Abstract
Chlamydial heat shock proteins have important roles in Chlamydia infection and immunopathogenesis. Transcription of chlamydial heat shock genes is controlled by the stress response regulator HrcA, which binds to its cognate operator CIRCE, causing repression by steric hindrance of RNA polymerase. All Chlamydia spp. encode an HrcA protein that is larger than other bacterial orthologs because of an additional, well-conserved C-terminal region. We found that this unique C-terminal tail decreased HrcA binding to CIRCE in vitro as well as HrcA-mediated transcriptional repression in vitro and in vivo. When we isolated HrcA from chlamydiae, we only detected the full-length protein, but we found that endogenous HrcA had a higher binding affinity for CIRCE than recombinant HrcA. To examine this difference further, we tested the effect of the heat shock protein GroEL on the function of HrcA since endogenous chlamydial HrcA has been previously shown to associate with GroEL as a complex. GroEL enhanced the ability of HrcA to bind CIRCE and to repress transcription in vitro, but this stimulatory effect was greater on full-length HrcA than HrcA lacking the C-terminal tail. These findings demonstrate that the novel C-terminal tail of chlamydial HrcA is an inhibitory region and provide evidence that its negative effect on repressor function can be counteracted by GroEL. These results support a model in which GroEL functions as a corepressor that interacts with HrcA to regulate chlamydial heat shock genes.
INTRODUCTION
Chlamydia is a genus of obligate intracellular bacteria that causes a number of significant human diseases. Chlamydia trachomatis is the etiologic agent for the most prevalent bacterial sexually transmitted infection in the United States (40) and the most common form of infectious blindness in the world (9). A related species, Chlamydia pneumoniae, causes community-acquired pneumonia (18). More cases of chlamydial infections are reported to the CDC each year than any other notifiable infectious disease (10).
The major chlamydial heat shock proteins, DnaK, GroEL, and GroES (also known as Hsp70, Hsp60, and Hsp10, respectively), have been proposed to play an important role in the host immune response to chlamydial infection (20). GroEL and DnaK are highly immunogenic in patients infected with C. trachomatis (38), and GroEL and GroES have been serologically linked to severe sequelae of C. trachomatis infection (3, 7, 20, 30). In addition, GroEL can induce a number of host processes, such as inflammation and apoptosis, through the Toll-like receptor TLR4 (8, 11, 39). It has also been suggested that the immunopathogenesis of chronic chlamydial infection is due to cross-reactivity between conserved epitopes in chlamydial heat shock proteins and their human homologs (20).
Heat shock proteins include molecular chaperones and proteases that help to refold or degrade proteins during cellular stress (13, 24). Expression of heat shock proteins is maintained at baseline levels under normal conditions but is transiently upregulated in response to cellular stressors such as elevated temperature (2). This conserved heat shock response can be controlled at the transcriptional level by different regulatory mechanisms (28). In Chlamydia and many other bacteria, expression of heat shock genes is negatively regulated by the transcriptional repressor HrcA through binding to an operator called CIRCE (controlling inverted repeat of chaperone expression) (26, 34, 41, 45, 51). HrcA-CIRCE interactions prevent RNA polymerase from binding to heat shock promoters through steric hindrance, resulting in the repression of heat shock genes (55).
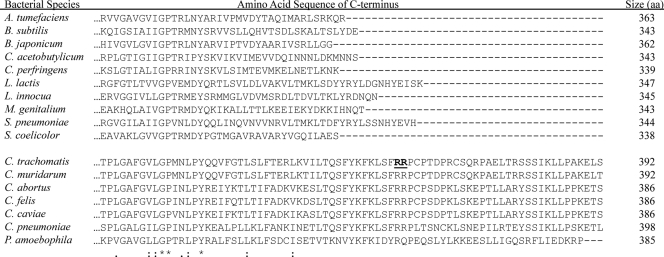
HrcA orthologs in bacteria are generally proteins of 39 to 40 kDa in molecular size, with conservation in two N-terminal regions that make up the DNA-binding domain and a C-terminal region of unknown function (21, 34, 41, 47). However, we noted that HrcA encoded by all Chlamydia spp. are about 10% larger because of additional sequence at their C termini. This extra C-terminal tail is well conserved in all Chlamydia spp. but not present in HrcA from other bacteria, and its functional significance is unknown (Fig. 1).
Fig. 1.
Chlamydial HrcA contains an additional C-terminal tail. Alignment of the C-terminal sequence of HrcA from six Chlamydia spp. (C. trachomatis, C. muridarum, C. abortus, C. felis, C. caviae, and C. pneumoniae), the related organism Protochlamydia amoebophila, and selected bacteria in which HrcA has been previously studied and/or identified. The amino acid sequence was inferred from each nucleotide sequence and analyzed with ClustalW software at http://www.ebi.ac.uk/Tools/clustalw2/index.html. Amino acid residues in the C terminus that are identical, strongly similar, or weakly similar among all HrcA orthologs are marked by an asterisk, colon, or period, respectively. The total number of amino acids (aa) in each HrcA ortholog is listed. The tandem arginine residues of C. trachomatis HrcA implicated in the premature termination of translation in E. coli are bolded and underlined.
In this report, we examined whether the Chlamydia-specific C-terminal tail affects the function of HrcA as a transcriptional regulator. Unexpectedly, we found that this additional C-terminal region reduced the ability of HrcA to bind its CIRCE operator and repress chlamydial heat shock genes. In addition, we present evidence that this effect of the C-terminal tail can be modulated by the heat shock protein GroEL. These findings support a model in which chlamydial HrcA is dependent upon GroEL interactions to regulate heat shock genes in Chlamydia trachomatis.
MATERIALS AND METHODS
Cloning of overexpression plasmids.
Plasmid pMT1133 contains the entire wild-type coding sequence of C. trachomatis serovar L2 hrcA, as previously described (51). Since pMT1133 expresses both a full-length and a truncated form of HrcA in Escherichia coli, plasmid pMT1214 was constructed to express only full-length recombinant HrcA. pMT1214 contains the coding region for C. trachomatis hrcA with sequence changes at nucleotides 1080 to 1085 (AGAAGA to CGTCGC), which substitute alternative arginine codons without changing the amino acid sequence of the expressed protein. To clone pMT1214, an upstream portion of hrcA was amplified from pMT1133 by PCR with Tgo DNA polymerase (Roche) using a T7 promoter primer (5′-TGAATTGTAATACGACTCACTATAGGG) and primer T507 (5′-GGGTCGGTCGGGCAAGGGCGACGGAATGACAATTTAAACTTG). In addition, a downstream portion of hrcA was amplified from pMT1133 using primers T472 (5′-TGCCCGACCGACCCTAGA) and T123 (5′-CCGGTACCTCATGATAGCTCCTTAGCGGGTAAT). The upstream PCR product digested with XbaI and the downstream PCR product digested with EcoRI were ligated together at their respective blunt ends to form a 1,330-bp ligation product. This 1,330-bp insert was then ligated into pRSET-C (Invitrogen) digested with XbaI and EcoRI.
Plasmid pMT1215 expresses a truncated form of C. trachomatis rHrcA, from amino acids 1 to 360, that lacks the Chlamydia-specific C-terminal tail. Nucleotides 1 to 1080 of hrcA were amplified by PCR from pMT1133 with Tgo DNA polymerase using the T7 promoter primer, described above, and primer T356 (5′-AGCGGTACCTCAGAATGACAATTTAAACTTGTAAAA). This PCR product and pRSET-C were digested with XbaI and EcoRI and then ligated together.
