25N, D99C) | ||
| pKP1204 (D25N, T100C) | ||
| pKP1260 (D25N, T101C) | ||
| pKP1189 (D25N, I102C) | ||
| pKP1051 (D25N, F103C) | ||
| pKP1226 (D25N, F104C) | ||
| pKP1227 (D25N, R105C) | ||
| pKP1228 (D25N, A106C) | ||
| pKP1261 (D25N, D107C) | ||
| pKP1270 (D25N, K108C) | ||
| pKP1052 (D25N, T109C) | ||
| pKP1237 (D25N, V110C) | ||
| pKP1262 (D25N, D111C) | ||
| pKP1269 (D25N, Y112C) | ||
| pKP1234 (D25N, E113C) | ||
| pkP1274 (D25N, T114C) | ||
| pKP1053 (D25N, L115C) | ||
| pKP1429 (D25N, M116C) | ||
| pKP1290 (D25N, K117C) | ||
| pKP1291 (D25N, V118C) | ||
| pKP1294 (D25N, M119C) | ||
| pKP1297 (D25N, D120C) | ||
| pKP1082 (D25N, T121C) |
The plasmids listed below pKP999 and pKP1064 are derivatives of pKP999 and pKP1064, respectively, unless otherwise noted in Materials and Methods. The ExbD substitutions expressed from each plasmid are listed in parentheses.
To construct pKP1005, pKP1011, pKP1017, and pKP1023, forward and reverse primers were designed to amplify the last 22 codons of exbB through the stop codon of exbD from a pKP885 (exbB exbD K97C), pKP899 (exbB exbD F103C), pKP905 (exbB exbD T109C), or pKP911 (exbB exbD L115C) template, respectively, introducing flanking NcoI sites. The PCR-amplified, NcoI-digested fragment was cloned into the unique NcoI site in pPro24 (34). Proper orientation was determined by FspI digestion. The sequences of the exbB segment and the exbD gene were confirmed by DNA sequencing at the Penn State Genomics Core Facility, University Park, PA.
Media and culture conditions.
Luria-Bertani (LB), tryptone (T), and M9 minimal salts were prepared as previously described (35, 44). Liquid cultures and agar plates were supplemented with 100 μg ml−1 ampicillin and plasmid-specific levels of sodium propionate, pH 8, as needed for expression of ExbD (see Table S1 in the supplemental material). When coexpression of plasmid-encoded TonB(C18G, A150C) was examined, cultures and agar plates were also supplemented with 34 μg ml−1 chloramphenicol and plasmid-specific levels of l-arabinose as needed for TonB expression. M9 salts were supplemented with 0.5% glycerol, 0.4 μg ml−1 thiamine, 1 mM MgSO4, 0.5 mM CaCl2, 0.2% Casamino Acids, 40 μg ml−1 tryptophan, and 37 μM FeCl3·6H2O. Cultures were grown with aeration at 37°C. All assays were performed using mid-exponential-phase cells (A550 ≈ 0.43 to 0.5, as measured on a Spectronic 20 with a path length of 1.5 cm).
Activity assays.
Initial rates of [55Fe]ferrichrome uptake were determined as described previously (32, 40). For assays where cultures were treated with copper-(1,10-phenanthroline)3 (CuoP), harvested cells were pelleted, resuspended in 1× M9 supplemented as described above (no sodium propionate added), and treated with an equal volume of 0.06 mM CuoP (0.03 mM [final]) or 0.5 mM sodium phosphate buffer, pH 7.4 (buffer only) for 5 min at 37°C with aeration. Cells were pelleted, and the standard iron transport protocol was followed, starting with resuspension in the assay medium.
In initial assays testing CuoP treatment, it was found that resuspension of harvested cells in unsupplemented 1× M9 and subsequent treatment with 0.03 mM CuoP decreased iron transport of a wild-type strain to about 25%. However, treatment with the same concentration, 0.03 mM CuoP, in supplemented 1× M9 did not inhibit the activity of a wild-type strain. In all assays, the CuoP solution was removed and the assays were carried out in identical media, but the medium present during the 5-min CuoP treatment was important and differentially affected the activity of a wild-type strain.
In vivo disulfide cross-linking.
Saturated LB overnight cultures were subcultured 1:100 in T broth. Equivalent A550-ml (total amount of cells; obtained by multiplying the A550 of the culture by the volume of culture used) of mid-exponential-phase cultures were harvested and precipitated by addition of an equal volume of 20% trichloroacetic acid (TCA). Cell pellets were solubilized in nonreducing Laemmli sample buffer (26) containing 50 mM iodoacetamide, as previously described (11). Samples were resolved on nonreducing 15%, 13%, or 11% SDS-polyacrylamide gels and immunoblotted with ExbD-specific polyclonal antibodies (16) or TonB-specific monoclonal antibodies (31). Disulfide-linked complexes still formed when samples were not TCA precipitated (see Fig. S3 in the supplemental material). For reasons that were unclear, anti-ExbD immunoblots from nonreducing gels consistently showed lower detection than reducing gels. Typically, a 10-min exposure from a nonreducing gel immunoblot was comparable in intensity to a 1-min exposure from a reducing gel immunoblot. To determine total protein expression levels, cell pellets of TCA-precipitated samples harvested at the same time as the cultures described above were solubilized in reducing Laemmli sample buffer containing β-mercaptoethanol (β-ME), resolved on reducing SDS-polyacrylamide gels, and immunoblotted as described above.
For assays catalyzing oxidative cross-linking with copper-(1,10-phenanthroline)3, saturated LB overnight cultures were subcultured 1:100 in M9 minimal medium. The harvested cells were resuspended in unsupplemented 1× M9 and treated with an equal volume of 0.06 mM CuoP (0.03 mM [final]) or 0.5 mM sodium phosphate buffer, pH 7.4 (buffer only) for 5 min at 37°C with aeration. Following treatment, samples were precipitated by addition of an equal volume of 20% TCA. All cell pellets were resuspended in nonreducing Laemmli sample buffer containing 50 mM iodoacetamide. At the time of initial harvest, samples from each culture were also TCA precipitated, and the pellets were resuspended in reducing Laemmli sample buffer containing β-ME. These served as the reduced sample controls for total protein expression levels. Equal A550-ml were loaded for all samples. Immunoblotting was performed as described above.
In vivo formaldehyde cross-linking following oxidative cross-linking.
Saturated LB overnight cultures were subcultured 1:100 into M9 minimal medium supplemented with sodium propionate (see Table S1 in the supplemental material for induction levels). At mid-exponential phase, cells were harvested and treated with an equal volume of 0.6 mM CuoP (0.3 mM [final]) for 5 min at 37°C with aeration. The cell pellets were washed once with 1× M9 and then resuspended in sodium phosphate buffer, pH 6.8, and treated with formaldehyde as previously described (17). Half of the samples were resuspended in nonreducing Laemmli sample buffer containing 50 mM iodoacetamide and half in reducing Laemmli sample buffer containing β-ME. The samples were resolved on 13% nonreducing or reducing SDS-polyacrylamide gels, respectively, and immunoblotted with ExbD-specific polyclonal antibodies.
RESULTS
Cysteine substitutions from ExbD residue 92 through residue 121 form spontaneous disulfide-linked dimers.
