Abstract
The cotG and cotH genes of Bacillus subtilis encode two previously characterized spore coat proteins. The two genes are adjacent on the chromosome and divergently transcribed by σK, a sporulation-specific σ factor of the RNA polymerase. We report evidence that the cotH promoter maps 812 bp upstream of the beginning of its coding region and that the divergent cotG gene is entirely contained between the promoter and the coding part of cotH. A bioinformatic analysis of all entirely sequenced prokaryotic genomes showed that such chromosomal organization is not common in spore-forming bacilli. Indeed, CotG is present only in B. subtilis, B. amyloliquefaciens, and B. atrophaeus and in two Geobacillus strains. When present, cotG always encodes a modular protein composed of tandem repeats and is always close to but divergently transcribed with respect to cotH. Bioinformatic and phylogenic data suggest that such genomic organizations have a common evolutionary origin and that the modular structure of the extant cotG genes is the outcome of multiple rounds of gene elongation events of an ancestral minigene.
INTRODUCTION
Endospore-forming bacteria are Gram-positive microorganisms belonging to different genera and including more than 200 species (13). The common feature of these organisms is the ability to form a quiescent cellular type called an endospore (spore) in response to harsh environments. The spore can survive in this dormant state for long periods, resisting a vast range of stresses, such as high temperature, dehydration, absence of nutrients, and presence of toxic chemicals. When the environmental conditions ameliorate, the spore germinates, originating a vegetative cell able to grow and to sporulate again. Spore resistance is made possible by the presence of the spore coat, a multilayered structure composed by more than 70 proteins synthesized in the mother cell compartment of the sporangium and assembled around the forming spore (16). Coat formation is finely controlled through various processes acting at the transcriptional or posttranslational level. The synthesis of coat proteins is regulated by a cascade of at least five transcription factors: σE and σK (two mother cell-specific σ factors of the RNA polymerase), SpoIIID and GerE (two transcriptional regulators acting in conjunction with σE and σK, respectively) (18), and GerR (initially found to control at least 14 σE genes [6] and more recently identified as affecting directly or indirectly also some σK-dependent genes [3]). The assembly of coat components on the surface of the forming spore is governed by a subset of morphogenetic proteins that guide the correct packaging process (16). The main morphogenetic factors are SpoIVA, CotE, and SafA (25). SpoIVA (5, 33) is assembled into the basement layer of the coat and is anchored to the outer membrane of the forespore through its C terminus that contacts SpoVM, a small, amphipathic peptide embedded in the forespore membrane (24, 30, 31). SpoIVA controls the assembly of most coat components either directly or through SafA and CotE, proposed as key regulators of the inner coat and the outer coat, respectively (25). CotE self-interacts (23) and assembles into a ring that surrounds the SpoIVA basement structure (40). The inner layer of the coat is then formed between the SpoIVA layer and the CotE ring, while the outer coat is formed outside the CotE ring (25, 40). SafA and CotE have been proposed to interact with most coat components based on the results of a fluorescence microscopy analysis of a collection of strains carrying cot-gfp fusions (21, 25). Biochemical experiments have confirmed the direct interaction of CotE with two outer coat components and have revealed the essential role of CotE in mediating their interaction (20).
Other morphogenetic proteins include CotH and CotG (16). CotH plays a role in the assembly of at least 9 other coat components, including CotG (21), and in the development of lysozyme resistance of the mature spore (28, 41). CotG is needed for the conversion of CotB from an immature 44-kDa form into a mature 66-kDa form (42). The structural genes coding for CotH and CotG are clustered together on the Bacillus subtilis chromosome but are divergently transcribed (28). While CotH is a 42.8-kDa protein found in several Bacillus species and also in some Clostridium species (16), CotG is a 24-kDa protein containing nine tandem repeats of a 13-amino-acid stretch in its central part (34) and so far has been found only in B. subtilis (16).