Plasmid pMT1620 expresses full-length rHrcA without an affinity tag and was used for size comparison to endogenous HrcA purified from C. trachomatis. To clone pMT1620, the coding region for hrcA from pMT1214 was amplified by PCR with Tgo DNA polymerase using primers T1182 (5′-CGCCATATGGAAAATAGAATAGAAATGTCCC) and T1183 (5′-TCATGATAGCTCCTTAGCGGG). The PCR product was digested with NdeI and ligated into the pET21a overexpression vector (Novagen) between NdeI and blunted XhoI sites. Plasmid pMT1621, which expresses truncated rHrcA without an affinity tag, was cloned in the same manner as pMT1620 except that primer T1184 (5′-TCAGAATGACAATTTAAACTTGTAAAAACTTTGAG) was used instead of T1183.
Plasmid pMT1494 expresses recombinant DcrA and contains the coding sequence for C. trachomatis CT296 cloned into pRSET-C. To clone pMT1494, the coding sequence for CT296 was amplified from C. trachomatis L2 genomic DNA by PCR with Pfu DNA polymerase using primers T1177 (5′-GATCCTCGAGATATGAGGGCAGTTTTACACCTAGAGCACAAGCGTTATTTC) and T1174 (5′-GATCCTGCAGTTAGTTAGGAAATCCCGCTGAGGAGAACCTAAG). The PCR product was digested with PstI and XhoI and ligated into pRSET-C between PstI and XhoI sites.
Overexpression and purification of recombinant proteins.
All His6-tagged recombinant proteins were overexpressed in E. coli BL21(DE3) and cells were lysed as previously described (51). rHrcA was purified with metal affinity chromatography with slight variations in the purification scheme, as described below, to optimize the purification of full-length and truncated forms of rHrcA. For some experiments, we further purified rHrcA to isolate the active fraction of rHrcA on the basis of specific binding to the cognate CIRCE operator (also described below).
Full-length rHrcA was purified with nickel affinity chromatography as previously described (51) with minor changes: the nickel column was washed with 10 column volumes of buffer N (10 mM Tris-HCl [pH 8.0], 300 mM NaCl, 10 mM 2-mercaptoethanol) containing 100 mM imidazole, and protein was eluted with 5 column volumes of buffer N containing 250 mM imidazole. Eluted proteins were further purified as described below or dialyzed overnight against storage buffer (10 mM Tris-HCl [pH 8.0], 10 mM MgCl2, 100 μM EDTA, 10 mM 2-mercaptoethanol, 100 mM NaCl, 30% glycerol) and again for 4 h.
Truncated rHrcA for binding assays with DNA affinity beads was purified with nickel affinity chromatography as described above except that the nickel column was washed with 20 column volumes of buffer N containing 20 mM imidazole. For electrophoretic mobility shift assays (EMSA) and in vitro transcription assays, truncated rHrcA was purified with cobalt affinity chromatography using TALON metal affinity resin (Clontech) instead of nickel to remove nonspecific nucleases that copurified with the recombinant protein. The cobalt column was washed with 500 column volumes of buffer N containing 10 mM imidazole prior to elution with 5 column volumes of buffer N containing 250 mM imidazole.
To isolate the active fraction of rHrcA, we performed an additional DNA affinity purification step using CIRCE DNA conjugated to magnetic beads (53). Full-length rHrcA purified by nickel affinity chromatography was diluted in bead buffer (10 mM Tris-HCl [pH 8.0], 10 mM 2-mercaptoethanol) to a NaCl concentration of 50 mM and incubated with the CIRCE DNA beads for 1 h at 4°C with gentle rocking. The beads were then washed 3 times with bead buffer containing 50 mM NaCl and eluted with bead buffer containing 1 M NaCl. Eluted proteins were dialyzed overnight against storage buffer and again for 4 h. The active fraction of truncated rHrcA was purified in a similar manner except that the buffer used to wash the beads contained 200 mM NaCl.
Recombinant GroEL was purified using nickel affinity chromatography as previously described for rHrcA (51) with minor changes: the nickel column was washed with 10 column volumes of buffer N containing 40 mM imidazole prior to elution with 5 column volumes of buffer N containing 250 mM imidazole. All protein samples were >90% pure as assayed by silver staining on SDS-PAGE gels. Protein concentrations were determined by the Bio-Rad protein assay.
Purification of C. trachomatis RNA polymerase.
RNA polymerase was partially purified from C. trachomatis serovar L2 at 24 h postinfection by heparin agarose chromatography as previously described (48).
Binding assays with DNA affinity beads.
Full-length and truncated forms of rHrcA purified with nickel affinity chromatography were diluted in bead buffer to a NaCl concentration of 100 mM. Diluted protein was incubated with CIRCE DNA magnetic beads for 20 min at room temperature (RT) with gentle rocking. Beads were then washed with bead buffer containing 150 mM NaCl for 5 min at RT, and wash fractions were collected. Washes were repeated with bead buffer containing 200, 250, 300, 350, 400, 450, and 800 mM NaCl. Wash fractions were resolved on SDS-PAGE and analyzed by silver staining.
EMSA.
DNA probes containing the C. trachomatis dnaK promoter region were prepared for EMSA as previously described (51). EMSA reactions with rHrcA were performed and the data were analyzed as previously described (51). For these experiments, we used rHrcA that had been purified by metal affinity chromatography only as well as active fractions of rHrcA that had been isolated with an additional DNA affinity purification step, as indicated in the text. The dissociation constants for binding reactions were calculated using Lineweaver-Burk analysis as previously described (51). For EMSA experiments with rGroEL and rHrcA, a modified 1× binding buffer (40 mM Tris-HCl [pH 8.0], 135 μM EDTA, 100 μM dithiothreitol, 7.5% glycerol) was used. rGroEL was preincubated with rHrcA for 10 min at RT prior to addition to the EMSA reaction mixture. Reaction mixtures were then incubated for an additional 10 min at RT. Samples were then loaded onto a 7% polyacrylamide EMSA gel. Anti-GroEL (A57-B9) antibodies (a generous gift of Richard Morrison, University of Arkansas for Medical Sciences) and anti-myc antibodies (Invitrogen) were used for supershift experiments.
In vitro transcription assays.
In vitro transcription reactions with rHrcA, partially purified C. trachomatis RNA polymerase, and transcription templates containing the dnaK promoter were performed and the data were analyzed as previously described (51). These experiments were performed with rHrcA that had been purified by metal affinity chromatography only as well as active fractions of rHrcA that had been further purified by a DNA affinity purification step, as indicated in the text. In vitro transcription experiments with rGroEL and rHrcA were performed in a modified reaction mixture containing 400 μM ATP, 400 μM UTP, 1.2 μM CTP, 0.21 μM [α-32P]CTP (800 Ci/mmol), 100 μM 3′-O-methylguanosine 5′-triphosphate Na salt, 18 U of rRNasin, 5% glycerol, and 15 nM supercoiled DNA template. For these reactions, rGroEL and rHrcA were first preincubated for 10 min at RT. These recombinant proteins were then added to the reaction mixture, which was incubated for an additional 10 min at RT prior to the addition of 0.02 U of E. coli RNA polymerase holoenzyme (Epicentre).
Cloning of lacZ reporter fusions.