In vivo, ExbD A92C, a periplasmic domain substitution, forms disulfide-linked homodimers (38). In vitro, the ExbD periplasmic domain forms higher-order homomultimers through residues 104 to 116 (10). Our unpublished in vivo studies suggested a 30-residue region of the ExbD periplasmic domain, from residues 92 to 121, was important in mediating protein-protein interactions of ExbD (A. A. Ollis, A. Kumar, and K. Postle, unpublished data). To investigate the role of this region of ExbD in homodimer formation in vivo, cysteine substitutions were individually constructed at each of the remaining 29 residues. ExbD has no native cysteine residues, so wild-type ExbD encoded on plasmid pKP999 was used as the template.
Each plasmid-encoded substitution was expressed in strain RA1045 (ΔexbD ΔtolQR), where deletion of tolQR prevented the activity attributable to cross talk of TonB with this homologous system (3). Attempts were made to express each ExbD Cys substitution at levels equal to those of native ExbD when analyzed under reducing conditions. Immunoblots with an ExbD-specific polyclonal antibody of steady-state levels of TCA-precipitated proteins resolved on SDS-polyacrylamide gels showed all but one of the substitutions could be stably expressed (Fig. 1, reduced). ExbD I102C was proteolytically unstable and only faintly detected on longer exposures, even when induced with the highest concentration of sodium propionate. The instability was not specific to substitution of cysteine, since ExbD I102A was also proteolytically unstable (data not shown). Cys substitutions at T94, K98, and D107 also decreased ExbD stability, but these substitutions could be expressed to native ExbD levels with higher concentrations of sodium propionate (see Table S1 in the supplemental material). ExbD L93C showed aberrant migration, migrating more slowly than wild-type ExbD. ExbD L93A also exhibited slower migration (data not shown). Single-amino-acid substitutions can alter protein mobility on SDS-polyacrylamide gels (36).
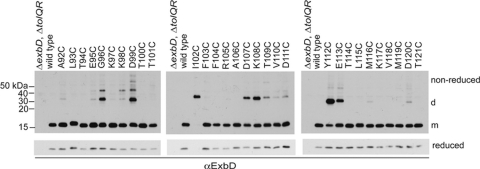
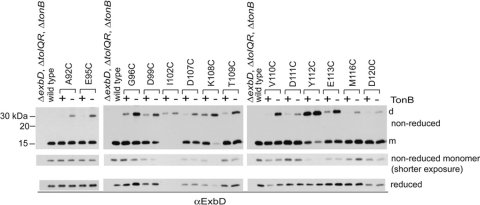
Fig. 1.
Cysteine substitutions in the ExbD periplasmic domain form spontaneous disulfide-linked dimers in vivo. TCA-precipitated proteins from strains expressing chromosomally encoded wild-type ExbD (W3110) or a ΔexbD ΔtolQR strain (RA1045) expressing plasmid-encoded ExbD variants near native ExbD levels (see Table S1 in the supplemental material for induction levels) were resolved on nonreducing or reducing 15% SDS-polyacrylamide gels and immunoblotted with ExbD-specific polyclonal antibodies. Reduced and nonreduced samples came from the same culture. d indicates the position of the homodimer, and m indicates the position of the monomer. The positions of nonreducing molecular mass standards are indicated on the left.
To see if any of the ExbD periplasmic domain Cys substitutions were capable of forming spontaneous disulfide-linked homodimers, samples were prepared under nonreducing conditions. Bacterial cell pellets were solubilized in sample buffer containing iodoacetamide to alkylate free cysteines and prevent dimer formation after cell lysis. ExbD homodimers would theoretically migrate at about 31 kDa. Accordingly, ExbD-specific immunoblots of nonreducing SDS-polyacrylamide gels showed a number of ExbD Cys substitutions formed disulfide-linked homodimers, with the highest level of complex formation observed for G96C, D99C, I102C, D107C, K108C, Y112C, and E113C (Fig. 1, nonreduced). Notably, dimer formation appeared to stabilize ExbD I102C. Reducing conditions completely hydrolyzed the homodimer, yet only a small amount of monomeric ExbD I102C was detected on long exposures of immunoblots (see Fig. S1 in the supplemental material). Lower levels of dimer formation were observed for A92C, E95C, K98C, T109C, V110C, D111C, M116C, and D120C. For a few substitutions, a higher complex was formed, migrating around 40 kDa and most significantly detected with G96C, K98C, and D99C. This complex was still detected in a strain lacking ExbB, ruling out the possibility that it was a disulfide-linked complex between ExbD and ExbB, which has a single native cysteine (see Fig. S2 in the supplemental material; data not shown). The identity of this complex is unknown.
It is important to note that, for certain Cys substitutions, the nature of the native side chain would not favor interaction with the same residue of another ExbD. Asp99, for example, formed prominent homodimeric interactions when replaced with Cys, but two side chains with the same charge are unlikely to interact. Disulfide-linked interactions served as evidence that the regions of ExbD where these substitutions are located came into close contact in vivo. Disulfide bonds may trap normally transient conformations in addition to any native interfaces.
ExbD residues 92 through 121 are tolerant of replacement with cysteine.
The ability of each ExbD with a Cys substitution to support ferrichrome-mediated iron transport under nonreducing conditions was determined, with each substitution expressed to native ExbD levels (Fig. 2, lower immunoblots). The corresponding degree of dimer formation for each strain assayed was also determined. All 30 substitutions were active, with Y112C supporting the lowest initial rate of transport at about 80% the rate of wild-type plasmid-encoded ExbD (Fig. 2C).
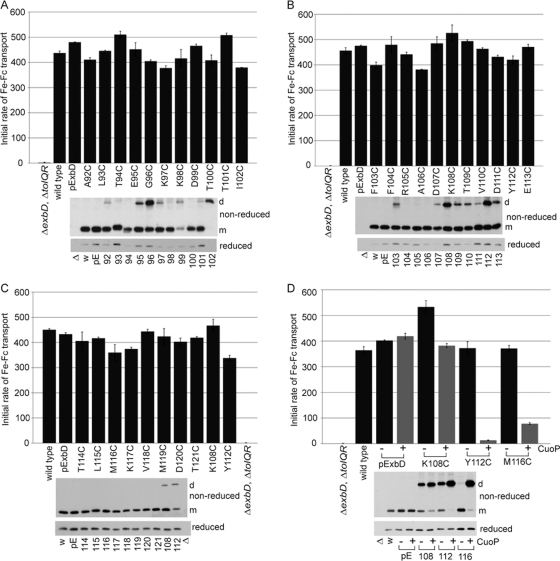
Fig. 2.
