Abstract
Homologous recombination between repeated DNA elements in the genomes of Mycoplasma species has been hypothesized to be a crucial causal factor in sequence variation of antigenic proteins at the bacterial surface. To investigate this notion, studies were initiated to identify and characterize the proteins that form part of the homologous DNA recombination machinery in Mycoplasma pneumoniae as well as Mycoplasma genitalium. Among the most likely participants of this machinery are homologs of the Holliday junction migration motor protein RuvB. In both M. pneumoniae and M. genitalium, genes have been identified that have the capacity to encode RuvB homologs (MPN536 and MG359, respectively). Here, the characteristics of the MPN536- and MG359-encoded proteins (the RuvB proteins from M. pneumoniae strain FH [RuvBFH] and M. genitalium [RuvBMge], respectively) are described. Both RuvBFH and RuvBMge were found to have ATPase activity and to bind DNA. In addition, both proteins displayed divalent cation- and ATP-dependent DNA helicase activity on partially double-stranded DNA substrates. The helicase activity of RuvBMge, however, was significantly lower than that of RuvBFH. Interestingly, we found RuvBFH to be expressed exclusively by subtype 2 strains of M. pneumoniae. In strains belonging to the other major subtype (subtype 1), a version of the protein is expressed (the RuvB protein from M. pneumoniae strain M129 [RuvBM129]) that differs from RuvBFH in a single amino acid residue (at position 140). In contrast to RuvBFH, RuvBM129 displayed only marginal levels of DNA-unwinding activity. These results demonstrate that M. pneumoniae strains (as well as closely related Mycoplasma spp.) can differ significantly in the function of components of their DNA recombination and repair machinery.
INTRODUCTION
A significant portion of the genomes of Mycoplasma pneumoniae and Mycoplasma genitalium consists of repeated DNA elements. This is remarkable as the genomes of these closely related human pathogens are relatively small, with lengths of 816 kb and 580 kb, respectively (6, 8). In M. pneumoniae, the repeated DNA elements are termed RepMP elements (29, 37, 44), whereas in M. genitalium, they are referred to as MgPa repeats (MgPars) (6, 26, 27). Although the RepMP elements and MgPa repeats do not have significant sequence homology, they do share two important features: (i) the different variants of these elements are similar in sequence but not identical, and (ii) one or more of these variants form part of open reading frames (ORFs) that code for antigenic surface proteins, such as the P1 protein in M. pneumoniae and the MgPa protein in M. genitalium. Because both P1 and MgPa can display sequence variation in the regions encoded by the RepMP or MgPar elements, it has been hypothesized that this variation is caused by homologous DNA recombination between the different variants of the RepMP or MgPar elements (10–11, 15, 17, 35, 36). Obviously, these recombination processes may supply a plethora of sequence variation to the P1 and MgPa genes, which can subsequently lead to amino acid sequence variation of the encoded antigenic proteins. Homologous DNA recombination between the repeated DNA elements in both Mycoplasma species may thus contribute significantly to evasion of these pathogens from the host's immune system.
It is probable that the enzymatic machinery that governs recombination between repeated DNA elements in M. pneumoniae and M. genitalium largely overlaps with the machinery that is involved in homologous DNA recombination and DNA repair in these bacteria. Sequence analysis of the genomes of both M. pneumoniae and M. genitalium has revealed the presence of a distinct and limited set of ORFs that may encode the core proteins involved in DNA recombination pathways (2, 6, 8). The activities of some of these putative core proteins have already been investigated. These proteins include RuvA and a single-stranded DNA (ssDNA)-binding protein (SSB) from M. pneumoniae (9, 32) and the RecA and RecU proteins from both M. pneumoniae and M. genitalium (31, 33, 34). While the first three proteins exhibited activities that are similar to those of their counterparts from other bacteria, the RecU proteins from both Mycoplasma species displayed unusual and unique characteristics. First, while RecU from M. genitalium (RecUMge) was found to bind and cleave Holliday junction (HJ) substrates in a specific fashion, the RecU protein from M. pneumoniae (RecUMpn) did not possess obvious DNA-binding or -cleavage activities. The inactivity of RecUMpn was found to be caused by the presence of a glutamic acid residue at position 67 of the protein (33), which is not conserved in RecUMge or any other known RecU-like sequence. In addition, RecUMpn can be expressed only by a subgroup of M. pneumoniae strains (the so-called subtype 2 strains) and not by other strains (subtype 1 strains) as the latter strains contain a premature TAA translation termination codon within the RecU gene (33). The apparent lack of a functional RecU protein in M. pneumoniae was hypothesized to be a possible cause of the relatively low level of homologous DNA recombination events in this bacterial species (33).
To further delineate the composition and characteristics of the DNA recombination machinery of M. pneumoniae and M. genitalium, the current study focused on the proteins encoded by ORFs MPN536 and MG359 from M. pneumoniae and M. genitalium, respectively. These ORFs were previously reported to show sequence similarity to the RuvB HJ branch migration motor proteins from other bacterial species (6, 8). Here, we show that the MPN536-derived protein from M. pneumoniae strain FH (RuvBFH) is a potent DNA helicase, whose activity is dependent on ATP and divalent cations (Mg2+ or Mn2+). Interestingly, the characteristics of RuvBFH differ significantly from those of the MPN536-encoded protein from M. pneumoniae strain M129 (RuvBM129) and from the MG359-encoded protein from M. genitalium (RuvBMge).
MATERIALS AND METHODS
Strains.
All M. pneumoniae strains used in this study, including reference strains FH (ATCC 15531), M129 (ATCC 29342), and PI 1428 (ATCC 29085), as well as M. genitalium strain G37 (ATCC 33530), were cultured in Mycoplasma medium, as described previously (18).
Cloning of the M. pneumoniae MPN536 gene and M. genitalium MG359 gene.
Bacterial DNA was purified from cultures of M. pneumoniae and M. genitalium following standard procedures (32). The MPN536 ORF of M. pneumoniae strain FH was amplified by PCR. Although this ORF has recently been designated MPNE_0635 in the complete genome sequence of strain FH (GenBank accession number CP002077.1), we have maintained the name MPN536, which is derived from the published annotation of the genome sequence of strain M129 (3, 8). The PCR was performed using the following primers: RuvB_FW (5′-GGTCGTCATATGAAGTTACAAATAAAACCG-3′, which overlaps with the translation initiation codon [underlined] of MPN536) and primer RuvB_RV (5′-GCAGCC-GGATCCTTAGCAGCTAGTTAAATAATTAC-3′, which overlaps with the antisense sequence of the translation termination codon [underlined] of the gene). The resulting 0.9-kbp (kb) PCR fragment was digested with NdeI and BamHI (the recognition sites for these enzymes are indicated in italics in the sequences of primers RuvB_FW and RuvB_RV, respectively) and cloned into NdeI- and BamHI-digested vector pET-11c, resulting in plasmid pET-11c-RuvBFH. In this plasmid, the MPN536 ORF is cloned such as to express the M. pneumoniae FH MPN536-encoded protein (RuvBFH) in its native form in Escherichia coli.
The cloning of the M. genitalium MG359 ORF was performed using the following primers: RuvBmgpET_fw (5′-GCTGACATATGAAATTACAAATAAAACCGCCT-3′ which includes an NdeI restriction site [in italics] and the translation initiation codon of MG359 [underlined]) and RuvBmgpEt_rv (5′-CAAGCGGATCCGCTAATAAGCTTAAAAGTTAAC-3′, which includes a BamHI site [in italics] and the antisense sequence of the translation termination codon [underlined] of the gene). The 0.9-kb PCR product was digested with NdeI and BamHI and ligated into NdeI- and BamHI-digested vector pET-11c, resulting in expression vector pET-11c-RuvBMge. From this construct, the M. genitalium MG359-encoded protein (RuvBMge) was expressed in its native form.
Generation of MBP fusion constructs.