Here we report that cotH expression depends on a newly identified promoter located 812 nucleotides upstream of the coding region. The long sequence at 5′ end of cotH is most likely not translated and completely overlaps the divergent cotG gene (see Fig. 1A). The apparent lack of function of the long 5′ untranslated region along with the evidence that the cotG-cotH genome organization is not conserved in Bacillus species prompted us to investigate the evolutionary origin of the cotG gene and of the cotH-cotG gene organization.
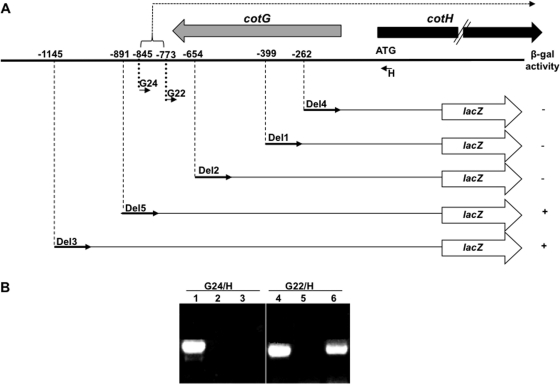
Fig. 1.
(A) Deletion analysis of the DNA region upstream of the cotH coding part. Numbers indicate positions on the DNA sequence, considering the first base of the translation start site as +1. The lacZ gene of E. coli is fused in frame to the cotH coding part as described by Baccigalupi et al. (2). β-gal, β-galactosidase. Thin, short arrows indicate the positions of the oligonucleotides used in the RT-PCR experiment corresponding to panel B, while thick gray and black arrows indicate the coding parts of cotG and cotH, respectively. The dashed arrow indicates the mRNA produced from the cotH promoter. (B) Agarose gel electrophoresis used to analyze the extension products of an RT-PCR experiment. Total RNA was extracted from sporulating cells 5 h after the onset of sporulation from a wild-type (PY79) strain. cDNA synthesis was primed with oligonucleotide H (panel A; Table 1), while the amplification reactions were primed with oligonucleotide pairs H/G22 and H/G24 (panel A; Table 1). Control experiments were performed using chromosomal DNA as a template (lanes 1 and 4) or mRNA without the addition of the RT enzyme (lanes 2 and 5). cDNA was used as a template in the reactions of lanes 3 and 6.
MATERIALS AND METHODS
Bacterial strains and transformation.
B. subtilis PY79 (39) was used as recipient strain in transformation procedures. Plasmid amplification for DNA sequencing, subcloning experiments, and transformation of Escherichia coli competent cells were performed with E. coli strain DH5α (36). Bacterial strains were transformed by previously described procedures, i.e., CaCl2-mediated transformation of E. coli competent cells (36) and two-step transformation of B. subtilis (4).
Genetic and molecular procedures.
Isolation of plasmids, restriction digestion, and ligation of DNA, were carried out by standard methods (36). Chromosomal DNA from B. subtilis was isolated as described elsewhere (4).
Deletion analysis, transcriptional gene fusions, and β-galactosidase assays.
Deletion mutants of DNA upstream of the cotH coding part were constructed starting from B. subtilis strain GC237, containing a cotH::lacZ translational fusion inserted at the cotH locus (2). Deletion mutants were obtained by PCR using chromosomal DNA of strain GC237 as a template. PCRs were carried out with oligonucleotide lacZ2 (5′-GAATTCTATTTTTGACACCAGACC-3′ [underlined is the EcoRI site]), annealing at the 3′ end of the lacZ gene, and one of the following oligonucleotides: Del4, Del1, Del2, Del5, and Del3 (see Table S1 in the supplemental material). Amplified fragments were subcloned in pGEM-T Easy vector, sequenced to ensure there were no mutations (BMR Genomics), digested with BamHI/EcoRI, and cloned in the integrative vector pDG364 (4), previously restricted with the same enzymes. Gene fusions were then inserted at the amyE locus of a wild-type (PY79) strain of B. subtilis by a double reciprocal crossing-over event.