Plasmid pMT1622 contains a fusion of the C. trachomatis dnaK promoter and the coding sequence of E. coli lacZ, cloned into plasmid pK-184 (17). The E. coli lacZ coding sequence was excised from plasmid pRS415 (44) by digestion with BamHI and SnaBI. This lacZ coding sequence was ligated into pK-184 between BamHI and blunted EcoRI sites to generate plasmid pMT1619. The C. trachomatis dnaK promoter was amplified from plasmid pMT1161 (51) by PCR with Tgo DNA polymerase using primers T1417 (5′-ATAGCATGCCCTATAAATTGATCATTGGGAAGTCTTTCC) and T1391 (5′-AAGTTGGTGTCATTATAGGAAAACCGGAG). The PCR product was digested with SphI and ligated into pMT1619 between SphI and blunted BamHI sites to generate plasmid pMT1622.
β-Galactosidase assays.
T7 Express (New England BioLabs) is an E. coli strain with the gene encoding T7 RNA polymerase stably integrated into the chromosomal lac operon, resulting in a nonfunctional lacZ gene. T7 Express cells were cotransformed with pMT1622 and one of four overexpression vectors: pRSET-C, pMT1214, pMT1215, or pMT1494. Overnight cultures were diluted 1:100 in Luria broth containing 200 μM isopropyl β-d-1-thiogalactopyranoside (IPTG) and grown at 37°C to an optical density at 600 nm of 0.8. β-Galactosidase activity was assayed as previously described (56) with minor changes: 2 or 4 μl of each culture was mixed with permeabilization buffer (0.8 mg/ml hexadecyltrimethylammonium bromide, 0.4 mg/ml sodium deoxycholate, 100 mM Na2HPO4, 20 mM KCl, 2 mM MgSO4, 100 mM 2-mercaptoethanol) in a total volume of 100 μl and incubated at 30°C for 30 min. Then, 600 μl of substrate solution (60 mM Na2HPO4, 40 mM NaH2PO4, 4 mg/ml o-nitrophenyl-β-galactoside) was added to each sample to initiate the reaction. Reactions were terminated with 700 μl of 1 M Na2CO3, and samples were centrifuged at 16,000 × g for 10 min at RT to pellet debris. The optical densities of each sample at 420 nm and 550 nm were measured, and β-galactosidase activity was calculated in Miller units (25). Overexpression of recombinant proteins in T7 Express cells was monitored by Western blotting with anti-His antibodies (GE Healthcare) as previously described (53).
Isolation of endogenous HrcA.
Murine L929 host cells (7.5 × 108) grown in suspension were infected with C. trachomatis serovar L2 at a multiplicity of infection of 3. Infected cells were harvested at 24 h postinfection, partially purified, and lysed as previously described (48). This lysate of chlamydial reticulate bodies was diluted in bead buffer to a NaCl concentration of 50 mM. Diluted lysate was incubated with CIRCE DNA magnetic beads for 15 min at 4°C with gentle rocking. Beads were then washed with bead buffer containing 50 mM NaCl for 10 min at 4°C, and wash fractions were collected. Washes were repeated with bead buffer containing increasing NaCl concentrations (100, 150, 200, 250, 300, and 350 mM NaCl). Wash fractions were resolved on SDS-PAGE and analyzed by Western blotting with anti-HrcA antibodies as previously described (53). Lysates of E. coli BL21(DE3) overexpressing untagged forms of either full-length or truncated rHrcA were included as size comparisons for endogenous HrcA. In parallel experiments, full-length rHrcA purified by nickel affinity chromatography was bound to the CIRCE DNA magnetic beads, and wash fractions were analyzed by Western blotting with anti-His antibodies.
RESULTS
Chlamydial HrcA contains a unique C-terminal tail.
When we compared the predicted amino acid sequence of HrcA from Chlamydia spp. and those of other bacteria, we noted that the chlamydial HrcA proteins have approximately 30 to 40 amino acids of additional sequence at their C termini (Fig. 1). The additional C-terminal sequence is well conserved among all Chlamydia spp. (71 to 97% similarity) but not present in HrcA from other bacteria (Fig. 1). This C-terminal tail accounts for the 10% larger size of chlamydial HrcA compared to its bacterial orthologs. It appears to be a unique sequence, since it did not have significant sequence similarity to other proteins in a BLAST search (data not shown). Interestingly, HrcA from the related bacterium Protochlamydia amoebophila, an endosymbiont of amoeba that has a Chlamydia-like developmental cycle, also contains an extended tail at its C terminus that is 64% similar to the C-terminal tail from C. trachomatis (Fig. 1).
Full-length recombinant HrcA binds its CIRCE operator with lower affinity than a truncated form lacking the C-terminal tail.
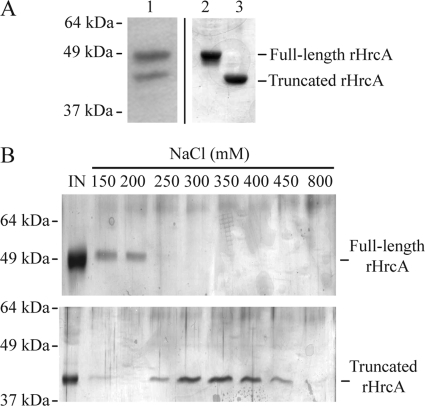
To examine whether this Chlamydia-specific C-terminal tail has functional significance, we generated recombinant forms of C. trachomatis HrcA containing or lacking this region. We engineered a truncated form of rHrcA that does not have the C-terminal tail by introducing a stop codon to prevent translation of this region (Fig. 2A, lane 3). We had to take extra steps to generate full-length rHrcA because expression of the wild-type C. trachomatis hrcA sequence in E. coli also produced a shorter form of the recombinant protein (Fig. 2A, lane 1). We reasoned that this shorter form was due to premature termination of translation in E. coli because of tandem arginine codons AGA-AGA at amino acid positions 361 to 362. AGA is a rare codon in E. coli (14) but not in Chlamydia (36), and tandem AGA codons are known to cause premature translational termination (12, 15, 35). By replacing these tandem AGA codons with the silent mutations CGT-CGC, we were able to produce full-length rHrcA alone (Fig. 2A, lane 2).
Fig. 2.
Full-length and truncated forms of rHrcA differ in their binding affinity for the CIRCE operator. (A) Silver stains of 12% SDS-PAGE showing purified wild-type chlamydial rHrcA (lane 1) utilized in previous studies (51–53), and full-length (lane 2) and truncated forms (lane 3) of recombinant HrcA used for in vitro assays in this report. (B) Recombinant forms of full-length and truncated HrcA that had been purified by metal affinity chromatography were bound to CIRCE DNA affinity beads in the presence of 100 mM NaCl. The beads were then sequentially washed with increasing concentrations of NaCl over a range from 150 to 800 mM NaCl. Wash fractions were resolved by 12% SDS-PAGE, and proteins were visualized by silver staining. IN, input sample.
We tested and compared purified forms of full-length and truncated rHrcA to determine whether binding of HrcA to its CIRCE operator is altered by the presence of the C-terminal tail. We measured their respective binding affinity for the CIRCE operator located on a DNA fragment conjugated to magnetic beads. Full-length rHrcA eluted at 150 to 200 mM NaCl, compared to 250 to 450 mM NaCl for truncated rHrcA (Fig. 2B). These unexpected results suggest that full-length chlamydial rHrcA has a lower binding affinity for its operator than a truncated form lacking the C-terminal tail.