The plasmid expressing a maltose-binding protein (MBP)-RuvBFH fusion was generated as follows. First, a PCR was done using primers ruvb_pmal_fw (5′-GGTGAATTCATGAAGTTACAAATAAAACCG-3′, which contains an EcoRI site [in italics] immediately 5′ of the start codon [underlined] of MPN536) and ruvb_pmal_rv 5′-GCACTGCAGTTAGCAGCTAGTTAAATAATTAC-3′, which contains a PstI site [in italics] and the antisense sequence of the translation termination codon [underlined] of MPN536, using plasmid pET-11c-RuvBFH as a template. The resulting fragment was digested with EcoRI and PstI and ligated into the EcoRI- and PstI-digested vector pMAL-c (New England BioLabs), yielding plasmid pMALc-RuvBFH. This plasmid was employed for the expression of RuvBFH as a carboxyl-terminal fusion to MBP (MBP-RuvBFH). By using a similar protocol, expression plasmid pMALc-RuvBM129 was constructed. In this plasmid, the TGA codon that encodes Trp140 of RuvBM129 was modified into a TGG codon by a PCR-based procedure; this change was required for expression of the full-length protein in E. coli.
Plasmid pMALc-RuvBMge was generated by a method similar to that used for pMALc-RuvBFH and pMALc-RuvBM129. In the initial PCR, plasmid pET-11c-RuvBMge was used as template DNA in combination with primers RuvBmgpMAL_fw (5′-GCTGAGAATTCATGAAATTACAAATAAAACCGCCT-3′) and RuvBmgpMAL_rv (5′-CAAGCCTGCAGGCTAATAAGCTTAAAAGTTAAC-3′).
DNA sequencing.
The integrity of all DNA constructs used in this study was checked by dideoxy sequencing, as described before (36). The complete sequences of the MPN536 ORFs from three M. pneumoniae subtype 1 strains (Mp72, Mp4817, and PI 1428) and three subtype 2 strains (Mp5181, Ofo, and R003), were determined using previously described procedures (33).
Expression and purification of RuvBFH, RuvBMge, and MBP fusion proteins.
Constructs pET-11c-RuvBFH and pET-11c-RuvBMge were introduced into E. coli BL21(DE3), and the resulting strains were grown overnight at 37°C in LB medium containing 100 μg/ml ampicillin. Protein expression was induced as described before (34).
RuvBFH was purified as follows. The frozen bacterial pellet was resuspended in 20 ml of buffer A (20 mM Tris-HCl [pH 7.4], 0.1 M NaCl, 0.1 mM EDTA, 1 mM dithiothreitol [DTT]) containing 0.5 mg/ml of lysozyme. The suspension was sonicated on ice and clarified by centrifugation for 20 min at 12,000 × g (4°C). All subsequent purification steps were performed either on ice or at 4°C. The pellet, which contained ∼60% of the expressed RuvBFH proteins, was resuspended in 20 ml of buffer B (20 mM Tris-HCl [pH 7.4], 1 M NaCl, 0.1 mM EDTA, 1 mM DTT). Then, the suspension was homogenized on ice using a Heidolph Potter homogenizer. After incubation on a roller bench for 1 h, the suspension was centrifuged for 20 min at 12,000 × g. The supernatant (20 ml) was dialyzed overnight to buffer A, after which the sample was diluted with buffer A to 45 ml. The sample was then added to 3 ml of Q Sepharose Fast Flow (GE Healthcare), which was previously equilibrated in buffer A. The suspension was incubated on a roller bench for 2.5 h, after which the unbound fraction, which contained the RuvBFH proteins, was subjected to affinity chromatography using Heparin Sepharose 6 Fast Flow (GE Healthcare) and single-stranded DNA-cellulose (Worthington Biochemical Corp., Lakewood, NJ). The RuvBFH-containing fractions were pooled, dialyzed against a solution of 20 mM Tris-HCl (pH 7.4), 0.2 M NaCl, 0.1 mM EDTA, 1 mM DTT, and 50% glycerol and stored at −20°C. RuvBMge was purified in a similar fashion as RuvBFH.
The expression and purification of MBP fusion proteins have previously been described (31, 33). The purification of RuvAMpn was reported by Ingleston and coworkers (9); this protein does not vary in sequence among M. pneumoniae subtype 1 and subtype 2 strains. The expression and purification of RuvAMge will be reported elsewhere.
SDS-PAGE.
Proteins were separated on SDS-polyacrylamide gels, essentially as described before (16). After electrophoresis, gels were stained with Coomassie brilliant blue (CBB), destained in 40% methanol–10% acetic acid, and photographed using a GelDoc XR system (Bio-Rad). Digital images were processed using Quantity One 1-D Analysis Software (Bio-Rad).
ATPase assays.
The ATPase activities of the RuvB proteins were determined by using a β-NAD reduced form (NADH)-coupled assay on a VersaMax Tunable Microplate Reader (Molecular Devices), as described previously (19, 34).
DNA substrates.
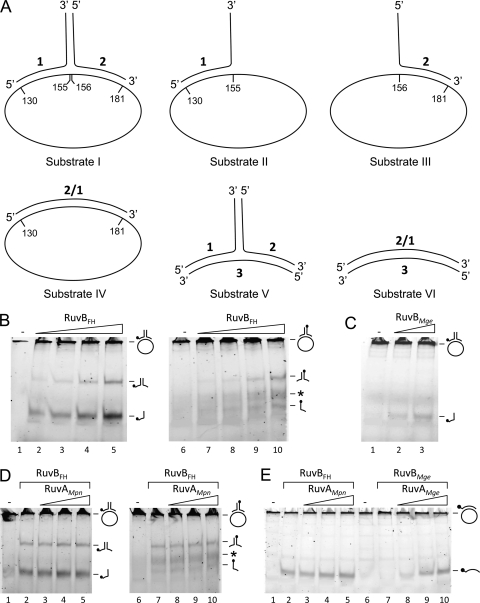
The DNA substrates that were used in the DNA helicase assays (see below) were previously described by Tsaneva and coworkers (40). The structures of these substrates are schematically depicted in Fig. 3A. They are composed of synthetic oligonucleotides alone (substrates V and VI) or of a combination of oligonucleotides and single-stranded, circular 5,386-bp ϕX174 DNA (substrates I to IV). The oligonucleotides were purchased from Eurogentec. The ϕX174 virion DNA was obtained from New England BioLabs.
Fig. 3.
DNA helicase activity of RuvBFH and RuvBMge. (A) Schematic illustration of the DNA substrates that were used in the DNA helicase assays. The substrates are composed of oligonucleotides (substrates V and VI) or a combination of oligonucleotides and single-stranded, circular 5,386-bp ϕX174 DNA (substrates I to IV). The ϕX174 DNA is pictured as an ellipse and is not drawn to scale with respect to the oligonucleotides. The sequences of the oligonucleotides (oligonucleotides 1, 2, 2/1, and 3) were described by Tsaneva et al. (40). (B) The activity of RuvBFH on substrate I. Substrate I, 6-FAM labeled at the 5′ end of either oligonucleotide 1 (lanes 1 to 5) or oligonucleotide 2 (lanes 6 to 10), was incubated with either 0 μM (−; lanes 1 and 6), 0.1 μM (lanes 2 and 7), 0.3 μM (lanes 3 and 8), 0.9 μM (lanes 4 and 9), or 2.7 μM (lanes 5 and 10) RuvBFH in the presence of Mg2+ (10 mM) and ATP (2 mM). After the reaction (5 min at 37°C), the samples were deproteinized, electrophoresed through native 12% polyacrylamide gels, and analyzed by fluorometry. The position of the substrate, which is too large to enter the gel, as well as the positions of the oligonucleotide reaction products, is indicated at the right-hand side of the gels by schematic illustrations. In these illustrations, the position of the 6-FAM label is indicated by a black circle. The asterisk points to DNA products in the gels that represent incorrectly annealed duplexes of unlabeled oligonucleotide 1, which is preferentially produced in the helicase reactions with substrate I, and labeled oligonucleotide 2 (see text). (C) The activity of RuvBMge on substrate I. Substrate I, 6-FAM labeled at the 5′ end of oligonucleotide 1, was incubated with either 0 μM (−; lane 1), 0.9 μM (lane 2), or 2.7 μM (lane 3) of RuvBMge. The other reaction parameters were similar to those described for panel A. (D) RuvAMpn does not influence the DNA helicase activity of RuvBFH. Substrate I, 6-FAM labeled at the 5′ end of either oligonucleotide 1 (lanes 1 to 5) or oligonucleotide 2 (lanes 6 to 10), was incubated without protein (lanes 1 and 6) or with 2.7 μM RuvBFH in the presence of either 0 nM (lanes 2 and 7), 19 nM (lanes 3 and 8), 56 nM (lanes 4 and 9), or 167 nM (lanes 5 and 10) RuvAMpn. The other reaction parameters were similar to those described for panel A. The asterisk points to DNA products in the gels that represent incorrectly annealed duplexes of unlabeled oligonucleotide 1 and labeled oligonucleotide 2. (E) RuvAMpn-independent helicase activity of RuvBFH and RuvAMge-dependent helicase activity of RuvBMge. The activities of RuvBFH (lanes 2 to 5) and RuvBMge (lanes 7 to 10) were tested on substrate IV in the presence of various concentrations of either RuvAMpn or RuvAMge (as indicated above the lanes) as follows: lanes 2 and 7, 0 nM; lanes 3 and 8, 56 nM; lanes 4 and 9, 167 nM; and lanes 5 and 10, 0.5 μM. The samples loaded in lanes 1 and 6 were incubated in the absence of any protein (−). The other reaction parameters were similar to those described in panel A.