The integrative vector pSN32 (26) was used to obtain a transcriptional fusion of the cotH promoter to the lacZ gene of E. coli. A genomic fragment containing the cotH promoter was PCR amplified by using chromosomal DNA of strain PY79 as a template and oligonucleotides Del3 and G27 as primers (see Table S1 in the supplemental material). Purified fragments were cloned into pGEM-T Easy vector (Promega), excised by EcoRI/BamHI digestion and cloned upstream of the lacZ gene into the pSN32 vector (26) restricted with the same enzymes. The resulting plasmid was linearized and used to transform competent cells of B. subtilis strain PY79. The obtained strain, AZ530, contained the cotH::lacZ transcriptional fusion at the amyE locus of the chromosome. The fusion was then moved by chromosome-mediated transformation into an isogenic strain carrying null mutations in either spoIVB (spoIVB::erm) or gerE (gerE36). Specific β-galactosidase activity was determined using o-nitrophenol-β-d-galactoside (ONPG) as the substrate (4). Samples of cells (1 ml each) bearing the fusion were collected, during sporulation, at the indicated times and assayed as previously described (4).
Construction of a cotH internal deletion mutant.
DNA coding for the 5′ part of cotH mRNA extending from nucleotide −843 to nucleotide −34 (with respect to the first base of cotH coding sequence) was deleted using the gene splicing by overlap extension (SOEing) technique (17). Briefly, two PCR products were obtained with oligonucleotide pairs Del3/H33s (to amplify the cotH promoter region) and H32s/H13 (extending 34 bases upstream of the ATG to the unique EcoRI site internal to the cotH coding sequence) (see Table S1 in the supplemental material). The obtained products were used as templates to prime a third PCR with the external primers Del3 and H13 (Table S1). The modified version of cotH was cloned into the vector pER19 (32), and the correct gene fusion was verified by sequencing reactions. The resulting plasmid, pRG25, was introduced by single reciprocal (Campbell-like) recombination at the cotH locus of the B. subtilis chromosome. Several chloramphenicol-resistant clones were analyzed by PCR to select the clone containing the modified cotH promoter sequence upstream the entire cotH gene.
Primer extension analysis.
Total RNA was extracted from the wild-type strain PY79 and the isogenic mutant strains carrying null mutations in spoIVB (spoIVB::erm) or gerE (gerE36), 5 h after the onset of sporulation using the Qiagen minikit (Qiagen, Milan, Italy) according to the manufacturer's instructions. Total RNAs were dissolved in 50 μl of RNase-free water and stored at −80°C. The final concentration and quality of the RNA samples were estimated spectrophotometrically and by agarose gel electrophoresis with ethidium bromide staining. Total RNAs were treated with RNase-free DNase (1 U/μg of total RNA; Fermentas) for 30 min at 37°C, and the reaction was stopped with DNase inactivation reagent. For primer extension experiments, 10 μg of total RNA was used with [γ-32P]dATP (GE Healthcare)-labeled oligonucleotide G25 (see Table S1 in the supplemental material), deoxynucleoside triphosphates (dNTPs), and reverse transcriptase (Stratagene) to prime cDNA synthesis as previously described (28) The reaction products were fractionated on 6 M urea–6% polyacrylamide gels, along with DNA sequencing reactions using pNC12 (carrying the cotH-cotG chromosomal fragment) as a template primed with the same oligonucleotide.
For reverse transcription-PCR (RT-PCR) analysis a sample containing 2 μg of DNase-treated RNA was incubated with oligonucleotide H at 65°C for 5 min and slowly cooled to room temperature to allow the primer annealing. The mixture was incubated at 50°C for 1 h in the presence of 1 μl AffinityScript multiple-temperature reverse transcriptase (Stratagene), 4 mM dNTPs, reaction buffer (Stratagene), and 10 mM dithiothreitol (DTT). The enzyme was inactivated at 70°C for 15 min. One-tenth of the reaction mix was used as a template in PCRs using oligonucleotide H coupled with oligonucleotides Del4, G22, G24, and Del5 (see Table S1 in the supplemental material). For a control, PCRs were carried out with RNA alone to exclude the possibility that the amplification products could derive from contaminating genomic DNA.