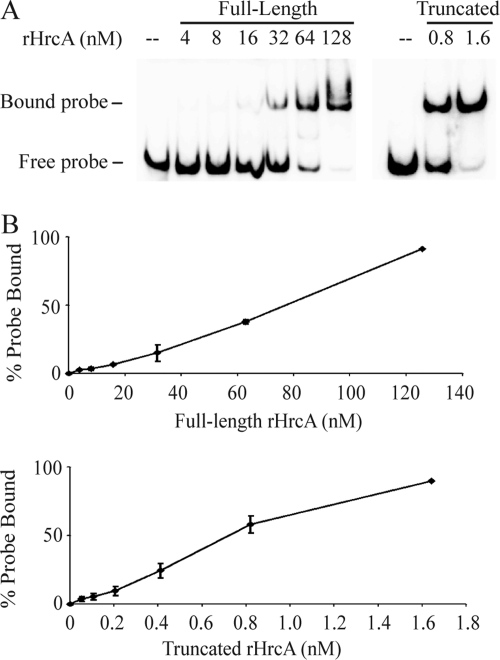
We used an electrophoretic mobility shift assay (EMSA) to quantitate this paradoxical increase in binding affinity when the C-terminal tail was deleted. The lower binding affinity of full-length HrcA could not be explained by differences in solubility because full-length rHrcA was much more soluble than truncated rHrcA (data not shown). Nevertheless, we isolated the active fraction of each recombinant HrcA protein by performing an additional affinity purification step with CIRCE DNA to remove misfolded or inactive protein that did not bind the CIRCE operator. In EMSA experiments, we obtained a 90% shift of the CIRCE DNA probe with 128 nM this active fraction of full-length rHrcA compared to only 1.6 nM active truncated rHrcA (Fig. 3). From these in vitro binding studies, we calculated an apparent dissociation constant (KD) of 49 nM for full-length rHrcA and 0.89 nM for truncated rHrcA, which is a >50-fold difference. These results demonstrate the striking effect of the Chlamydia-specific C-terminal tail in decreasing the affinity of HrcA for its CIRCE operator.
Fig. 3.
EMSA experiments quantitating the lower binding affinity of full-length rHrcA for the CIRCE operator compared to truncated rHrcA. (A) Representative EMSA in which active fractions of full-length and truncated rHrcA that had been purified with DNA affinity chromatography were incubated with a DNA probe containing the C. trachomatis dnaK promoter and its CIRCE operator (51). Full-length rHrcA was tested over a range of concentrations from 4 to 128 nM, while truncated rHrcA was tested at a lower concentration range, up to 1.6 nM, which was sufficient to produce an almost complete gel shift. The locations of the free and bound probes on the gel are marked. (B) Graphs showing quantification of the EMSA results over a range of rHrcA concentrations. Note the different scales for the x axes of the two graphs. The values shown are the mean of results of three independent experiments, and the error bars represent standard deviations.
Full-length recombinant HrcA does not repress transcription in vitro.
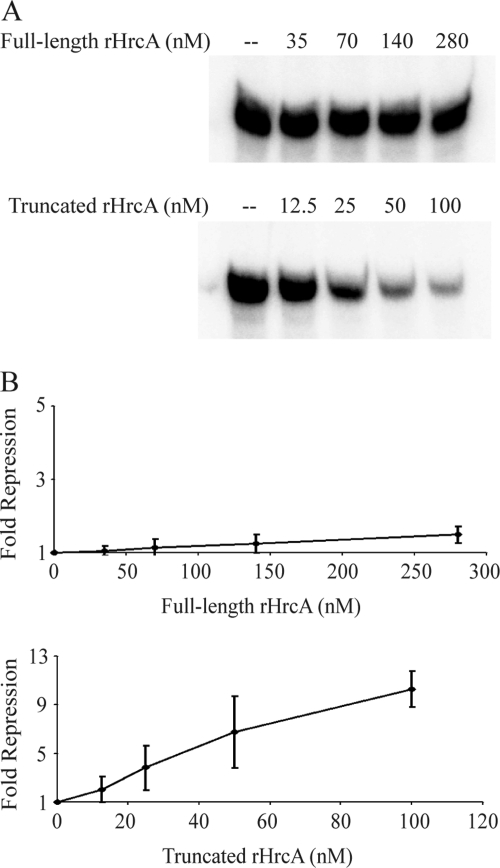
We next tested if this decrease in operator binding leads to decreased transcriptional repression of a heat shock promoter. We compared the ability of active full-length and truncated rHrcA to repress the C. trachomatis dnaK promoter, which is an HrcA-regulated promoter that contains a CIRCE operator (51). Active full-length rHrcA did not repress dnaK transcription in vitro, even at concentrations as high as 280 nM (Fig. 4). In contrast, the active fraction of truncated rHrcA decreased dnaK transcription in a concentration-dependent manner, up to a maximum of 10-fold at 100 nM protein (Fig. 4). Taken together, these in vitro binding and transcription studies indicate that the C-terminal tail of chlamydial HrcA is an inhibitory region that reduces the ability of HrcA to bind to its operator and to repress transcription.
Fig. 4.
Only truncated and not full-length rHrcA repressed transcription of the dnaK promoter in vitro. (A) In vitro transcription of the C. trachomatis dnaK promoter with C. trachomatis RNA polymerase in the presence of rHrcA over a range of concentrations from 35 to 280 nM for full-length rHrcA and 12.5 to 100 nM for truncated rHrcA. These experiments were performed with active fractions of rHrcA that had been purified with DNA affinity chromatography. (B) Graphs showing quantification of the transcription results as measured by phosphorimager analysis. Note the different scales for the x and y axes of the two graphs. Reactions were performed in triplicate, and the amount of repression is reported as an average fold decrease relative to transcription in the absence of rHrcA. Error bars represent standard deviations.
At first glance, this inability of full-length rHrcA to repress the dnaK promoter stands in contrast to our previous studies that showed that C. trachomatis HrcA is a transcriptional repressor (51–53). However, those studies were performed with a chlamydial rHrcA preparation that also contained truncated protein lacking the C-terminal tail (Fig. 2A, lane 1) as an artifact of premature translational termination in E. coli, as described above. In light of our new findings showing that full-length rHrcA has minimal repressor activity, it is likely that the repressor activity that we measured in our previous studies was due to the presence of truncated HrcA in our rHrcA preparation.
Full-length HrcA causes less repression in vivo than truncated HrcA.
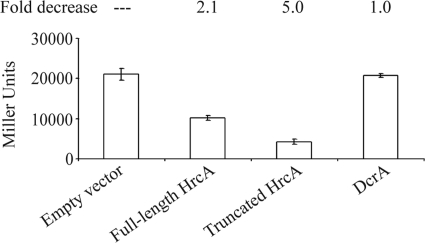
We used a β-galactosidase reporter assay to test if the presence of the C-terminal tail also decreases HrcA-mediated repression in vivo. Since no experimental genetic system exists for Chlamydia, we performed these experiments in E. coli as a heterologous host that lacks its own HrcA homolog (5). Overexpression of truncated HrcA caused a 5-fold decrease in β-galactosidase activity from the C. trachomatis dnaK promoter, while full-length HrcA decreased reporter activity by 2-fold (Fig. 5). This increased repression with truncated HrcA could not be accounted for by differences in protein overexpression alone because truncated HrcA was expressed at 2-fold-lower levels than full-length HrcA by Western blot analysis (data not shown). These effects on reporter activity were specific, since there was no repression with C. trachomatis DcrA, which is a metal ion-responsive regulator (31, 54). These results provide support for our in vitro findings by verifying that the presence of the Chlamydia-specific C-terminal tail decreases the ability of HrcA to repress transcription in vivo. These experiments, however, raise the question of how full-length chlamydial HrcA is able to function as a repressor in vivo but not in vitro.