DNA-binding assays.
Binding of the RuvB proteins to supercoiled pBluescript SK− DNA (Stratagene) and EcoRI-linearized pBluescript SK− was carried out in 10-μl volumes and included 20 mM Tris-acetate (OAc), pH 7.5, 1 mM DTT, 20 ng of DNA, 1 mM ATPγS, and various concentrations of RuvB proteins. The binding to single-stranded oligonucleotide 1 (see Fig. 3A), double-stranded oligonucleotide substrate VI (see Fig. 3A), and Holliday junction (HJ) substrate HJ1.1 (33), which were each 5′-end labeled on a single strand with 6-carboxyfluorescein (6-FAM), was done similarly as described above, using a DNA concentration of 12.3 nM. After incubation for 30 min at 37°C, 1 μl was added of a solution containing 40% glycerol and 0.25% bromophenol blue. Then, the reaction mixtures were electrophoresed through either 0.6% agarose gels (when plasmid substrates were used) or 5% polyacrylamide gels (when oligonucleotide substrates were used) in 0.5× TBE buffer (45 mM Tris, 45 mM boric acid, 1 mM EDTA). Following electrophoresis, the agarose gels were stained with ethidium bromide and photographed using the GelDoc XR system. The polyacrylamide gels were analyzed by fluorometry, using a Typhoon Trio 9200 Variable Mode Imager (GE Healthcare). Digital images were converted into TIFF files using the Typhoon Scanner Control, version 4.0, software (Amersham Bioscience) and processed using Quantity One 1-D Analysis Software.
DNA helicase assays.
DNA helicase assays were performed with 5′ fluorescently (6-FAM)-labeled DNA substrates (see Fig. 3A for the structures of these substrates) and were analyzed by native polyacrylamide gel electrophoresis. Standard reaction mixtures (10 μl) contained 20 mM Tris-HCl, pH 7.5, 1 mM DTT, 50 ng/μl bovine serum albumin (BSA), 10 mM MgCl2, 2 mM ATP, either 11.5 nM (for substrates I to IV) (see Fig. 3A) or 8 nM (for substrates V and VI) (see Fig. 3A) substrate DNA, and various concentrations of RuvB and RuvA proteins. Reactions were carried out for 5 min at 37°C, after which the reactions were terminated by the addition of 1 μl of Termination Mix (100 mM Tris-HCl, pH 7.4, 5% SDS, 0.2 M EDTA) and 1 μl of proteinase K (at 10 mg/ml). After deproteinization for 15 min at 37°C, 1.5 μl of loading dye (40% glycerol, 0.25% bromophenol blue) was added, and the samples were loaded onto a native 12% polyacrylamide–1× TBE mini-gel. Following electrophoresis, gels were analyzed by fluorometry, using the Typhoon Trio 9200 Variable Mode Imager. Digital images were processed as described above.
RESULTS
M. pneumoniae MPN536 and M. genitalium MG359 encode RuvB homologs.
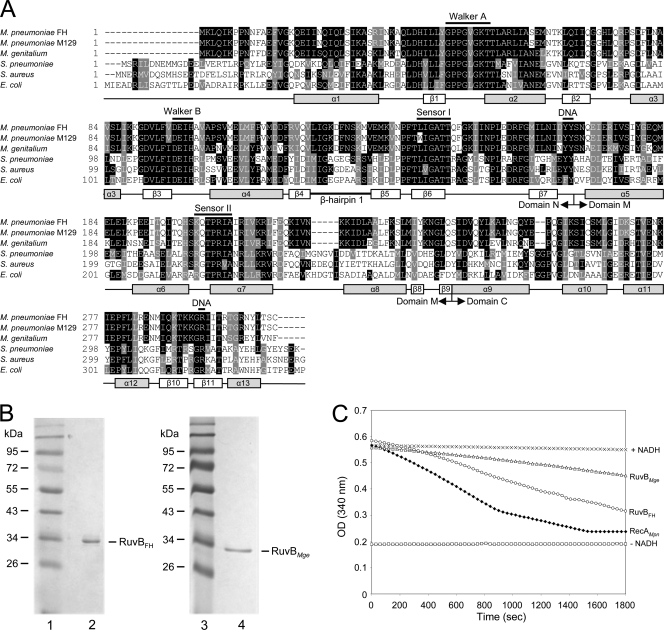
The MPN536 ORF of M. pneumoniae and MG359 ORF of M. genitalium have previously been annotated as genes that potentially encode RuvB homologs (6, 8, 27). The similarity between the amino acid sequences encoded by MPN536 and MG359 and those of (putative) RuvB proteins from other bacterial species can readily be observed in a multiple sequence alignment (Fig. 1A). The similarity between the sequences from M. pneumoniae and M. genitalium is very high (84.4% identity). A significantly lower similarity is seen between the sequences from the Mycoplasma spp. and those from other bacterial species (Fig. 1A). Protein regions that are highly conserved among the RuvB proteins are found in the amino-terminal (N) and central (M) domains and include the Walker A and B motifs and the sensor I and II motifs (23, 45). These motifs are characteristic for proteins that belong to the so-called AAA+ protein superfamily (ATPases associated with various cellular activities) (22). The Walker and sensor motifs are involved in the interaction with, and hydrolysis of, ATP (4). The RuvB region that was previously characterized as a β-hairpin 1 motif (Fig. 1A) is not highly conserved between the Mycoplasma RuvB sequences and the sequences of other RuvB proteins. In E. coli RuvB (RuvBEco), this motif was found to be important for the interaction of the protein with RuvA (7).
Fig. 1.