ExbD residues 92 through 121 are tolerant of replacement with cysteine, but disulfide-linked dimer formation can inhibit activity. A strain expressing chromosomally encoded wild-type ExbD (W3110) or a ΔexbD ΔtolQR strain (RA1045) expressing plasmid-encoded wild-type ExbD (pExbD) or ExbD Cys substitutions near native ExbD levels (see Table S1 in the supplemental material, 1× M9, for induction levels) was assayed for the ability to support transport of iron-loaded ferrichrome (Fe-Fc) as described in Materials and Methods. The results presented are representative data where activity was within 5% across at least two sets of triplicate assays. The y axis indicates the initial rate of transport. The ExbD variants assayed are indicated along the x axis. TCA-precipitated samples harvested just prior to each assay were resolved on 15% nonreducing or reducing SDS-polyacrylamide gels and immunoblotted with ExbD-specific polyclonal antibody. Δ indicates the ΔexbD ΔtolQR strain (RA1045), w indicates wild-type strain W3110, and pE indicates plasmid-encoded wild-type ExbD. The residue numbers indicate the positions of the ExbD Cys substitution. On the right, d indicates the position of the homodimer, and m indicates the position of the monomer. (A, B, and C) Sets of activity assays performed on the same days. (D) Activity assays for select ExbD homodimers catalyzed by CuoP treatment. − and + indicate buffer only and 0.03 mM CuoP treatment, respectively, prior to assay. The gray bars highlight CuoP-treated strains. The error bars indicate standard errors of the means.
The 80% activity of ExbD I102C, for which a monomer was not detected, suggested that ExbD was active as a homodimer (Fig. 2A). ExbD G96C, K108C, and Y112C also had significant levels of disulfide-linked dimers present (Fig. 2A and B). Dimers observed here for F103C were detected only when this substitution was overexpressed. ExbD G96C, with ∼2/3 present as a homodimer and 1/3 present as a monomer, supported 85% activity, also suggesting that the dimeric form was active. ExbD K108C and Y112C, which were initially expressed at higher levels than native ExbD and exhibited dimer levels equal to or greater than monomer levels, supported 110% and 90% activity, respectively (Fig. 2B). When inducer was decreased to achieve the desired native ExbD levels of expression for these substitutions, significantly less dimer was present for K108C and activity was the same, near 110%. Y112C now showed equal dimer and monomer levels, and activity decreased to 80% (Fig. 2C). The activities of these substitutions could be attributed to the significant level of monomer present.
ExbD homodimers differentially affect ExbD activity.
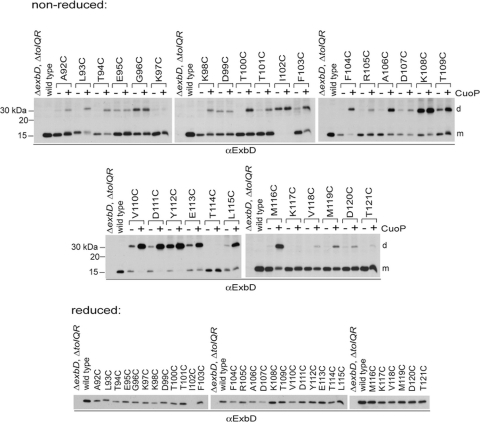
To increase the ratio of dimer to monomer so that the activity of the dimer could be estimated, strains expressing ExbDs with Cys substitutions were treated with the oxidizing agent CuoP as described in Materials and Methods. After treatment, only 3 substitutions, K97C, T114C, and K117C, still showed no homodimer formation (Fig. 3). For E95C, G96C, D99C, and I102C, homodimer levels remained unchanged after CuoP treatment. Significant increases in homodimers were observed for T109C through E113C, L115C, and M116C.
Fig. 3.
Addition of an oxidizing agent increases the number of ExbD Cys substitutions trapped in disulfide-linked homodimers. (Nonreduced) ΔexbD ΔtolQR strains (RA1045) expressing plasmid-encoded ExbD variants near native ExbD levels (see Table S1 in the supplemental material, 1× M9, for induction levels) were treated with buffer only (−) or 0.03 mM CuoP (+) for 5 min at 37°C, with aeration. TCA-precipitated samples were resolved on nonreducing 15% SDS-polyacrylamide gels and immunoblotted with ExbD-specific polyclonal antibodies. The positions of nonreducing molecular mass standards are indicated on the left. d indicates the position of the homodimer, and m indicates the position of the monomer. (Reduced) TCA-precipitated samples taken from the same cultures prior to treatment were resolved on reducing 15% SDS-polyacrylamide gels and immunoblotted with ExbD-specific polyclonal antibodies.
The degree of dimer formation could be gauged only by the decrease in monomer levels. This was due to the fact that, although treated and untreated samples came from the same culture, the detectability of ExbD in the dimer band had greatly increased—exemplified by D111C, Y112C, and E113C—suggesting a change in conformation that allowed greater access to antibody binding. Only a slight increase in total ExbD levels was observed when CuoP-treated samples of these Cys substitutions were reduced after treatment (data not shown). Inducer was absent during the 5-min CuoP treatment, ruling out an actual increase in the amount of protein synthesized, which would otherwise have been evident in all treated samples.
Since CuoP treatment catalyzed almost 100% dimer formation for some of the Cys substitutions, attempts were made to determine the activities of the dimeric forms. It was necessary to adapt standard CuoP treatments to the demands of the iron transport assays because the standard treatment inhibited iron transport in wild-type cells (reference 27 and data not shown). To retain full iron transport activity of a wild-type strain after CuoP treatment, the concentration of CuoP was reduced 10-fold to 0.03 mM, and cells had to be suspended in fully supplemented M9 medium as described in Materials and Methods (Fig. 2D, pExbD).
Unfortunately, this treatment no longer catalyzed high levels of dimer formation for the majority of ExbD Cys substitutions (data not shown). Only Y112C and M116C exhibited nearly 100% homodimers. K108C also showed a significant increase in homodimers, though monomer was still present (Fig. 2D, nonreduced). This limited our ability to examine the activity or inactivity of observed ExbD homodimers to these few substitutions.
For both Y112C and M116C, the dimeric form inhibited iron transport activity. CuoP-treated ExbD Y112C and M116C supported only 5% and 20% activity, respectively, compared to about 90% for samples treated with buffer only (Fig. 2D). In contrast, ExbD K108C exhibited monomer levels similar to those of ExbD M116C after CuoP treatment but was still at least 70% active. This was similar to the result, described above, where ExbD I102C was highly active as a spontaneously formed homodimer.
ExbD dimers are assembled with ExbB.
It was not known if the inactive ExbD disulfide-linked dimers were inhibited in their ability to assemble as part of a normal complex with ExbB, such as if nonnative conformations were trapped after translocation of the periplasmic domain across the CM or if dimer formation blocked later conformational changes after assembly. Both active and inactive ExbD (ExbD D25N or wild-type ExbD after the collapse of PMF) can be cross-linked with formaldehyde to ExbB (38). Since CuoP catalyzed almost complete dimerization for certain ExbD Cys substitutions, attempts were made to formaldehyde cross-link these disulfide-linked dimers to ExbB to determine if the trapped ExbD homodimeric conformations could still assemble with ExbB.