Fig. 5.
Full-length HrcA produced less repression in vivo compared to truncated HrcA. β-Galactosidase assays in which E. coli was cotransformed with a reporter plasmid containing a C. trachomatis dnaK promoter-lacZ transcriptional fusion and an expression plasmid encoding a C. trachomatis transcription factor (full-length HrcA, truncated HrcA, or DcrA as a negative control). The results are reported in Miller units, together with the average fold decrease relative to control levels obtained with an empty expression vector. The assays were performed in triplicate, and error bars represent standard deviations.
Full-length HrcA is the form of HrcA present in Chlamydia.
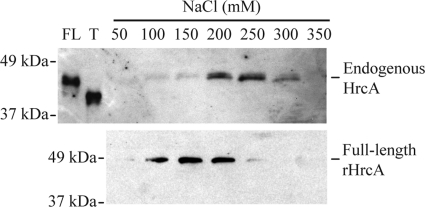
To determine if endogenous HrcA in C. trachomatis contains the inhibitory C-terminal tail, we attempted to purify HrcA from a lysate of chlamydial reticulate bodies. However, we did not detect HrcA in this lysate by Western blot analysis with polyclonal antibodies against HrcA, which is likely due to its low abundance (53). As an alternative, we used DNA affinity chromatography to isolate HrcA from the lysate on the basis of specific binding to the CIRCE operator and then detected the protein by Western blot analysis. Using this approach, we recovered a single form of endogenous HrcA that comigrated with untagged full-length rHrcA by SDS-PAGE (Fig. 6). We did not detect any endogenous protein corresponding to truncated HrcA, even though this form would be predicted to bind the CIRCE DNA beads with higher affinity. Intriguingly, endogenous HrcA eluted from the DNA affinity beads at 200 to 300 mM NaCl, in contrast to full-length rHrcA, which eluted at 100 to 200 mM NaCl (Fig. 6). These results indicate that endogenous HrcA in C. trachomatis is the full-length protein, although we detected a difference in its binding affinity for the CIRCE operator compared to recombinant full-length protein.
Fig. 6.
Endogenous HrcA from C. trachomatis is the full-length protein. Western blot showing endogenous HrcA purified from a lysate of chlamydial reticulate bodies (top panel, probed with anti-HrcA antibodies) or full-length His6-tagged rHrcA overexpressed and purified from E. coli (bottom panel, probed with anti-His antibodies). Each form of HrcA was incubated with CIRCE DNA beads at 50 mM NaCl and then washed in a stepwise manner with increasing concentrations of NaCl from 50 to 350 mM as marked. Wash fractions were then resolved by 12% SDS-PAGE. Samples of untagged full-length (FL) and truncated (T) rHrcA are shown as size markers.
GroEL enhances binding of rHrcA to the CIRCE operator.
One explanation for the apparent higher binding activity of endogenous full-length HrcA is that its repressor activity is modulated by an additional factor. We focused our efforts on GroEL since this heat shock protein has been shown to copurify as a complex with endogenous HrcA from a C. trachomatis lysate (53). Furthermore, GroEL has been shown to promote the binding of HrcA to CIRCE in vitro for a number of bacterial species (23, 27, 37, 47). This interaction with GroEL has been proposed to increase the binding affinity of HrcA for CIRCE by inducing an active conformation of the repressor (27).
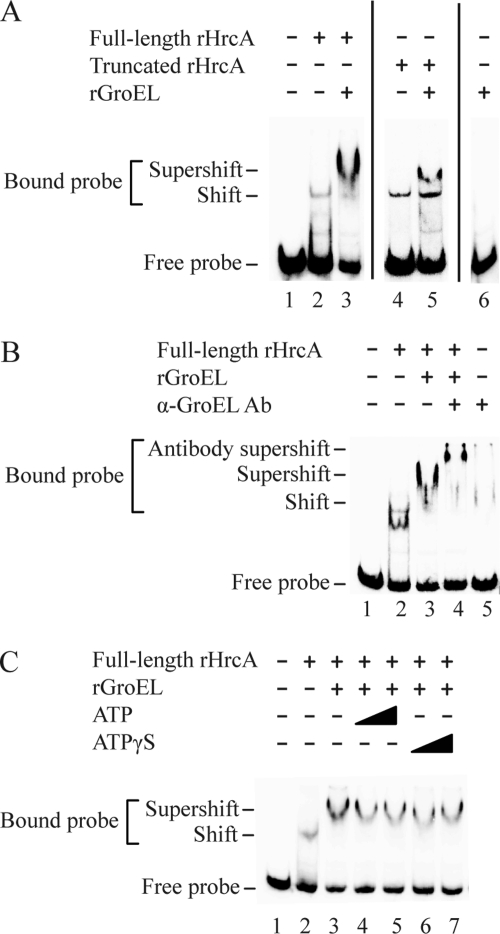
We performed EMSA experiments to test if GroEL can increase the binding of full-length chlamydial HrcA to a DNA probe containing the CIRCE operator. For these binding experiments, and subsequent transcription studies, we reasoned that this putative effect of GroEL would be best measured on rHrcA that had not been enriched for high binding affinity with the additional DNA affinity chromatography step. When we preincubated 120 nM C. trachomatis rGroEL with 350 nM full-length rHrcA, the proportion of bound probe increased by 6.4-fold to 58%, compared to 9% for rHrcA alone, and produced an additional slower-migrating supershifted band (Fig. 7A, compare lanes 2 and 3). This extra band represents a higher-order complex that also contains rGroEL, since the addition of anti-GroEL antibodies caused a further retardation of this band on the gel (Fig. 7B, lane 4, marked as antibody supershift). The antibody supershift was not seen with anti-myc antibodies (data not shown). These complexes were specific for the dnaK promoter and its CIRCE operator, as no gel shifts were observed with a DNA probe containing the C. trachomatis troA promoter, which is a non-heat shock promoter (data not shown). rGroEL by itself did not bind the CIRCE DNA probe (Fig. 7A, lane 6), which is consistent with published reports (53). These results demonstrate that GroEL interacts with the HrcA-CIRCE complex and enhances the binding of full-length HrcA to its CIRCE operator.
Fig. 7.
GroEL enhances rHrcA binding to the CIRCE operator. Representative EMSA in which rHrcA was preincubated with rGroEL prior to binding to a DNA probe containing the C. trachomatis dnaK promoter and its CIRCE operator. The rHrcA preparations used for this assay were purified by metal affinity chromatography without further purification by DNA affinity chromatography. DNA probes shifted by rHrcA alone or supershifted by rHrcA and rGroEL are marked. (A) rGroEL (120 nM) had a greater effect on 350 nM full-length rHrcA (lane 3) than on 2 nM truncated rHrcA (lane 5), which are concentrations of full-length and truncated rHrcA that produced similar baseline proportions of shifted probe in the absence of rGroEL (lanes 2 and 4). (B) Specificity control showing that anti-GroEL antibody further retarded the mobility of the supershifted band produced by incubation of 120 nM rGroEL with 350 nM full-length rHrcA. (C) Addition of ATP and ATPγS at 50 or 100 μM had no effect on the enhancement caused by 120 nM rGroEL when it was preincubated with 350 nM full-length rHrcA.