Multiple alignment of RuvB sequences and ATPase activity of RuvBFH and RuvBMge. (A) A multiple alignment was generated with amino acid sequences predicted to be encoded by the following ORFs (with GenBank accession numbers in parentheses): M. pneumoniae FH MPN536 (ADK87167), M. pneumoniae M129 MPN536 (AAB95954), M. genitalium G37 MG359 (ZP_05405689), Streptococcus pneumoniae ruvB (Q97SR6), Staphylococcus aureus ruvB (NP_374754), and E. coli ruvB (P0A812). Predicted secondary structural features of the RuvB proteins are shown below the alignment and are based on the crystal structures of the RuvB proteins from Thermus thermophilus HB8 (45) and Thermotoga maritima (28). The annotation of the (predicted) α helices and β strands, as well as the boundaries of the amino-terminal (N), central (M), and carboxyl-terminal domains, is derived from Yamada et al. (45). The positions of crucial, conserved motifs of AAA+ proteins, i.e., Walker A, Walker B, and sensor I and sensor II motifs, are indicated above the sequences. Amino acids potentially involved in DNA or nucleotide binding are also indicated (DNA) (12, 45).The multiple alignment was performed using Clustal W (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The program BOXSHADE, version 3.21 (http://www.ch.embnet.org/software/BOX_form.html), was used to generate white letters on black boxes (for residues that are identical in at least three out of six sequences) and white letters on gray boxes (for similar residues). (B) Purification of RuvBFH and RuvBMge. Samples of purified RuvBFH (left panel, lane 2) and RuvBMge (right panel, lane 4) were analyzed by SDS-PAGE (12%) and Coomassie brilliant blue (CBB) staining. The sizes of protein markers (lanes 1 and 3; PageRuler Prestained Protein Ladder [Fermentas]) are shown on the left-hand side of each panel in kDa. (C) ATPase activity of RuvBFH and RuvBMge. ATP hydrolysis by RuvBFH (○) and RuvBMge (Δ) was measured at a protein concentration of 0.5 μM in the presence of Mg2+ (1 mM) and ϕX174 single-stranded DNA (1.5 nM). The ATPase activity was determined using an NADH-coupled assay. Using this assay, the activity is calculated from the stationary velocities of ATP hydrolysis as determined by monitoring the absorption of NADH at 340 nm (19, 34). The RecAMpn protein, characterized previously by Sluijter et al. (34), was taken along as a positive control at a concentration of 0.5 μM (♦). Control reactions were performed in the absence of any protein (+NADH, ×) and in the absence of both NADH and protein (−NADH, □). OD, optical density.
It is interesting that the MPN536-encoded amino acid sequences of M. pneumoniae reference strains M129 and FH differ by a single residue, at position 140 (Fig. 1A); at this position a tryptophan (Trp) residue is present within the M129 RuvB protein (RuvBM129), and a leucine (Leu) is present within the FH protein (RuvBFH). Although sequence differences can be observed throughout the genomes of these two strains, which are representative of the two major genotypes of M. pneumoniae (subtypes 1 and 2), the presence of a Trp residue at position 140 of RuvBM129 is remarkable as all other known RuvB sequences contain a Leu at this position. Moreover, this Leu residue forms part of the highly conserved sensor II sequence (Fig. 1A). To investigate whether the sequences of RuvBFH and RuvBM129 are representative for subtype 2 and subtype 1 strains, respectively, we determined the sequence of the MPN536 ORF from three additional subtype 2 strains (Mp5181, Ofo, and R003) and three subtype 1 strains (Mp72, Mp4817, and PI 1428) (36). While the sequences from the subtype 2 strains were all found to be identical to the MPN536 sequence from strain FH, the sequences from the subtype 1 strains were all identical to the M129 sequence. Due to the deviant nature of the RuvB sequence from subtype 1 strains, we initially decided to focus our attention on characterization of the RuvB variant that is expressed by subtype 2 strains (RuvBFH).
Expression and purification of RuvBFH and RuvBMge.
The MPN536 and MG359 ORFs were amplified by PCR and cloned into protein expression vector pET-11c. This vector was used for the expression of RuvBFH and RuvBMge in their native forms in Escherichia coli. Both proteins were purified to near homogeneity using similar procedures (Fig. 1B). The estimated molecular masses of the purified proteins corresponded to their theoretical molecular masses of 35.0 kDa for both RuvBFH (Fig. 1B, lane 2) and RuvBMge (lane 4).
RuvBFH and RuvBMge possess ATPase activity.
As described above, the predicted amino acid sequences of RuvBFH and RuvBMge show all the characteristics of ATPases belonging to the AAA+ protein family. To test whether these proteins can be classified as ATPases, their ability to hydrolyze ATP was tested in the presence of Mg2+ and single-stranded DNA. Although both RuvBFH and RuvBMge were, indeed, found to possess ATPase activity under these conditions, the RuvBFH protein appeared to have a higher activity than RuvBMge (Fig. 1C). Although the ATPase activity of the RuvB proteins is likely to depend on the nature and concentration of DNA and other reaction constituents, as was demonstrated previously for other RuvB proteins such as E. coli RuvB (RuvBEco) (13, 21), the influence of these factors was not investigated further.
DNA-binding activity of RuvBFH and RuvBMge.
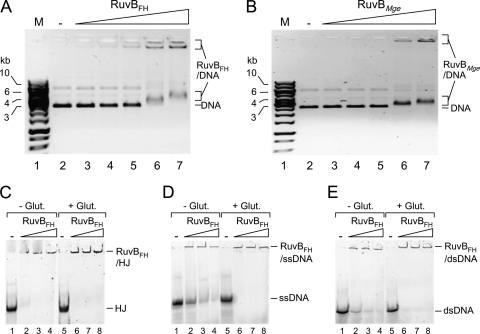
The ability of RuvBFH and RuvBMge to bind DNA was first investigated using supercoiled plasmid DNA in an agarose-based electrophoretic mobility shift assay. As shown in Fig. 2, both proteins formed large DNA-protein complexes in a protein concentration-dependent manner in the presence of ATPγS. These complexes, some of which did not enter the gel (Fig. 2A and B, lanes 6 and 7), were similarly formed in the presence of ATP or in the absence of any nucleotide cofactor (data not shown). Moreover, the addition of divalent cations to the binding reaction mixtures (10 mM Mg2+ or 5 mM Mn2+) did not significantly influence the binding characteristics of RuvBFH and RuvBMge (data not shown). While the use of the cross-linking agent glutaraldehyde was shown to be required for stabilization and, as a consequence, detection of the complexes between E. coli RuvB and DNA (21), the use of glutaraldehyde did not have a significant effect on the nature of the complexes that were generated between plasmid DNA and either RuvBFH or RuvBMge.
Fig. 2.
DNA-binding activity of RuvBFH and RuvBMge. (A) Binding of RuvBFH to supercoiled plasmid DNA. The DNA-binding reactions were performed as indicated in Materials and Methods. In short, reactions were performed in volumes of 10 μl and contained 20 ng of DNA, 1 mM ATPγS, and either 0 nM (−; lane 2), 340 nM (lane 3), 680 nM (lane 4), 1.35 μM (lane 5), 2.7 μM (lane 6), or 5.4 μM (lane 7) RuvBFH. The protein-DNA mixtures were separated by native 0.6% agarose gel electrophoresis, followed by staining with ethidium bromide. A black/white inverted image of the stained gel is shown. The positions of the unbound DNA (DNA) and RuvBFH-DNA complexes are indicated at the right-hand side of the gel. The sizes of DNA marker fragments (M, lane 1; SmartLadder [Eurogentec]) are shown on the left-hand side of the gel in kb. (B) Binding of RuvBMge to supercoiled plasmid DNA. Reactions were performed and analyzed using a method similar to that described for panel A and contained either 0 nM (−; lane 2), 340 nM (lane 3), 680 nM (lane 4), 1.35 μM (lane 5), 2.7 μM (lane 6), or 5.4 μM (lane 7) of RuvBMge. (C) Binding of RuvBFH to (6-FAM-labeled) HJ substrate HJ1.1 (33). Reactions were performed using a method similar to that described for panel A and contained either 0 nM (−; lanes 1 and 5), 1.35 μM (lanes 2 and 6), 2.7 μM (lanes 3 and 7), or 5.4 μM (lanes 4 and 8) of RuvBFH. The reaction products were separated on a 5% polyacrylamide gel and analyzed by fluorometry. The reactions shown in lanes 5 to 8 (+Glut.) were treated with 0.25% glutaraldehyde (30 min at 37°C) after the binding reactions. The reactions shown in lanes 1 to 4 were not treated with glutaraldehyde (−Glut.) (D) Binding of RuvBFH to ssDNA (5′ 6-FAM-labeled oligonucleotide 1) (Fig. 3A). Reactions were performed using a method similar to that described in panel C. (E) Binding of RuvBFH to dsDNA (substrate VI; 5′ 6-FAM labeled on oligonucleotide 2/1) (Fig. 3A). Reactions were performed using a method similar to that described in panel C.