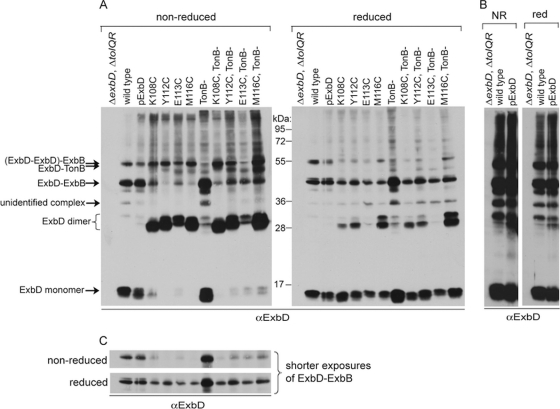
Cultures expressing wild-type ExbD or the 4 ExbD Cys substitutions that showed the highest dimer levels (Fig. 3), K108C, Y112C, E113C, and M116C, were treated with 0.3 mM CuoP for 5 min and then cross-linked with formaldehyde under nonreducing conditions. Cross-linked samples were divided in half. One half was reduced with β-mercaptoethanol to hydrolyze disulfide cross-links, and the other half remained nonreduced. All were solubilized at 60°C to retain formaldehyde cross-links. Samples were resolved on reducing or nonreducing SDS-polyacrylamide gels and immunoblotted with ExbD-specific polyclonal antibodies (Fig. 4). An ExbD disulfide-linked homodimer (∼31 kDa) formaldehyde cross-linked to ExbB (∼26 kDa) would theoretically migrate at approximately 57 kDa. Due to migration similar to that of a heterodimeric complex of ExbD and TonB (52 kDa), each substitution was also cross-linked in a ΔtonB background (KP1509) to rule out those complexes due to TonB-ExbD cross-links.
Fig. 4.
ExbD disulfide-linked homodimers assemble with ExbB. Strains expressing chromosomally encoded wild-type ExbD (W3110) in the absence of TonB (KP1344) or a ΔexbD ΔtolQR strain (RA1045) or a ΔexbD ΔtolQR ΔtonB (KP1509) strain expressing plasmid-encoded wild-type ExbD (pExbD) or ExbD Cys substitutions near native ExbD levels (see Table S1 in the supplemental material for induction levels) were treated with CuoP, washed, and then cross-linked with formaldehyde as described in Materials and Methods. (A) Samples were resolved on 13% nonreducing or reducing SDS-polyacrylamide gels and immunoblotted with ExbD-specific polyclonal antibodies. The positions of monomeric ExbD and ExbD-specific cross-linked complexes are indicated on the left. The positions of molecular mass standards are indicated between the immunoblots. (B) Long exposures of the immunoblots in panel A showing only the strains expressing wild-type ExbD. NR indicates nonreduced. Red indicates reduced. (C) Short exposures of the immunoblots in panel A, cropped to the region of the ExbB-ExbD heterodimer, are shown for better comparison of relative levels of the complex under nonreducing or reducing conditions.
As seen previously under reducing conditions (38), here, wild-type monomeric ExbD formaldehyde cross-linked into homodimers and heterodimeric complexes with ExbB and TonB, which were detected under both nonreducing and reducing conditions (Fig. 4A). The formaldehyde-cross-linked homodimer is typically the complex with the lowest abundance (38) and was apparent here on long exposures (Fig. 4B). Under nonreducing conditions, cross-linking profiles for ExbD Cys substitutions had much lower levels of the ExbD-ExbB complex and appeared to show a TonB-ExbD cross-link (Fig. 4A, left immunoblot). However, the ∼57-kDa bands were still present in a ΔtonB strain, ruling out a cross-link to TonB. The size of the complex suggested it could represent a novel complex of an ExbD disulfide-linked homodimer that was formaldehyde cross-linked to ExbB (ExbD2-ExbB). Due to the CuoP treatment, much higher levels of homodimers and no monomers were detected. It was not clear why ExbD homodimer bands appeared as a doublet; however, both bands decreased in the presence of reducing agent, suggesting disulfide-linked dimers were in both bands. Doublet bands were also apparent for the ExbD monomer, including cysteineless wild-type ExbD. These may represent two conformations of ExbD, an unknown modification, or a degradation product.
The same samples were also immunoblotted under reducing conditions that hydrolyzed the disulfide bond of the ExbD homodimer and retained ExbD monomers formaldehyde cross-linked to other proteins (Fig. 4A, right immunoblot). Under these conditions, levels of the suspected ExbD2-ExbB heterotrimer were greatly lowered with concomitant restoration of the typical ExbD-ExbB heterodimer and ExbD monomer band (Fig. 4A and C). This confirmed the identity of this 57-kDa band as an ExbD2-ExbB heterotrimer, demonstrating that the ExbD disulfide-linked dimers assembled with ExbB in vivo. This also confirmed the supposition, based on data from the Mot and Tol systems, that dimeric ExbD interacts with ExbB (4, 6, 23). The absence of TonB had no effect on the cross-linking profiles in the reduced samples, indicating that while ExbD was trapped as a disulfide-linked homodimer, it could not formaldehyde cross-link to TonB.
ExbD homodimer formation does not require ExbB or ExbD D25.
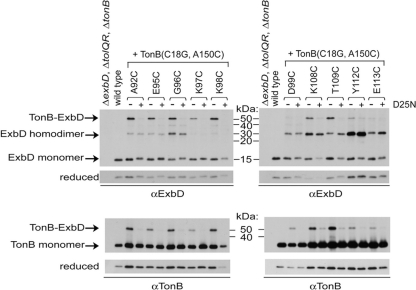
While it was observed that select ExbD disulfide-linked homodimers could still complex with ExbB, it was not known if they formed before or after interaction with ExbB. Since ExbD homodimers formaldehyde cross-link in the absence of ExbB, it was possible disulfide-linked dimer formation occurred before interaction with ExbB (38). The abilities of the Cys substitutions to form spontaneous disulfide-linked dimers were compared in a ΔexbBD ΔtolQRA strain (RA1017). All dimers still formed, and no new dimers were detected in the absence of ExbB (see Fig. S2 in the supplemental material and data not shown). Apparent decreases in the levels of dimers mostly reflected variations in ExbD expression, since ExbB stabilizes ExbD (9). For ExbD D111C, Y112C, and E113C, the decrease in dimers suggested that they interacted with ExbB.
D25 is an essential residue in the ExbD transmembrane domain, and a D25N mutation prevents energized interaction between TonB and ExbD periplasmic domains (38). A functional ExbD TMD, however, was not important for homodimer formation. Inactive ExbD D25N can be cross-linked into homodimers with formaldehyde (38), and accordingly, all ExbD D25N Cys substitutions still formed disulfide-linked dimers (see Fig. S2 in the supplemental material). No new dimers were detected in the presence of the D25N mutation (data not shown).
ExbD homodimers increase in the absence of TonB.
The results discussed above, where formaldehyde cross-linking of disulfide-linked ExbD dimers was analyzed, suggested that the ExbD periplasmic domain could form disulfide-linked homodimers or formaldehyde-cross-linked TonB-ExbD heterodimers, but not both at the same time. For the four Cys substitutions examined in that assay, dimer formation was efficiently catalyzed even in the absence of TonB. To determine if TonB was important for spontaneous ExbD homodimer formation, disulfide-linked dimer formation was examined in a ΔexbD ΔtolQR ΔtonB strain (KP1509). All 14 dimer-forming Cys substitutions showed increased disulfide-linked dimers in the absence of TonB, suggesting possible competition existed between homodimers and heterodimers and a TonB-dependent change in the dynamics of the ExbD periplasmic domain (Fig. 5).
Fig. 5.