We also found that GroEL enhanced the binding of truncated chlamydial HrcA lacking the C-terminal tail, although the effect was more modest. Since truncated rHrcA binds to CIRCE with a higher affinity than full-length rHrcA (Fig. 3), we titrated the concentration of truncated rHrcA in this experiment in order to start with a level of baseline binding (6%) that was similar to that of full-length rHrcA (Fig. 7A, lane 2). rGroEL (120 nM) caused the proportion of probe bound by truncated rHrcA to increase by 3-fold, from 6% to 18%, while also producing a supershifted band (Fig. 7A, compare lanes 4 and 5). These results indicate that GroEL also interacts with truncated chlamydial HrcA in vitro and enhances its binding to CIRCE. The greater effect of GroEL on full-length HrcA than on truncated HrcA (Fig. 7A, compare lanes 3 and 5) suggests that at least some of the enhancement by GroEL is mediated through the Chlamydia-specific C-terminal tail of HrcA. These results are consistent with a role for GroEL in counteracting the inhibitory effect of the C-terminal tail on HrcA repressor activity.
We tested if GroEL enhances the binding of full-length HrcA to the CIRCE operator via its conventional role as a molecular chaperone that refolds proteins in an ATP-dependent process (6). To prevent GroEL chaperone activity, we performed EMSA experiments in the presence of ATPγS, a nonhydrolyzable form of ATP. Addition of 50 or 100 μM ATPγS to reaction mixtures containing 120 nM rGroEL and 350 nM full-length rHrcA did not decrease the amount of bound probe (Fig. 7C, lanes 6 and 7). There was also no effect if we added 50 or 100 μM ATP (Fig. 7C, lanes 4 and 5) instead of ATPγS. These results indicate that GroEL enhances the binding of full-length HrcA to its operator in an ATP-independent manner.
GroEL enhances transcriptional repression by full-length but not truncated rHrcA in vitro.
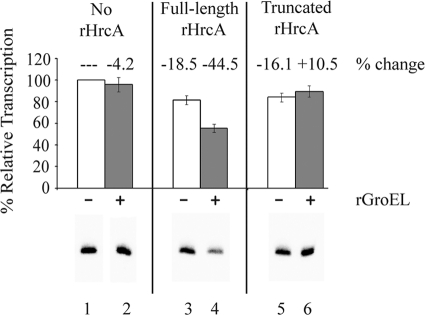
We tested if the stimulatory effect of GroEL on HrcA-CIRCE binding also causes increased HrcA-mediated repression of the C. trachomatis dnaK promoter in vitro. Since our C. trachomatis RNA polymerase preparation is partially purified and contains unidentified chlamydial proteins (48), we performed these transcription assays with commercially purified E. coli RNA polymerase to ensure that we were measuring the effect of GroEL, and not those of other proteins, on HrcA. The addition of 170 nM rGroEL more than doubled the level of repression by 350 nM full-length rHrcA from 18.5% to 44.5% (Fig. 8, compare lanes 3 and 4). To compare the effect of GroEL on truncated HrcA, which is a more effective repressor by itself (Fig. 4), we titrated the concentration of truncated rHrcA in this experiment so that we started with a level of repression (16.1%) similar to that of full-length rHrcA (Fig. 8, lane 3). In contrast to the results with full-length rHrcA, 170 nM rGroEL did not enhance this repression by truncated rHrcA (Fig. 8, compare lanes 5 and 6). These results demonstrate that GroEL augments HrcA-mediated repression, which agrees with previous reports (53). However, these EMSA and transcription studies indicate that GroEL has a greater effect when the C-terminal tail of HrcA is present. These results are consistent with a model in which the auto-inhibitory effect of the C-terminal tail is counteracted by GroEL, thereby facilitating the ability of full-length chlamydial HrcA to function as a repressor.
Fig. 8.
GroEL enhanced transcriptional repression by full-length but not truncated rHrcA in vitro. In vitro transcription of the C. trachomatis dnaK promoter with E. coli RNA polymerase in the presence of 350 nM full-length rHrcA or 100 nM truncated rHrcA, which produced similar baseline repression in the absence of rGroEL (lanes 3 and 5). rGroEL (170 nM) was added to reactions in lanes 2, 4, and 6. These rHrcA preparations were purified by metal affinity chromatography without further purification by DNA affinity chromatography. The graph shows quantification of the transcription results as measured by phosphorimager analysis. Transcription levels were normalized to the amount of transcription produced by RNA polymerase alone, which was defined as 100%, and the percentage change in transcription due to the addition of rHrcA and/or rGroEL is reported. Reactions were performed in triplicate, and error bars represent standard deviations.
DISCUSSION
Our study demonstrates that the C-terminal tail of C. trachomatis HrcA has an inhibitory effect on the repressor function of this stress response regulator. This additional region is a distinguishing feature of chlamydial HrcA that is not present in HrcA from other bacteria (Fig. 1). It is well conserved in orthologs from six Chlamydia spp. and also present in HrcA from a related intracellular bacterium, Protochlamydia amoebophila, which suggests that it may have a conserved evolutionary function (4, 16, 32, 33, 46, 49). We found that the C-terminal tail decreased binding of chlamydial HrcA to its cognate CIRCE operator in vitro. The presence of this Chlamydia-specific region also reduced the ability of chlamydial rHrcA to repress transcription in vitro and in vivo.
The inhibitory effect of the C-terminal tail was not recognized in previous studies that utilized recombinant chlamydial HrcA (51–53). The likely reason is that expression of chlamydial rHrcA in E. coli artifactually produces a truncated product lacking the C-terminal tail (Fig. 2A, lane 1) that binds CIRCE and represses heat shock promoters with higher efficiency than the full-length protein. This truncated recombinant protein appears to be the result of premature termination of translation in E. coli (14), since we were able to prevent it from being produced by replacing tandem AGA arginine codons in the hrcA sequence with silent mutations. By performing experiments with 100% full-length C. trachomatis rHrcA, the current study demonstrates that the C-terminal tail has a negative effect on DNA binding and repression. We detected full-length HrcA only in chlamydiae and found no evidence that C. trachomatis overcomes the inhibitory effect of the C-terminal tail by generating a truncated form of HrcA.
There is precedent for the DNA-binding activity of a protein to be altered by an inhibitory C-terminal tail. E. coli single-stranded DNA-binding protein (SSB) and T7 bacteriophage gene 2.5 protein (gp2.5) each contain a C-terminal tail that inhibits binding to single-stranded DNA (ssDNA) (19, 22). Inhibitory domains are also common on eukaryotic transcription factors, such as p63, FoxM1, and Pitx2 (1, 29, 43). In some cases, this auto-inhibition can be regulated, as illustrated by Pit-1, a factor that enhances the DNA-binding activity of Pitx2 by counteracting the inhibitory effect of its C-terminal tail (1).
The inhibitory C-terminal tail provides a second example of how HrcA-CIRCE binding has been weakened in Chlamydia. The CIRCE sequence is normally well conserved in bacteria (42), but the CIRCE operator of the C. trachomatis groESL operon is unusual in having 5/18 mismatches with the consensus CIRCE sequence (52). We have previously demonstrated that chlamydial HrcA showed reduced binding to this diverged operator compared to binding to the conserved operator of the C. trachomatis dnaK operon (52). Thus, it appears that Chlamydia has utilized genetic changes in both the repressor and the operator to reduce HrcA-CIRCE interactions.