Both RuvBFH and RuvBMge were also found to bind to linearized plasmid DNA (data not shown) and to single-stranded (ssDNA), double-stranded (dsDNA), and Holliday junction (HJ) substrates; with each of these substrates, large DNA-protein complexes were produced that did not enter the gels (Fig. 2C to E and data not shown). Binding of RuvBFH to the HJ substrate resulted in the most stable complexes; cross-linking with glutaraldehyde did not have a significant effect on these complexes (Fig. 2C). In contrast, cross-linking did have a stabilizing effect on complexes formed between RuvBFH and either ssDNA (Fig. 2D) or dsDNA (Fig. 2E). Similar results were obtained with the RuvBMge protein (data not shown).
With respect to the binding to oligonucleotide substrates, the activities of RuvBFH and RuvBMge differ in several important aspects from those of RuvBEco. First, in contrast to RuvBFH and RuvBMge, RuvBEco requires RuvAEco as well as Mg2+ for binding to HJs; this binding could be detected only after cross-linking with glutaraldehyde and resulted in large protein-DNA complexes that did not enter the polyacrylamide gels (25). Second, RuvBEco did not appear to bind to other oligonucleotide substrates, such as duplex oligonucleotides, irrespective of the presence of RuvAEco and divalent cations in the binding reactions (25).
RuvBFH and RuvBMge have DNA helicase activity.
To test the putative DNA helicase activity of RuvBFH and RuvBMge, both proteins were incubated with different types of DNA substrates. Schematic representations of the substrates, which have been described previously by Tsaneva et al. (40), are depicted in Fig. 3A. RuvBFH was found to readily unwind a DNA substrate consisting of a large single-stranded portion (ϕX174 virion DNA) to which a double-stranded oligonucleotide was annealed (Fig. 3A, substrate I). This activity was RuvBFH concentration, ATP, and Mg2+ dependent (see below) and resulted in the production of single-stranded oligonucleotides as well as (partially) double-stranded oligonucleotides (consisting of oligonucleotides 1 and 2) (Fig. 3B). The release of oligonucleotide 1 from the substrate was more efficient than that of oligonucleotide 2 (Fig. 3B, compare lanes 2 to 5, where oligonucleotide 1 of the substrate is labeled, to lanes 7 to 10, where oligonucleotide 2 is labeled). This result indicated that the DNA unwinding activity of RuvBFH has a preference regarding the polarity of the DNA substrate (see also below). Because RuvBFH was unable to unwind the (partial) duplexes consisting of oligonucleotides 1 and 2 in separate experiments (data not shown), we conclude that these duplexes represent end products from the unwinding of substrate 1 and cannot be processed further into the separate single-stranded oligonucleotides.
Apart from the discrete DNA products representing oligonucleotide 2 and the duplex of oligonucleotides 1 and 2, other, faint DNA products were generated with substrate I when oligonucleotide 2 was labeled (Fig. 3B, lanes 7 to 10, asterisk). The exact nature of these products, which migrated in the gel at a position intermediate to that of the single- and double-stranded oligonucleotide products, is not known. However, as their formation depended on the presence of unlabeled oligonucleotide 1 in the substrate, it is likely that these products represent incorrectly annealed duplexes of unlabeled oligonucleotide 1, which is preferentially produced in the helicase reaction, and labeled oligonucleotide 2. This notion was corroborated by the observation that similar products were formed after labeled oligonucleotide 2 was mixed with unlabeled oligonucleotide 1 at relatively low temperatures (≤37°C) (data not shown).
Like the RuvBFH protein, RuvBMge also displayed DNA unwinding activity. This activity, however, was significantly lower than that of its M. pneumoniae ortholog (Fig. 3C, lanes 2 and 3). This finding corresponded with the results from the ATPase assay, in which RuvBMge was found to have a lower activity than RuvBFH (Fig. 1C).
The DNA helicase activities of both RuvBFH and RuvBMge are remarkable as the counterpart of these proteins from E. coli, RuvBEco, was previously reported to be incapable of DNA unwinding by itself (39). Although the RuvBEco protein does have intrinsic DNA helicase activity, the protein is able to exert this activity only in the presence of RuvAEco (39). To investigate whether the RuvA homologues from M. pneumoniae and M. genitalium (RuvAMpn and RuvAMge, respectively) can influence the activities of RuvBFH and RuvBMge, various concentrations of the RuvA proteins were included in helicase assays using a range of DNA substrates. As shown in Fig. 3D, the helicase activity of RuvBFH on substrate I was not changed significantly by RuvAMpn. We were also unable to detect a significant effect of RuvAMpn on RuvBFH activity with other DNA substrates and at different RuvBFH concentrations (Fig. 3E, lanes 2 to 5, and data not shown). In contrast, the RuvBMge protein did show an RuvAMge-dependent effect although this effect was observed only with one of the substrates that was employed in this study, i.e., substrate IV. While RuvBMge alone was unable to unwind this substrate (Fig. 3E, lane 7), the protein displaced the substrate in the presence of RuvAMge. This stimulatory effect of RuvAMge on RuvBMge was RuvAMge concentration dependent (Fig. 3E, lanes 7 to 10). Like RuvAMpn and RuvAEco (39), RuvAMge alone was unable to catalyze DNA strand displacement (data not shown). Thus, while the DNA helicase activity of RuvBFH is not dependent on RuvAMpn, the helicase activity of RuvBMge is RuvAMge dependent on specific DNA substrates. In this respect, the activity of RuvBMge plus RuvAMge resembles that of the RuvAB helicase from E. coli (RuvABEco) (39).
Both RuvBFH and RuvBMge were unable to unwind HJ substrates and double-stranded oligonucleotide substrates, such as substrates V and VI (Fig. 3A), irrespective of the presence of RuvAMpn or RuvAMge in the reaction mixture (data not shown).
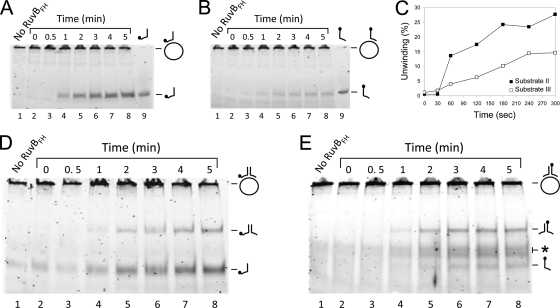
Time course of the RuvBFH-catalyzed helicase reaction.
As the RuvBFH protein appears to have unique characteristics in comparison to RuvBMge and RuvBEco, we investigated the time course as well as the reaction requirements of the helicase activity of this protein on substrates I, II, and III. Substrate II, consisting of oligonucleotide 1 annealed to ϕX174 single-stranded DNA, was most efficiently unwound by RuvBFH (Fig. 4A). Under the specific test conditions, maximal levels of 25 to 30% were reached after 3 min of incubation (Fig. 4A and C). Substrate III was less efficiently unwound by RuvBFH than substrate II: approximately 15% of this substrate was displaced after 4 min of incubation (Fig. 4B and C). These results corresponded with the results obtained with substrate I, from which oligonucleotide 1 was more efficiently displaced than oligonucleotide 2 (compare Fig. 4D and E). As expected, the efficiency by which the partial duplex of oligonucleotides 1 and 2 was displaced from substrate I was not influenced by the position of the fluorescent label: a similar time course of production of the duplex was seen when either oligonucleotide 1 (Fig. 4D) or oligonucleotide 2 (Fig. 4E) was labeled. The preferential displacement of oligonucleotide 1 from substrate I by RuvBFH is similar to that reported for RuvABEco. The preferred polarity of the helicase activity of the latter enzyme complex was found to be 5′ → 3′ with respect to the (long) single-stranded part of a branched or partially double-stranded substrate (39, 40).
Fig. 4.