ExbD disulfide-linked dimers increase in the absence of TonB. TCA-precipitated samples of wild-type chromosomally encoded ExbD (W3110) and a ΔexbD ΔtolQR strain (RA1045) or a ΔexbD ΔtolQR ΔtonB strain (KP1509) expressing plasmid-encoded ExbD Cys substitutions near native ExbD levels (see Table S1 in the supplemental material for induction levels) were resolved on nonreducing or reducing 15% SDS-polyacrylamide gels and immunoblotted with ExbD-specific polyclonal antibodies. Reduced and nonreduced samples came from the same culture. + indicates the presence (RA1045), and − indicates the absence (KP1509) of TonB. ExbD Cys substitutions are indicated across the top. The positions of nonreducing molecular mass standards are indicated on the left. d indicates the position of the homodimer, and m indicates the position of the monomer. The middle set of immunoblots is a shorter exposure of the top set showing the monomer band only.
Most ExbD Cys substitutions that can dimerize also form heterodimers with TonB with a Cys substitution.
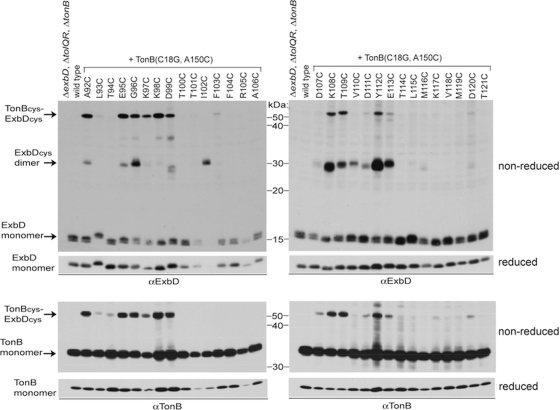
It was previously observed that ExbD A92C forms a homodimer and a disulfide-linked heterodimer with TonB(C18G, A150C) (38). A150 is a nonessential residue of the TonB carboxy terminus (residues 150 to 239), a functionally essential region of the TonB periplasmic domain (30, 41). To determine if any of the other ExbD residues with Cys substitutions could be trapped in specific interaction with TonB, each was coexpressed with TonB(C18G, A150C), and disulfide-linked complex formation was examined for both ExbD and TonB. A number of ExbD Cys substitutions were trapped in disulfide-linked heterodimers with TonB (Fig. 6). There was a strong correspondence between those Cys substitutions that participated in ExbD-TonB heterodimer formation and those that formed homodimers: A92C, E95C through D99C, K108C, T109C, Y112C, and E113C. Both interactions, however, were not observed through all sites. ExbD I102C and V110C formed homodimers but not ExbD-TonB heterodimers. K97C and K98C formed heterodimers but not homodimers.
Fig. 6.
ExbD cysteine substitutions share common interfaces between homodimeric and heterodimeric interactions. TCA-precipitated samples of strains expressing chromosomally encoded wild-type ExbD and TonB (W3110) or a ΔexbD ΔtolQR ΔtonB strain (KP1509) coexpressing plasmid-encoded ExbD Cys substitutions and TonB(C18G, A150C) near native levels were resolved on nonreducing or reducing 13% and 11% SDS-polyacrylamide gels and immunoblotted with ExbD-specific polyclonal antibodies or TonB-specific monoclonal antibodies. Reduced and nonreduced samples came from the same culture. The positions of the monomeric proteins or disulfide-cross-linked complexes are indicated on the left. L93C, T94C, D107C, D111C, and D120C ExbD-TonB heterodimers were detected on longer exposures (data not shown). The positions of nonreducing molecular mass standards are indicated between the immunoblots. The strain designations across the top apply to both immunoblots below. All samples were processed on the same day.
ExbD D25 is specifically important for disulfide-linked TonB-ExbD heterodimers.
We had previously observed that ExbD D25N still formed disulfide-linked homodimers but did not form the ExbD A92C-TonB A150C heterodimers (38). To determine if a functional ExbD TMD was also important for the other observed disulfide-linked heterodimers, ExbD Cys substitutions with a wild-type TMD or containing a D25N TMD substitution were coexpressed with TonB(C18G, A150C) and examined under nonreducing conditions. ExbD Cys substitutions formed homodimers with or without residue D25, but significant interaction with TonB A150C required D25 (Fig. 7). In the presence of wild-type TonB, ExbD D25N Cys homodimers were increased compared to substitutions with a wild-type TMD, as expected if disulfide-linked homodimer formation prevented subsequent interactions with TonB (see Fig. S2 in the supplemental material). In studies of these same substitutions with the coexpression of TonB A150C, the differences between ExbD Cys and ExbD D25N Cys homodimers were not as obvious (Fig. 7). Disulfide-linked heterodimer formation of the wild-type ExbD Cys with TonB A150C may also have resulted in increased ExbD homodimers, similar to the effect of the D25N substitution, possibly by blocking further cycles of ExbD-TonB associations.
Fig. 7.
ExbD D25 is important for ExbD-TonB disulfide-linked heterodimer formation. TCA-precipitated samples of strains expressing chromosomally encoded wild-type ExbD and TonB (W3110) or a ΔexbD ΔtolQR ΔtonB strain (KP1509) coexpressing plasmid-encoded TonB(C18G, A150C) and ExbD Cys substitutions without (−) or with (+) a D25N TMD substitution near native levels were resolved on nonreducing or reducing 13% and 11% SDS-polyacrylamide gels and immunoblotted with ExbD-specific polyclonal antibodies or TonB-specific monoclonal antibodies. Reduced and nonreduced samples came from the same culture. The positions of the monomeric proteins or disulfide-cross-linked complexes are indicated on the left. The positions of nonreducing molecular mass standards are indicated between the immunoblots. The strain designations across the top apply to both immunoblots below. All samples were processed on the same day.
DISCUSSION
ExbD can formaldehyde cross-link in vivo into a homodimer or a heterodimer with TonB through its periplasmic domain. ExbD also formaldehyde cross-links efficiently to ExbB through unknown regions. ExbB and PMF are required to form the ExbD-TonB heterodimer, but not the ExbD homodimer. The functional significance of the ExbD homodimer is unknown. ExbD A92C was previously demonstrated to be a site of in vivo ExbD homodimeric and ExbD A92C-TonB A150C heterodimeric interaction. ExbD A92C formed homodimers even if the inactivating D25N TMD mutation was present, but the heterodimeric complex with TonB A150C required functional TMDs of both proteins (38).
The same periplasmic ExbD residues mediate in vivo interactions between ExbD dimers and ExbD-TonB heterodimers.
We recently identified a 30-residue region of ExbD that was important in supporting ExbD formaldehyde-cross-linked protein-protein interactions, including homodimers and ExbD-TonB heterodimers (Ollis et al., unpublished). To determine if this region was directly involved in these interactions, ExbD residues 92 through 121 were scanned with Cys substitutions for analysis of in vivo disulfide cross-linking with another ExbD and with TonB A150C.
Here, we showed that most of the 14 ExbD Cys substitutions that mediated disulfide cross-linking of ExbD homodimers also mediated formation of heterodimers with TonB. This suggested that ExbD periplasmic domain homodimeric interactions were not formed through static interfaces, since a permanent interaction would have prevented interaction with TonB in that region. Consistent with this idea, the absence of competing TonB led to an increase in ExbD homodimerization.