We propose that the inhibitory effect of the C-terminal tail on the DNA binding and repressor functions of HrcA is reversible and can be regulated by a cofactor such as the heat shock protein GroEL. We showed that rGroEL enhanced the ability of full-length rHrcA to bind the CIRCE operator and to repress transcription in vitro. rGroEL had a greater effect in stimulating CIRCE binding and transcriptional repression by full-length rHrcA than by truncated rHrcA, which suggests that GroEL interactions may specifically counteract the inhibitory effect of the C-terminal tail. This proposed role in modulating HrcA repressor activity does not appear to involve the canonical function of GroEL as a molecular chaperone, since the effect was ATP independent.
This proposed role for GroEL as a regulator of HrcA may explain some of our in vivo findings. For example, our ability to detect repression by full-length rHrcA in vivo, but not in vitro, may be due to the presence of endogenous E. coli GroEL in our in vivo reporter system (Fig. 5). In addition, the higher CIRCE binding affinity of HrcA isolated from chlamydiae compared to that of rHrcA may be due to the association of GroEL with endogenous HrcA, which has been previously shown (53).
A role for GroEL in counteracting the inhibitory effect of the C-terminal tail of HrcA is consistent with the GroEL titration model of heat shock regulation in bacteria (28). According to this model, GroEL promotes the binding of HrcA to its CIRCE operator when bacteria are in nonstress conditions. During cellular stress, however, GroEL is titrated away to correct protein misfolding, which reduces HrcA-CIRCE binding and causes derepression of the heat shock genes. While there is evidence that GroEL promotes HrcA-CIRCE binding (23, 27, 37, 47), the mechanism has not been determined experimentally. From in vitro studies, GroEL has been proposed to increase HrcA binding to its operator by correcting HrcA misfolding via its role as an ATP-dependent molecular chaperone (23, 27). Structural data, however, suggest that HrcA is intrinsically in an inactive conformation because residues in its C terminus interact with and inhibit the N-terminal CIRCE-binding domain (21). These findings have led to an alternative model in which GroEL enhances HrcA repressor function by preventing this auto-inhibition (21). Our studies provide the first experimental support for this auto-inhibitory model by showing that GroEL can counteract a C-terminal auto-inhibitory region of chlamydial HrcA. Although the C-terminal tail described in this report is unique to chlamydial HrcA, it may represent an exaggerated form of HrcA auto-inhibition that provides insight into the mechanism by which GroEL regulates HrcA-mediated repression of heat shock genes in other bacteria. However, we have not ruled out the possibility that the C-terminal tail of chlamydial HrcA may have an indirect effect on operator binding by disrupting HrcA dimerization (50).
In summary, the C-terminal tail of chlamydial HrcA represents a novel feature that may have implications for the heat shock response in Chlamydia. We propose that its inhibitory effect on HrcA-mediated repression leads to a greater dependence on GroEL as a corepressor during nonstress conditions. Consequently, when GroEL is titrated away in response to cellular stressors, such as elevated temperature, nutrient deprivation, or oxidative stress, there is increased derepression and higher expression of heat shock proteins. Thus, we propose that the Chlamydia-specific C-terminal tail of HrcA is an adaptation by this intracellular pathogen to alter heat shock protein expression in response to cellular stress.
ACKNOWLEDGMENTS
We thank Christopher Rosario, Johnny Akers, Eric Cheng, Kirsten Johnson, and Jennifer Lee for critical reading of the manuscript.
This work was supported by a grant from the NIH (AI 44198). M.T. was supported by an NIH Independent Scientist Award (AI 057563).
Footnotes
Published ahead of print on 30 September 2011.
REFERENCES
- 1. Amendt B. A., Sutherland L. B., Russo A. F. 1999. Multifunctional role of the Pitx2 homeodomain protein C-terminal tail. Mol. Cell. Biol. 19:7001–7010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arsene F., Tomoyasu T., Bukau B. 2000. The heat shock response of Escherichia coli. Int. J. Food Microbiol. 55:3–9 [DOI] [PubMed] [Google Scholar]
- 3. Ault K. A., et al. 1998. Antibodies to the chlamydial 60 kilodalton heat shock protein in women with tubal factor infertility. Infect. Dis. Obstet. Gynecol. 6:163–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Azuma Y., et al. 2006. Genome sequence of the cat pathogen, Chlamydophila felis. DNA Res. 13:15–23 [DOI] [PubMed] [Google Scholar]
- 5. Blattner F. R., et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1474 [DOI] [PubMed] [Google Scholar]
- 6. Bochkareva E. S., Lissin N. M., Girshovich A. S. 1988. Transient association of newly synthesized unfolded proteins with the heat-shock GroEL protein. Nature 336:254–257 [DOI] [PubMed] [Google Scholar]
- 7. Brunham R. C., Peeling R., Maclean I. K. M. L., Paraskevas M. 1992. Chlamydia trachomatis-associated ectopic pregnancy: serologic and histologic correlates. J. Infect. Dis. 165:1076–1081 [DOI] [PubMed] [Google Scholar]
- 8. Bulut Y., et al. 2009. Chlamydial heat shock protein 60 induces acute pulmonary inflammation in mice via the Toll-like receptor 4- and MyD88-dependent pathway. Infect. Immun. 77:2683–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burton M. J., Mabey D. C. 2009. The global burden of trachoma: a review. PLoS Negl. Trop. Dis. 3:e460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. CDC 2011. Summary of notifiable diseases—United States, 2009. MMWR Morb. Mortal. Wkly. Rep. 58:1–100 [PubMed] [Google Scholar]
- 11. Equils O., et al. 2006. Chlamydia heat shock protein 60 induces trophoblast apoptosis through TLR4. J. Immunol. 177:1257–1263 [DOI] [PubMed] [Google Scholar]
- 12. Gao W., Tyagi S., Kramer F. R., Goldman E. 1997. Messenger RNA release from ribosomes during 5′-translational blockage by consecutive low-usage arginine but not leucine codons in Escherichia coli. Mol. Microbiol. 25:707–716 [DOI] [PubMed] [Google Scholar]
- 13. Gragerov A., et al. 1992. Cooperation of GroEL/GroES and DnaK/DnaJ heat shock proteins in preventing protein misfolding in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 89:10341–10344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grantham R., Gautier C., Gouy M., Mercier R., Pave A. 1980. Codon catalog usage and the genome hypothesis. Nucleic Acids Res. 8:r49–r62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gutman G. A., Hatfield G. W. 1989. Nonrandom utilization of codon pairs in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 86:3699–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Horn M., et al. 2004. Illuminating the evolutionary history of Chlamydiae. Science 304:728–730 [DOI] [PubMed] [Google Scholar]
- 17. Jobling M. G., Holmes R. K. 1990. Construction of vectors with the p15a replicon, kanamycin resistance, inducible lacZ alpha and pUC18 or pUC19 multiple cloning sites. Nucleic Acids Res. 18:5315–5316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kauppinen M., Saikku P. 1995. Pneumonia due to Chlamydia pneumoniae: prevalence, clinical features, diagnosis, and treatment. Clin. Infect. Dis. 21:S244–252 [DOI] [PubMed] [Google Scholar]