Time course of the RuvBFH-catalyzed DNA helicase reaction on different DNA substrates. The substrates that were tested were substrate II (A), substrate III (B), and substrate I, which was 6-FAM labeled at the 5′ end of either oligonucleotide 1 (D) or oligonucleotide 2 (E). Reactions were performed as described in the legend of Fig. 3 and contained either 0 μM RuvBFH or 2.7 μM RuvBFH in the presence of Mg2+ (10 mM) and ATP (2 mM). Samples were taken at the time points indicated above the lanes of the figures. The labeled oligonucleotides 1 and 2 are taken along as makers in panel A (lane 9) and panel B (lane 9), respectively. (C) Comparison of the percentage of unwinding (the percentage of displaced oligonucleotide) of substrate II (■) with that of substrate III (□). The displaced oligonucleotides were measured from the gels shown in panels A and B as the percentage of released product relative to the total substrate in the reaction mixture. The labeling of substrate and reaction products is similar to that used in Fig. 3.
Reaction requirements of the RuvBFH-catalyzed DNA helicase reaction.
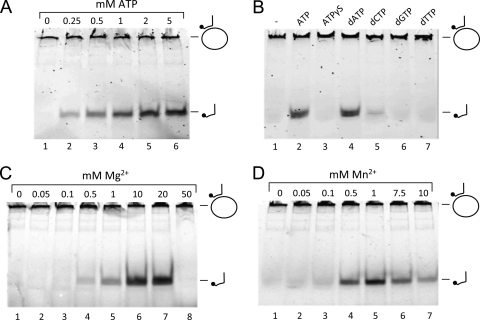
We previously found RuvBFH and RuvBMge to have ATPase activity (Fig. 1C). To investigate the ATP dependence of the helicase activity of RuvBFH, the protein's activity on substrate II was tested in the presence of various concentrations of ATP. As shown in Fig. 5A, the protein is inactive in the absence of ATP in the reaction mixture (lane 1). However, in the presence of ATP (0.25 mM or higher), RuvBFH readily unwound the substrate, with optimal activities reached at a concentration of ∼2 mM ATP. The dependence of RuvBFH activity on the hydrolysis of ATP is further demonstrated by the inactivity of the protein in the presence of the nonhydrolyzable analog of ATP, ATPγS [adenosine 5′-O-(3-thio)triphosphate] (Fig. 5B, lane 3). In the presence of dATP, however, the protein exhibited similar activities as in the presence of ATP (lane 4). Nucleotide cofactor dCTP was also able to support RuvBFH activity, albeit at a relatively low level (lane 5). The protein did not display significant activity in the presence of either dGTP or dTTP (Fig. 5B, lanes 6 and 7). This pattern of nucleotide cofactor dependence is highly similar to that reported for the HJ dissociation activity of RuvABEco (24).
Fig. 5.
Reaction requirements of the RuvBFH-catalyzed DNA helicase reaction. (A) ATP dependence of the DNA helicase activity of RuvBFH. Reactions with substrate II and 2.7 μM RuvBFH were performed in the absence (lane 1) or presence of ATP at a concentration of either 0.25, 0.5, 1, 2, or 5 mM (as indicated above the lanes). (B) Nucleotide cofactor dependence of the DNA helicase activity of RuvBFH. Reactions with substrate II and 2.7 μM RuvBFH were performed in the absence (lane 1) or presence of a 2 mM concentration of either ATP (lane 2), ATPγS (lane 3), dATP (lane 4), dCTP (lane 5), dGTP (lane 6), or dTTP (lane 7). (C) Mg2+ dependence of the DNA helicase activity of RuvBFH. Reactions with substrate II and 2.7 μM RuvBFH were performed in the absence (lane 1) or presence of various concentrations of Mg2+ (as indicated above the lanes). (D) Reactions with substrate II and 2.7 μM RuvBFH were performed in the absence (lane 1) or presence of various concentrations of Mn2+ (as indicated above the lanes). The labeling of substrate and reaction products is similar to that used in Fig. 3.
Like numerous other enzymes that interact with DNA, RuvBFH and RuvBMge require divalent cations for activity. RuvBFH was found to be active at ≥0.5 mM concentrations of either Mg2+ (Fig. 5C, lane 4) or Mn2+ (Fig. 5D, lane 4). The optimal concentrations of these divalent cations for stimulation of RuvBFH were ∼10 mM for Mg2+ (Fig. 5C, lane 6) and ∼1 mM for Mn2+ (Fig. 5D, lane 5).
Comparison of the DNA helicase activities of RuvBFH and RuvBM129.
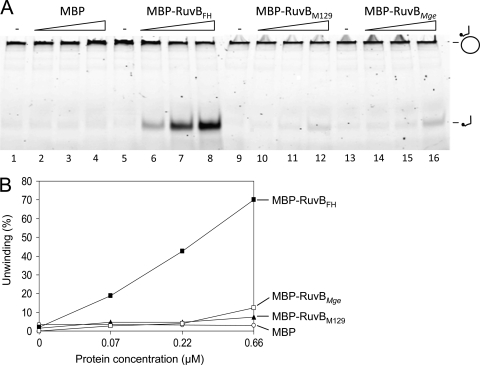
As described above, the RuvBFH protein can be expressed only by M. pneumoniae strains that belong to the subtype 2 genotype. Conversely, subtype 1 strains were found to express a RuvB version (RuvBM129) with a single amino acid residue substitution in comparison with RuvBFH (at position 140) (Fig. 1A). Because the Trp residue at position 140 of RuvBM129 is not conserved among bacterial RuvB sequences and, more importantly, because this residue is contained within the highly conserved sensor I motif of AAA+ proteins, we anticipated that RuvBM129 could have significantly different features than RuvBFH. To investigate this notion, we set out to purify RuvBM129 in its native state after overexpression of the protein in E. coli, in a similar fashion as described above for RuvBFH and RuvBMge. Nevertheless, while RuvBM129 could be expressed to high levels, the protein turned out to be extremely insoluble. After using different protocols of solubilization, including denaturation and renaturation procedures, we were unable to obtain sufficient amounts of soluble RuvBM129 to allow its purification. Because the expression of proteins as fusions to maltose-binding protein (MBP) can often resolve problems related to improper protein folding or insolubility (31, 33, 42–43), we expressed and purified RuvBM129, as well as RuvBFH and RuvBMge, as MBP fusion proteins. As shown in Fig. 6A (lanes 6 to 8), the fusion of MBP and RuvBFH (MBP-RuvBFH) displayed protein concentration-dependent DNA helicase activity similar to that of RuvBFH. Similarly, the activity of MBP-RuvBMge corresponded with that of its native counterpart, being significantly lower than that of MBP-RuvBFH (Fig. 6A, lanes 14 to 16, and B). Interestingly, MBP-RuvBM129 also displayed DNA helicase activity although this activity was only slightly higher than that observed for MBP, which served as a negative control (Fig. 6A, compare lanes 10 to 12 to lanes 2 to 4, and B). We conclude that RuvBM129 possesses DNA helicase activity even though this activity is significantly lower than that of RuvBFH.
Fig. 6.
DNA helicase activities of MBP fusion proteins of RuvBFH, RuvBM129, and RuvBMge. (A) DNA helicase activities of MBP-RuvBFH, MBP-RuvBM129, and MBP-RuvBMge. Reactions were performed with substrate II and various concentrations of MBP-β-galactosidase-α (MBP; lanes 2 to 4), MBP-RuvBFH (lanes 6 to 8), MBP-RuvBM129 (lanes 10 to 12), and MBP-RuvBMge (lanes 14 to 16). Each protein was tested at a concentration of 0.07, 0.22, and 0.66 μM (as indicated by the triangles above the lanes). (B) The percentage of unwinding (the percentage of displaced oligonucleotide) of substrate II by MBP-RuvBFH (■), MBP-RuvBMge (□), MBP-RuvBM129 (▴), and MBP (○). The displaced oligonucleotides were measured from the gel shown in panel A as the percentage of released product relative to the total substrate in the reaction mixture.