Because ExbD homodimers could associate with ExbB and because ExbD homodimers appeared to be the default arrangement for ExbD in the absence of other members of the complex, it appeared that the ExbD2-ExbB complex might be an initial assembly intermediate. TonB was also not required for the formation of the ExbD2-ExbB complex. It should be noted that Pramanik et al. recently identified an ExbB6-ExbD1 complex in vitro and could find no evidence for ExbD dimers (42). In that study, however, ExbD carried a StrepII affinity tag at its carboxy terminus and ExbB carried a His6 tag. Epitope tags have resulted in artifactual protein interactions or prevented native interactions, described in multiple protein systems where histidine tags were used (8, 19, 47). Within the TonB system, ExbD oligomerization states are affected by epitope tagging. A T7 epitope tag at the amino terminus of ExbD prevents in vivo formaldehyde cross-linking between ExbD and TonB or ExbB and results in artifactual formation of T7-ExbD homotrimers (38). It could be that the presence of the StrepII or His6 tag prevented detectable association of ExbD homodimers with ExbB in vitro.
The functional distinction between homodimeric ExbD and TonB-ExbD heterodimeric interactions became apparent when the ExbD D25N substitution was examined. Because the disulfide-linked complexes in this study occurred spontaneously, it was not possible to directly assess the role of PMF. Newly synthesized proteins require PMF for CM insertion, and disulfide-linked complexes would be preexisting in any cultures treated with a protonophore to collapse the PMF. ExbD TMD residue D25 is a candidate residue for promoting direct response of ExbD to PMF and is essential for energized ExbD-TonB periplasmic domain interaction (38). In this study ExbD D25N largely prevented the formation of ExbD-TonB heterodimers, which validated the biological relevance of these specific sites of disulfide-linked interaction.
A model is therefore proposed where ExbD homodimers constitute the initial interaction of the ExbD periplasmic domain. ExbD homodimers assembled with ExbB are able to interact with TonB. Homodimerization of ExbD appears to involve multiple interfaces, and specific regions of the ExbD homodimer transition to heterodimeric interactions with TonB. The ExbD TMD, specifically residue D25, is important for the conformational transitions of the ExbD periplasmic domain to these specific heterodimeric associations with the TonB carboxy terminus. The specific signal that initiates the transition is unknown.
The theme of homodimeric-to-heterodimeric transitions involving shared interfaces has been seen in other systems. In Agrobacterium tumefaciens, TraM inhibits the transcriptional activator TraR through formation of a heterodimeric antiactivation complex. Both TraM and TraR form homodimers, with the homodimeric interface of TraM also serving as its interactive site with TraR. Initial nonspecific binding of TraR to the TraM homodimer has been proposed to facilitate homodimer dissociation and formation of the inactive TraR-TraM heterodimer (7). If present, initial PMF-independent interaction between ExbD and TonB periplasmic domains could serve a similar purpose. In the case of the TonB system, however, it is likely that ExbD-TonB periplasmic domain interaction activates TonB, since TonB lacks TMD residues that could promote a direct, independent response of TonB to PMF (45). The TonB-ExbD periplasmic domain interaction, detected by formaldehyde cross-linking, requires PMF (38). Thus, an equilibrium between ExbD and TonB-ExbD interactions might be more like the yeast copper chaperone yCCS and copper, zinc superoxide dismutase SOD1. Both form homodimers, as do both TonB and ExbD (11, 38), with activation of SOD1 by yCCS occurring through heterodimeric interaction between conserved homodimer interface residues from both proteins (28). Copper binding to yCCS promotes and stabilizes heterodimeric interaction with SOD1 (46).
Similarities and differences between the solved structure of the ExbD periplasmic domain and in vivo results.
Previous in vitro studies using NMR diffusion or dynamic light-scattering assays with the isolated ExbD periplasmic domain (1 mM at pH 3.0 or 7.0) showed multimerization of 4 to 7 copies of the ExbD periplasmic domain. Based on amide chemical shifts, when the protein concentration was increased from 0.2 mM (monomeric) to 1 mM (multimeric), the region of residues 104 to 116 was proposed as the homomultimeric interface. The opposite end of the structure, including residues 92 to 103, was not involved in multimerization based on no change with increased protein concentration (10). The proposed region of homomultimerization was not investigated in vivo.
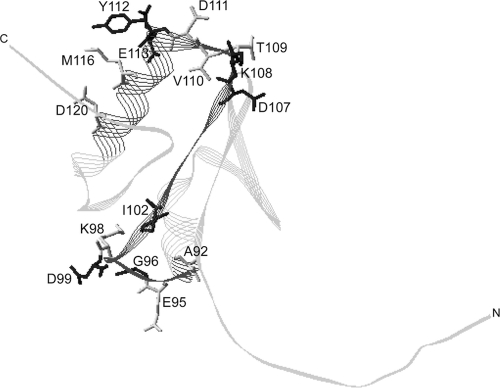
When all sites of spontaneous in vivo disulfide-linked dimer formation in this study were mapped onto the ExbD periplasmic domain NMR structure, homodimerization sites clustered at both ends of the structure (Fig. 8). These two distinct interactive domains suggested the possibility of at least two homodimeric interfaces in vivo. If these residues form a single interface, the interactive conformation is not represented by the solution structure, where residue side chains of even just the most prominent interaction sites face in multiple directions. One of the in vivo interactive regions, residues 106 to 116, overlapped most of the identified in vitro interactive region, residues 104 to 116. In vivo interaction through these substitution sites did not require function of the ExbD TMD, supporting the fact that this region supports homomultimeric associations of an inactive ExbD periplasmic domain, even lacking the entire TMD. In the monomeric NMR structure, this region has predominantly hydrophobic, surface-exposed residues, F104, A106, V110, L115, and M116, which could promote nonspecific aggregation of the 4 to 7 subunits. While in vivo results support interactions through this region, it is unknown if higher-order homomultimers, in addition to a homodimer, occur in vivo.
Fig. 8.
Sites of disulfide-forming Cys substitutions map to opposite ends of the ExbD periplasmic domain solution structure. Side chains of residues where Cys substitutions were trapped in disulfide-linked homodimers in vivo are mapped on the ExbD periplasmic domain NMR structure, Protein Data Bank (PDB) code 2pfu. The image was generated using Swiss-PdbViewer (14). The darker gray ribbon represents the Cys-scanned region examined in this study. The black side chains indicate significant spontaneous ExbD homodimer formation. The gray side chains indicate weak spontaneous homodimer formation. With the exception of I102, V110, and M116, the side chains pictured showed significant ExbD heterodimer formation with TonB A150C formation. C and N indicate the carboxy and amino termini of the domain, respectively.
A conformationally dynamic ExbD periplasmic domain.
This study provided evidence that the ExbD periplasmic domain conformation is dynamic in vivo. Interaction sites observed in vivo extend beyond those observed in vitro. ExbD I102, which was unstable as a Cys substitution unless trapped as a dimer, is a buried residue on the β4 strand of the NMR structure (Fig. 8). ExbD I102C was fully active as a dimer, suggesting that this region of the β4 strand is surface exposed in the monomers prior to forming the homodimer. The conformation of a monomeric unit of the active ExbD I102C dimer would require changes from the solved structure. Additional sites of interaction in vivo not observed in vitro clustered at the opposite end of the NMR structure from the identified sites and included A92, E95, G96, K98, and D99. Perhaps membrane insertion of ExbD is important for interaction through this region.