- 19. Kozlov A. G., Cox M. M., Lohman T. M. 2010. Regulation of single-stranded DNA binding by the C termini of Escherichia coli single-stranded DNA-binding (SSB) protein. J. Biol. Chem. 285:17246–17252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. LaVerda D., Kalayoglu M. V., Byrne G. I. 1999. Chlamydial heat shock proteins and disease pathology: new paradigms for old problems? Infect. Dis. Obstet. Gynecol. 7:64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu J., et al. 2005. Crystal structure of a heat-inducible transcriptional repressor HrcA from Thermotoga maritima: structural insight into DNA binding and dimerization. J. Mol. Biol. 350:987–996 [DOI] [PubMed] [Google Scholar]
- 22. Marintcheva B., Marintchev A., Wagner G., Richardson C. C. 2008. Acidic C-terminal tail of the ssDNA-binding protein of bacteriophage T7 and ssDNA compete for the same binding surface. Proc. Natl. Acad. Sci. U. S. A. 105:1855–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martirani L., Raniello R., Naclerio G., Ricca E., De Felice M. 2001. Identification of the DNA-binding protein, HrcA, of Streptococcus thermophilus. FEMS Microbiol. Lett. 198:177–182 [DOI] [PubMed] [Google Scholar]
- 24. Mathew A., Morimoto R. I. 1998. Role of the heat-shock response in the life and death of proteins. Ann. N. Y. Acad. Sci. 851:99–111 [DOI] [PubMed] [Google Scholar]
- 25. Miller J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 26. Minder A. C., Fischer H. M., Hennecke H., Narberhaus F. 2000. Role of HrcA and CIRCE in the heat shock regulatory network of Bradyrhizobium japonicum. J. Bacteriol. 182:14–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mogk A., et al. 1997. The GroE chaperonin machine is a major modulator of the CIRCE heat shock regulon of Bacillus subtilis. EMBO J. 16:4579–4590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Narberhaus F. 1999. Negative regulation of bacterial heat shock genes. Mol. Microbiol. 31:1–8 [DOI] [PubMed] [Google Scholar]
- 29. Park H. J., et al. 2008. An N-terminal inhibitory domain modulates activity of FoxM1 during cell cycle. Oncogene 27:1696–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peeling R. W., et al. 1998. Antibody response to the 60-kDa chlamydial heat-shock protein is associated with scarring trachoma. J. Infect. Dis. 177:256–259 [DOI] [PubMed] [Google Scholar]
- 31. Rau A., Wyllie S., Whittimore J., Raulston J. E. 2005. Identification of Chlamydia trachomatis genomic sequences recognized by chlamydial divalent cation-dependent regulator A (DcrA). J. Bacteriol. 187:443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Read T. D., et al. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Read T. D., et al. 2003. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res. 31:2134–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Roberts R. C., et al. 1996. Identification of a Caulobacter crescentus operon encoding hrcA, involved in negatively regulating heat-inducible transcription, and the chaperone gene grpE. J. Bacteriol. 178:1829–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Robinson M., et al. 1984. Codon usage can affect efficiency of translation of genes in Escherichia coli. Nucleic Acids Res. 12:6663–6671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Romero H., Zavala A., Musto H. 2000. Codon usage in Chlamydia trachomatis is the result of strand-specific mutational biases and a complex pattern of selective forces. Nucleic Acids Res. 28:2084–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Roncarati D., Danielli A., Spohn G., Delany I., Scarlato V. 2007. Transcriptional regulation of stress response and motility functions in Helicobacter pylori is mediated by HspR and HrcA. J. Bacteriol. 189:7234–7243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sanchez-Campillo M., et al. 1999. Identification of immunoreactive proteins of Chlamydia trachomatis by Western blot analysis of a two-dimensional electrophoresis map with patient sera. Electrophoresis 20:2269–2279 [DOI] [PubMed] [Google Scholar]
- 39. Sasu S., LaVerda D., Qureshi N., Golenbock D. T., Beasley D. 2001. Chlamydia pneumoniae and chlamydial heat shock protein 60 stimulate proliferation of human vascular smooth muscle cells via toll-like receptor 4 and p44/p42 mitogen-activated protein kinase activation. Circ. Res. 89:244–250 [DOI] [PubMed] [Google Scholar]
- 40. Schachter J. 1999. Infection and disease epidemiology, p. 139–169 In Stephens R. S. (ed.), Chlamydia: intracellular biology, pathogenesis, and immunity. American Society for Microbiology, Washington, DC [Google Scholar]
- 41. Schulz A., Schumann W. 1996. hrcA, the first gene of the Bacillus subtilis dnaK operon encodes a negative regulator of class I heat shock genes. J. Bacteriol. 178:1088–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Segal R., Ron E. Z. 1996. Regulation and organization of the groE and dnaK operons in Eubacteria. FEMS Microbiol. Lett. 138:1–10 [DOI] [PubMed] [Google Scholar]
- 43. Serber Z., et al. 2002. A C-terminal inhibitory domain controls the activity of p63 by an intramolecular mechanism. Mol. Cell. Biol. 22:8601–8611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Simons R. W., Houman F., Kleckner N. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85–96 [DOI] [PubMed] [Google Scholar]
- 45. Spohn G., et al. 2004. Dual control of Helicobacter pylori heat shock gene transcription by HspR and HrcA. J. Bacteriol. 186:2956–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stephens R. S., et al. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754–759 [DOI] [PubMed] [Google Scholar]
- 47. Susin M. F., Perez H. R., Baldini R. L., Gomes S. L. 2004. Functional and structural analysis of HrcA repressor protein from Caulobacter crescentus. J. Bacteriol. 186:6759–6767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tan M., Engel J. N. 1996. Identification of sequences necessary for transcription in vitro from the Chlamydia trachomatis rRNA P1 promoter. J. Bacteriol. 178:6975–6982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Thomson N. R., et al. 2005. The Chlamydophila abortus genome sequence reveals an array of variable proteins that contribute to interspecies variation. Genome Res. 15:629–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wiegert T., Schumann W. 2003. Analysis of a DNA-binding motif of the Bacillus subtilis HrcA repressor protein. FEMS Microbiol. Lett. 223:101–106 [DOI] [PubMed] [Google Scholar]
- 51. Wilson A. C., Tan M. 2002. Functional analysis of the heat shock regulator HrcA of Chlamydia trachomatis. J. Bacteriol. 184:6566–6571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wilson A. C., Tan M. 2004. Stress response gene regulation in Chlamydia is dependent on HrcA-CIRCE interactions. J. Bacteriol. 186:3384–3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wilson A. C., Wu C. C., Yates J. R., III, Tan M. 2005. Chlamydial GroEL autoregulates its own expression through direct interactions with the HrcA repressor protein. J. Bacteriol. 187:7535–7542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wyllie S., Raulston J. E. 2001. Identifying regulators of transcription in an obligate intracellular pathogen: a metal-dependent repressor in Chlamydia trachomatis. Mol. Microbiol. 40:1027–1036 [DOI] [PubMed] [Google Scholar]
- 55. Yuan G., Wong S.-L. 1995. Regulation of groE expression in Bacillus subtilis: the involvement of the σA-like promoter and the roles of the inverted repeat sequence (CIRCE). J. Bacteriol. 177:5427–5433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang X., Bremer H. 1995. Control of the Escherichia coli rrnB P1 promoter strength by ppGpp. J. Biol. Chem. 270:11181–11189 [DOI] [PubMed] [Google Scholar]