DISCUSSION
We have characterized the RuvB homologs from M. pneumoniae and M. genitalium and found these proteins to have several exceptional features compared to the well-characterized RuvB protein from E. coli. The most remarkable activity was exhibited by the RuvBFH protein. This protein was found to function as a potent divalent cation- and ATP-dependent DNA helicase in the absence of any protein cofactor, such as RuvA. In contrast, RuvBEco was previously found to mediate DNA unwinding exclusively in the presence of RuvAEco (39, 40). In this respect, the DNA helicase activity of RuvBEco differs from its HJ branch migration activity as the latter activity was also found to proceed in the absence of RuvA at high concentrations of both RuvBEco and Mg2+ (20, 38).
The DNA helicase activity of RuvBFH appeared to have a polarity similar to that of RuvABEco, which was reported to preferentially unwind DNA in the direction 5′ → 3′ with respect to the (relatively long) single-stranded portion of a partially double-stranded DNA substrate (39, 40). Interestingly, in contrast to RuvABEco (40), RuvBFH was unable to displace three-armed double-stranded oligonucleotide substrates, such as substrate V (Fig. 3A), in either the absence or the presence of RuvAMpn (data not shown). This indicated that RuvBFH is capable of unwinding only DNA substrates that are partially double stranded. Moreover, four-armed double-stranded DNA substrates that resembled HJs were also not processed by RuvBFH, irrespective of the presence of RuvAMpn in the reaction mixture (data not shown). Such substrates, however, were reported to be readily dissociated by RuvABEco through branch migration (14, 24, 25). The inability of RuvBFH to unwind HJ substrates may be caused by the apparent inability of the protein to interact directly with RuvAMpn. This view is based on the crucial role that is played by RuvAEco (as part of the RuvABEco complex) in the recognition and processing of HJs (14, 24, 30, 46).
Regarding the interaction between RuvB and RuvA proteins, it is important that the region of RuvBEco that was found to be crucial for binding to RuvAEco is localized between amino acid residues 134 and 153; this region consists of a β-hairpin (Fig. 1A, β-hairpin 1) that protrudes from the AAA+ ATPase domain of the protein (7, 45). As described in Results, the sequence of the β-hairpin 1 is not highly conserved between RuvBEco and the RuvB sequences from Mycoplasma spp. Also, two of the amino acid residues within the β-hairpin 1 of RuvBEco that were reported to be crucial for the functional and physical interaction of the protein with RuvAEco (Ile148 and Ile150) (7) are not conserved in RuvBFH, RuvBM129, or RuvBMge. While protein-protein interactions are not exclusively defined by primary protein structure, it is tempting to speculate that the incapacity of RuvBFH and RuvBM129 to be stimulated by RuvAMpn is determined by the inability of these proteins to physically interact. Obviously, this hypothetical failure to interact may be caused by sequences in both RuvB and RuvA. In this regard, it is interesting that RuvAMpn could not complement a ruvA-deficient E. coli mutant and did not stimulate the branch migration activity of RuvBEco in vitro (9). Nevertheless, in contrast to its M. pneumoniae orthologs, the M. genitalium RuvB protein did show RuvAMge-dependent DNA helicase activity on a specific DNA substrate (Fig. 3E). This result indicated that RuvBMge can functionally interact with RuvAMge. It is yet unclear, however, why this stimulatory effect of RuvAMge was observed with only a single substrate.
Another fascinating finding that came out of this study was the difference in characteristics between the RuvB protein expressed by subtype 2 M. pneumoniae strains (RuvBFH) and that expressed by subtype 1 strains (RuvBM129). While RuvBFH was expressed in E. coli as a soluble protein that could readily be purified, the RuvBM129 protein was found to be highly insoluble. As a consequence, we were unable to purify this protein. It could be purified, however, as an MBP fusion protein, in a similar fashion as RuvBFH and RuvBMge. Thus, we were able to demonstrate that the MBP-RuvBM129 protein does have DNA helicase activity, albeit dramatically less than that of MBP-RuvBFH (or RuvBFH). It is remarkable that the dissimilarity between RuvBFH and RuvBM129 is caused by a single residue difference in their amino acid sequences: RuvBFH has a Leu residue at position 140, whereas RuvBM129 carries a Trp residue at that position. Residue Leu140, which corresponds to Leu157 in RuvBEco, is highly conserved among virtually all known RuvB sequences and forms part of the (putative) sensor I sequence (Fig. 1A). The importance of this protein motif was previously demonstrated for RuvBEco by mutagenesis of the invariant sensor I residues Gly159, Ala160, T161, and T162 (12). Mutations in each of these residues resulted in proteins with significantly reduced in vivo DNA repair activities (12). We here show that another conserved residue from the putative sensor I sequence, Leu140 in RuvBFH, is also important for RuvB function, at least in vitro. By combining the in vivo properties of the RuvBEco sensor I mutants (12) with the characteristics of RuvBFH and RuvBM129 reported here, we would expect RuvB function to be considerably impaired in M. pneumoniae subtype 1 strains as opposed to subtype 2 strains. This could have a significant impact on the functionality or efficiency of the DNA recombination and repair machinery in subtype 1 strains. Whether subtype 1 and subtype 2 strains, indeed, differ in this respect is yet unknown. Moreover, we cannot rule out the possibility that native RuvBM129 is expressed in a soluble and active form in M. pneumoniae subtype 1 strains. Clearly, these notions should be investigated further by the generation of MPN536 knockouts and reciprocal knock-ins in strains that are derived from both genetic lineages of M. pneumoniae. Such experiments are currently being designed, despite the limited availability of tools for the genetic manipulation of M. pneumoniae.
In conclusion, this study once again underlines the major differences that exist between the components of the DNA recombination and repair machinery of Mycoplasma spp. and those of Gram-positive and Gram-negative bacteria. As a consequence, great care should be taken in the attribution of function to specific gene products on the sole basis of sequence similarities.
ACKNOWLEDGMENTS
We thank E. Spuesens for critically reading the manuscript.
A.M.C.V.R. is supported by grants of the European Society for Pediatric Infectious Diseases, ZonMW, and the Erasmus MC.
Footnotes
Published ahead of print on 23 September 2011.
REFERENCES
- 1. Reference deleted.
- 2. Carvalho F. M., et al. 2005. DNA repair in reduced genome: the Mycoplasma model. Gene 360:111–119 [DOI] [PubMed] [Google Scholar]
- 3. Dandekar T., et al. 2000. Re-annotating the Mycoplasma pneumoniae genome sequence: adding value, function and reading frames. Nucleic Acids Res. 28:3278–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Davey M. J., Jeruzalmi D., Kuriyan J., O'Donnell M. 2002. Motors and switches: AAA+ machines within the replisome. Nat. Rev. Mol. Cell Biol. 3:826–835 [DOI] [PubMed] [Google Scholar]
- 5. Reference deleted.