Specific regions of the ExbD periplasmic domain required conformational flexibility. Y112C, the site of highest dimer formation, and M116C, both located on the external face of the same helix in the NMR structure, were only active as monomers. Complete homodimer formation through these sites inhibited activity, indicating that these residues must be free to move in vivo. This could suggest that interaction at the interface of this helix is normally resolved within a cycle of conformational changes or that nonnative conformations trapped by disulfide bonds reflect a transient close association of the helices. ExbD K108C, like Y112C and M116C, formed both homodimers and heterodimers, yet high levels of K108C homodimers did not significantly inhibit TonB activity. K108C may be in a region that comes into close association during conformational changes but not itself an essential residue for direct interactions. Sites such as Y112 or M116 could still be accessible to TonB interaction in a disulfide-linked K108C homodimer. The fact that the trapped dimer of ExbD I102C was fully active indicated that homodimeric ExbD had a functional role and suggested some regions of ExbD remained associated as homodimers even during heterodimeric interaction with TonB.
In the presence of an oxidizing agent, only 3 of the 30 ExbD Cys substitutions were not trapped in homodimeric interaction—K97, T114, and K117. This suggested that ExbD dynamically sampled many environments prior to establishing its dimeric interfaces. Findings in this study were also consistent with earlier suggestions that ExbD guides the conformation of the TonB periplasmic domain, which itself is conformationally dynamic (12, 29, 33, 41).
Comparison to the TolR periplasmic domain.
The solution structure of residues 59 to 130 of the paralogous TolR periplasmic domain (residues 39 to 139) from Haemophilus influenzae is a homodimer, and the monomers have secondary and tertiary structures similar to the Escherichia coli ExbD monomer solution structure (39). ExbD I102C, which was active as a dimer and unstable as a monomer, is located on the β4 strand in the ExbD structure. In the TolR structure, the corresponding β4 strands of each monomer do form part of the homodimeric interface (39). However, the strands are oriented antiparallel, so Leu104, which corresponds to ExbD Ile102, is not in close interaction between monomers, with a Cβ-Cβ distance of about 16 Å (data not shown). It is unknown how the presence of the TolR C-terminal tail (residues 131 to 139) would alter the structure—in the ExbD monomeric structure, the corresponding tail region lies partially along the β4 strand. The TolR dimeric structure, however, supports the idea of homodimeric interaction involving the β4 strand of ExbD in vivo.
In vivo disulfide cross-linking of E. coli TolR Cys substitutions, which overlapped ExbD Cys substitutions from 111 to 121 in this study, shows similarities to ExbD homodimeric interactions, although TolR Y117C, corresponding to ExbD Y112C, formed the only spontaneous dimer. Like ExbD Y112C, TolR Y117C is nonfunctional as a homodimer and is hypothesized to block conformational changes of the TolR periplasmic domain (13). As reported, however, in the H. influenzae TolR dimer structure, the corresponding residue Y114, although located on a helix that forms part of the homodimer interface, is too distant for interaction between monomers (39). Overall, the in vitro and in vivo homodimeric interactions suggest the β4 strand and α2 helix form homodimeric interfaces for both the ExbD and TolR periplasmic domains. The structural similarities of these proteins suggest they may form very similar homodimers. The functional differences of these similar proteins may be defined by the way their homodimers change in response to TMD interactions with their respective CM complexes and/or PMF.
In summary, this work demonstrated the conformational plasticity of the ExbD periplasmic domain, as it interacted either with itself or with a TonB periplasmic domain in vivo. The results led to a model where some homodimeric interactions of ExbD must be released for subsequent heterodimeric interaction with TonB. Asp25 in the ExbD TMD was important to promote this transition. Sequestering specific ExbD periplasmic domain residues in homodimeric interactions could provide a level of regulation for TonB-ExbD interactions, restricting unproductive interactions of the ExbD periplasmic domain and controlling the transition with signals from the ExbD TMD.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mary Huber for construction of pKP885, pKP899, pKP905, and pKP911. We thank Ray Larsen and the Postle laboratory for critical reading of the manuscript.
This work was supported by grant GM42146 from the National Institute of General Medical Sciences to K.P.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 7 October 2011.
REFERENCES
- 1. Braun V. 2009. FhuA (TonA), the career of a protein. J. Bacteriol. 191:3431–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Braun V., et al. 1996. Energy-coupled transport across the outer membrane of Escherichia coli: ExbB binds ExbD and TonB in vitro, and leucine 132 in the periplasmic region and aspartate 25 in the transmembrane region are important for ExbD activity. J. Bacteriol. 178:2836–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Braun V., Herrmann C. 1993. Evolutionary relationship of uptake systems for biopolymers in Escherichia coli: cross-complementation between the TonB-ExbB-ExbD and the TolA-TolQ-TolR proteins. Mol. Microbiol. 8:261–268 [DOI] [PubMed] [Google Scholar]
- 4. Braun V., Herrmann C. 2004. Point mutations in transmembrane helices 2 and 3 of ExbB and TolQ affect their activities in Escherichia coli K-12. J. Bacteriol. 186:4402–4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brinkman K. K., Larsen R. A. 2008. Interactions of the energy transducer TonB with noncognate energy-harvesting complexes. J. Bacteriol. 190:421–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cascales E., Lloubes R., Sturgis J. N. 2001. The TolQ-TolR proteins energize TolA and share homologies with the flagellar motor proteins MotA-MotB. Mol. Microbiol. 42:795–807 [DOI] [PubMed] [Google Scholar]
- 7. Chen G., Malenkos J. W., Cha M. R., Fuqua C., Chen L. 2004. Quorum-sensing antiactivator TraM forms a dimer that dissociates to inhibit TraR. Mol. Microbiol. 52:1641–1651 [DOI] [PubMed] [Google Scholar]
- 8. Cheung K. L., Huen J., Kakihara Y., Houry W. A., Ortega J. 2010. Alternative oligomeric states of the yeast Rvb1/Rvb2 complex induced by histidine tags. J. Mol. Biol. 404:478–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fischer E., Günter K., Braun V. 1989. Involvement of ExbB and TonB in transport across the outer membrane of Escherichia coli: phenotypic complementation of exb mutants by overexpressed tonB and physical stabilization of TonB by ExbB. J. Bacteriol. 171:5127–5134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Garcia-Herrero A., Peacock R. S., Howard S. P., Vogel H. J. 2007. The solution structure of the periplasmic domain of the TonB system ExbD protein reveals an unexpected structural homology with siderophore-binding proteins. Mol. Microbiol. 66:872–889 [DOI] [PubMed] [Google Scholar]