- 6. Fraser C. M., et al. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397–403 [DOI] [PubMed] [Google Scholar]
- 7. Han Y. W., et al. 2001. A unique beta-hairpin protruding from AAA+ ATPase domain of RuvB motor protein is involved in the interaction with RuvA DNA recognition protein for branch migration of Holliday junctions. J. Biol. Chem. 276:35024–35028 [DOI] [PubMed] [Google Scholar]
- 8. Himmelreich R., et al. 1996. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 24:4420–4449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ingleston S. M., et al. 2002. Holliday junction binding and processing by the RuvA protein of Mycoplasma pneumoniae. Eur. J. Biochem. 269:1525–1533 [DOI] [PubMed] [Google Scholar]
- 10. Iverson-Cabral S. L., Astete S. G., Cohen C. R., Rocha E. P., Totten P. A. 2006. Intrastrain heterogeneity of the mgpB gene in Mycoplasma genitalium is extensive in vitro and in vivo and suggests that variation is generated via recombination with repetitive chromosomal sequences. Infect. Immun. 74:3715–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Iverson-Cabral S. L., Astete S. G., Cohen C. R., Totten P. A. 2007. mgpB and mgpC sequence diversity in Mycoplasma genitalium is generated by segmental reciprocal recombination with repetitive chromosomal sequences. Mol. Microbiol. 66:55–73 [DOI] [PubMed] [Google Scholar]
- 12. Iwasaki H., et al. 2000. Mutational analysis of the functional motifs of RuvB, an AAA+ class helicase and motor protein for Holliday junction branch migration. Mol. Microbiol. 36:528–538 [DOI] [PubMed] [Google Scholar]
- 13. Iwasaki H., Shiba T., Makino K., Nakata A., Shinagawa H. 1989. Overproduction, purification, and ATPase activity of the Escherichia coli RuvB protein involved in DNA repair. J. Bacteriol. 171:5276–5280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Iwasaki H., Takahagi M., Nakata A., Shinagawa H. 1992. Escherichia coli RuvA and RuvB proteins specifically interact with Holliday junctions and promote branch migration. Genes Dev. 6:2214–2220 [DOI] [PubMed] [Google Scholar]
- 15. Kenri T., et al. 1999. Identification of a new variable sequence in the P1 cytadhesin gene of Mycoplasma pneumoniae: evidence for the generation of antigenic variation by DNA recombination between repetitive sequences. Infect. Immun. 67:4557–4562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 17. Ma L., et al. 2007. Mycoplasma genitalium: an efficient strategy to generate genetic variation from a minimal genome. Mol. Microbiol. 66:220–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maquelin K., et al. 2009. Raman spectroscopic typing reveals the presence of carotenoids in Mycoplasma pneumoniae. Microbiology 155:2068–2077 [DOI] [PubMed] [Google Scholar]
- 19. Morimatsu K., Takahashi M., Norden B. 2002. Arrangement of RecA protein in its active filament determined by polarized-light spectroscopy. Proc. Natl. Acad. Sci. U. S. A. 99:11688–11693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Muller B., Tsaneva I. R., West S. C. 1993. Branch migration of Holliday junctions promoted by the Escherichia coli RuvA and RuvB proteins. I. Comparison of RuvAB- and RuvB-mediated reactions. J. Biol. Chem. 268:17179–17184 [PubMed] [Google Scholar]
- 21. Muller B., Tsaneva I. R., West S. C. 1993. Branch migration of Holliday junctions promoted by the Escherichia coli RuvA and RuvB proteins. II. Interaction of RuvB with DNA. J. Biol. Chem. 268:17185–17189 [PubMed] [Google Scholar]
- 22. Neuwald A. F., Aravind L., Spouge J. L., Koonin E. V. 1999. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9:27–43 [PubMed] [Google Scholar]
- 23. Ohnishi T., Hishida T., Harada Y., Iwasaki H., Shinagawa H. 2005. Structure-function analysis of the three domains of RuvB DNA motor protein. J. Biol. Chem. 280:30504–30510 [DOI] [PubMed] [Google Scholar]
- 24. Parsons C. A., Tsaneva I., Lloyd R. G., West S. C. 1992. Interaction of Escherichia coli RuvA and RuvB proteins with synthetic Holliday junctions. Proc. Natl. Acad. Sci. U. S. A. 89:5452–5456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Parsons C. A., West S. C. 1993. Formation of a RuvAB-Holliday junction complex in vitro. J. Mol. Biol. 232:397–405 [DOI] [PubMed] [Google Scholar]
- 26. Peterson S. N., et al. 1995. Characterization of repetitive DNA in the Mycoplasma genitalium genome: possible role in the generation of antigenic variation. Proc. Natl. Acad. Sci. U. S. A. 92:11829–11833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peterson S. N., Hu P. C., Bott K. F., Hutchison C. A., 3rd 1993. A survey of the Mycoplasma genitalium genome by using random sequencing. J. Bacteriol. 175:7918–7930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Putnam C. D., et al. 2001. Structure and mechanism of the RuvB Holliday junction branch migration motor. J. Mol. Biol. 311:297–310 [DOI] [PubMed] [Google Scholar]
- 29. Ruland K., Wenzel R., Herrmann R. 1990. Analysis of three different repeated DNA elements present in the P1 operon of Mycoplasma pneumoniae: size, number and distribution on the genome. Nucleic Acids Res. 18:6311–6317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shiba T., Iwasaki H., Nakata A., Shinagawa H. 1991. SOS-inducible DNA repair proteins, RuvA and RuvB, of Escherichia coli: functional interactions between RuvA and RuvB for ATP hydrolysis and renaturation of the cruciform structure in supercoiled DNA. Proc. Natl. Acad. Sci. U. S. A. 88:8445–8449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sluijter M., Aslam M., Hartwig N. G., van Rossum A. M., Vink C. 2011. Identification of amino acid residues critical for catalysis of Holliday junction resolution by Mycoplasma genitalium RecU. J. Bacteriol. 193:3941–3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sluijter M., Hoogenboezem T., Hartwig N. G., Vink C. 2008. The Mycoplasma pneumoniae MPN229 gene encodes a protein that selectively binds single-stranded DNA and stimulates recombinase A-mediated DNA strand exchange. BMC Microbiol. 8:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sluijter M., et al. 2010. The Mycoplasma genitalium MG352-encoded protein is a Holliday junction resolvase that has a non-functional orthologue in Mycoplasma pneumoniae. Mol. Microbiol. 77:1261–1277 [DOI] [PubMed] [Google Scholar]
- 34. Sluijter M., Spuesens E. B., Hartwig N. G., van Rossum A. M., Vink C. 2009. The Mycoplasma pneumoniae MPN490 and Mycoplasma genitalium MG339 genes encode RecA homologs that promote homologous DNA strand exchange. Infect. Immun. 77:4905–4911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Spuesens E. B., Hartwig N. G., van Rossum A. M., Vink C. 2010. Identification and classification of P1 variants of Mycoplasma pneumoniae. J. Clin. Microbiol. 48:680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Spuesens E. B., et al. 2009. Sequence variations in RepMP2/3 and RepMP4 elements reveal intragenomic homologous DNA recombination events in Mycoplasma pneumoniae. Microbiology 155:2182–2196 [DOI] [PubMed] [Google Scholar]
- 37. Su C. J., Chavoya A., Baseman J. B. 1988. Regions of Mycoplasma pneumoniae cytadhesin P1 structural gene exist as multiple copies. Infect. Immun. 56:3157–3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tsaneva I. R., Muller B., West S. C. 1992. ATP-dependent branch migration of Holliday junctions promoted by the RuvA and RuvB proteins of E. coli. Cell 69:1171–1180 [DOI] [PubMed] [Google Scholar]
- 39. Tsaneva I. R., Muller B., West S. C. 1993. RuvA and RuvB proteins of Escherichia coli exhibit DNA helicase activity in vitro. Proc. Natl. Acad. Sci. U. S. A. 90:1315–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tsaneva I. R., West S. C. 1994. Targeted versus non-targeted DNA helicase activity of the RuvA and RuvB proteins of Escherichia coli. J. Biol. Chem. 269:26552–26558 [PubMed] [Google Scholar]
- 41. Reference deleted.
- 42. Vink C., Oude Groeneger A. M., Plasterk R. H. 1993. Identification of the catalytic and DNA-binding region of the human immunodeficiency virus type I integrase protein. Nucleic Acids Res. 21:1419–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vink C., van der Linden K. H., Plasterk R. H. 1994. Activities of the feline immunodeficiency virus integrase protein produced in Escherichia coli. J. Virol. 68:1468–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wenzel R., Herrmann R. 1988. Repetitive DNA sequences in Mycoplasma pneumoniae. Nucleic Acids Res. 16:8337–8350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yamada K., et al. 2001. Crystal structure of the Holliday junction migration motor protein RuvB from Thermus thermophilus HB8. Proc. Natl. Acad. Sci. U. S. A. 98:1442–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yamada K., et al. 2002. Crystal structure of the RuvA-RuvB complex: a structural basis for the Holliday junction migrating motor machinery. Mol. Cell 10:671–681 [DOI] [PubMed] [Google Scholar]