- 11. Ghosh J., Postle K. 2005. Disulphide trapping of an in vivo energy-dependent conformation of Escherichia coli TonB protein. Mol. Microbiol. 55:276–288 [DOI] [PubMed] [Google Scholar]
- 12. Ghosh J., Postle K. 2004. Evidence for dynamic clustering of carboxy-terminal aromatic amino acids in TonB-dependent energy transduction. Mol. Microbiol. 51:203–213 [DOI] [PubMed] [Google Scholar]
- 13. Goemaere E. L., Devert A., Lloubes R., Cascales E. 2007. Movements of the TolR C-terminal domain depend on TolQR ionizable key residues and regulate activity of the Tol complex. J. Biol. Chem. 282:17749–17757 [DOI] [PubMed] [Google Scholar]
- 14. Guex N., Peitsch M. C. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723 [DOI] [PubMed] [Google Scholar]
- 15. Hannavy K., et al. 1990. TonB protein of Salmonella typhimurium. A model for signal transduction between membranes. J. Mol. Biol. 216:897–910 [DOI] [PubMed] [Google Scholar]
- 16. Higgs P. I., Larsen R. A., Postle K. 2002. Quantitation of known components of the Escherichia coli TonB-dependent energy transduction system: TonB, ExbB, ExbD, and FepA. Mol. Microbiol. 44:271–281 [DOI] [PubMed] [Google Scholar]
- 17. Higgs P. I., Myers P. S., Postle K. 1998. Interactions in the TonB-dependent energy transduction complex: ExbB and ExbD form homomultimers. J. Bacteriol. 180:6031–6038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hill C. W., Harnish B. W. 1981. Inversions between ribosomal RNA genes of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 78:7069–7072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jakovljevic V., Leonardy S., Hoppert M., Sogaard-Andersen L. 2008. PilB and PilT are ATPases acting antagonistically in type IV pilus function in Myxococcus xanthus. J. Bacteriol. 190:2411–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kampfenkel K., Braun V. 1992. Membrane topology of the Escherichia coli ExbD protein. J. Bacteriol. 174:5485–5487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kampfenkel K., Braun V. 1993. Topology of the ExbB protein in the cytoplasmic membrane of Escherichia coli. J. Biol. Chem. 268:6050–6057 [PubMed] [Google Scholar]
- 22. Karlsson M., Hannavy K., Higgins C. F. 1993. ExbB acts as a chaperone-like protein to stabilize TonB in the cytoplasm. Mol. Microbiol. 8:389–396 [DOI] [PubMed] [Google Scholar]
- 23. Kojima S., Blair D. F. 2004. Solubilization and purification of the MotA/MotB complex of Escherichia coli. Biochemistry 43:26–34 [DOI] [PubMed] [Google Scholar]
- 24. Krewulak K. D., Vogel H. J. 2011. TonB or not TonB: is that the question? Biochem. Cell Biol. 89:87–97 [DOI] [PubMed] [Google Scholar]
- 25. Kuehl C. J., Crosa J. H. 2010. The TonB energy transduction systems in Vibrio species. Future Microbiol. 5:1403–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 27. Lai W. C., Hazelbauer G. L. 2007. Analyzing transmembrane chemoreceptors using in vivo disulfide formation between introduced cysteines. Methods Enzymol. 423:299–316 [DOI] [PubMed] [Google Scholar]
- 28. Lamb A. L., Torres A. S., O'Halloran T. V., Rosenzweig A. C. 2001. Heterodimeric structure of superoxide dismutase in complex with its metallochaperone. Nat. Struct. Biol. 8:751–755 [DOI] [PubMed] [Google Scholar]
- 29. Larsen R. A., et al. 2007. His20 provides the sole functionally significant side chain in the essential TonB transmembrane domain. J. Bacteriol. 189:2825–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Larsen R. A., FosterHartnett D., McIntosh M. A., Postle K. 1997. Regions of Escherichia coli TonB and FepA proteins essential for in vivo physical interactions. J. Bacteriol. 179:3213–3221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Larsen R. A., et al. 1996. Identification of TonB homologs in the family Enterobacteriaceae and evidence for conservation of TonB-dependent energy transduction complexes. J. Bacteriol. 178:1363–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Larsen R. A., Postle K. 2001. Conserved residues Ser(16) and His(20) and their relative positioning are essential for TonB activity, cross-linking of TonB with ExbB, and the ability of TonB to respond to proton motive force. J. Biol. Chem. 276:8111–8117 [DOI] [PubMed] [Google Scholar]
- 33. Larsen R. A., Thomas M. G., Postle K. 1999. Protonmotive force, ExbB and ligand-bound FepA drive conformational changes in TonB. Mol. Microbiol. 31:1809–1824 [DOI] [PubMed] [Google Scholar]
- 34. Lee S. K., Keasling J. D. 2005. A propionate-inducible expression system for enteric bacteria. Appl. Environ. Microbiol. 71:6856–6862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miller J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N. Y [Google Scholar]
- 36. Noel D., Nikaido K., Ames G. F. 1979. A single amino acid substitution in a histidine-transport protein drastically alters its mobility in sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Biochemistry 18:4159–4165 [DOI] [PubMed] [Google Scholar]
- 37. Noinaj N., Guillier M., Barnard T. J., Buchanan S. K. 2010. TonB-dependent transporters: regulation, structure, and function. Annu. Rev. Microbiol. 64:43–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ollis A. A., Manning M., Held K. G., Postle K. 2009. Cytoplasmic membrane protonmotive force energizes periplasmic interactions between ExbD and TonB. Mol. Microbiol. 73:466–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Parsons L. M., Grishaev A., Bax A. 2008. The periplasmic domain of TolR from Haemophilus influenzae forms a dimer with a large hydrophobic groove: NMR solution structure and comparison to SAXS data. Biochemistry 47:3131–3142 [DOI] [PubMed] [Google Scholar]
- 40. Postle K. 2007. TonB system, in vivo assays and characterization. Methods Enzymol. 422:245–269 [DOI] [PubMed] [Google Scholar]
- 41. Postle K., Kastead K. A., Gresock M. G., Ghosh J., Swayne C. D. 2010. The TonB dimeric crystal structures do not exist in vivo. mBio 1:e00307–e00310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pramanik A., et al. 2011. Oligomeric structure of ExbB and ExbB-ExbD isolated from Escherichia coli as revealed by LILBID-mass spectrometry. Biochemistry 50:8950–8956 [DOI] [PubMed] [Google Scholar]
- 43. Roof S. K., Allard J. D., Bertrand K. P., Postle K. 1991. Analysis of Escherichia coli TonB membrane topology by use of PhoA fusions. J. Bacteriol. 173:5554–5557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shedlovsky A., Brenner S. 1963. A chemical basis for the host-induced modification of T-even bacteriophages. Proc. Natl. Acad. Sci. U. S. A. 50:300–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Swayne C., Postle K. 2011. Taking the Escherichia coli TonB transmembrane domain “offline”? Non-protonatable Asn substitutes fully for TonB His20. J. Bacteriol. 193:3693–3701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Torres A. S., Petri V., Rae T. D., O'Halloran T. V. 2001. Copper stabilizes a heterodimer of the yCCS metallochaperone and its target superoxide dismutase. J. Biol. Chem. 276:38410–38416 [DOI] [PubMed] [Google Scholar]
- 47. Wu J., Filutowicz M. 1999. Hexahistidine (His6)-tag dependent protein dimerization: a cautionary tale. Acta Biochim. Pol. 46:591–599 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.