Spore purification, extraction of spore coat proteins, and Western blot analysis.
Sporulation was induced in Difco sporulation medium (DSM; Difco) by the exhaustion method as described elsewhere (4). After a 30-h incubation at 37°C, the spores were collected, washed four times, and purified as described by Nicholson and Setlow (29) by using overnight incubation in H2O at 4°C to lyse residual sporangial cells. Spore coat proteins from B. subtilis PY79 and of the ΔcotH mutant (AZ535) were extracted either from a suspension of purified spores by using an SDS-DTT extraction buffer as previously described (29) or from sporulating cells harvested at various times after the onset of sporulation. In the latter case, sporulating cells were washed three times, suspended in 100 μl of lysozyme solution (25 mM Tris-HCl [pH 7.5], 50 mM glucose, 10 mM EDTA, 1% lysozyme) and incubated for 5 min on ice. The suspension was then boiled in 2% (vol/vol) SDS, 5% (vol/vol) 2-mercaptoethanol, 10% (vol/vol) glycerol, 62.5 mM Tris-HCl (pH 6.8), and 0.05% (wt/vol) bromophenol blue for 5 min. The concentration of extracted proteins was determined by using Bio-Rad DC protein assay kit (Bio-Rad), and 20-μg samples of total spore coat proteins were fractionated on 12.5% SDS-polyacrylamide gels and electrotransferred to nitrocellulose filters (Bio-Rad) for Western blot analysis following standard procedures. CotH-specific antibody was used at a working dilution of 1:5,000, and a horseradish peroxidase (HRP)-conjugated anti-rabbit antibody was utilized as secondary antibody (Santa Cruz). Western blot filters were visualized by the SuperSignal West Pico chemiluminescence (Pierce) method as specified by the manufacturer.
Homolog retrieval and phylogenetic analysis.
BLAST probing of the DNA and protein databases was performed with the BLASTn and BLASTp options of the BLAST program (1), using default parameters with no filters. Nucleotide sequences were retrieved from the GenBank and EMBL databases. The ClustalW program (38) was used to align the gene sequences obtained with the most similar ones retrieved from the databases. Each alignment was checked manually, corrected, and then analyzed using the neighbor-joining method (37) and the model of Kimura 2-parameter distances (22). Phylogenetic trees were constructed with the aligned sequences using Molecular Evolutionary Genetics Analysis 5 software (35). The robustness of the inferred trees was evaluated by 1,000 bootstrap resamplings.
RESULTS
A long upstream region is required for cotH expression.
It has been previously reported that a strain carrying a translational cotH::lacZ fusion placed at the cotH locus of the B. subtilis chromosome showed a sporulation-specific and GerE-dependent β-galactosidase activity starting from 4 h after the onset of sporulation (2). When the same gene fusion containing about 250 bp upstream of the first cotH codon was moved at the amyE locus of the B. subtilis chromosome, no β-galactosidase activity was observed (data not shown). To define the DNA region upstream of the first codon required for the ectopic expression of cotH, we performed a deletion analysis: DNA fragments containing the entire lacZ gene fused in frame at the beginning of the cotH coding region (2) and extending for 1145 (Del3), 891 (Del5), 654 (Del2), 399 (Del1), and 262 (Del4) bp upstream of the first cotH codon (Fig. 1A) were PCR amplified and cloned into plasmid pBK2, a pDG364 derivative (4). Plasmid DNA was then used to integrate all fusions at the amyE locus of the B. subtilis chromosome. As shown in Fig. 1A, only strains carrying the two longest fusions (Del3 and Del5) showed β-galactosidase activity. Those activities were indistinguishable from each other and from that observed with a strain carrying the translational fusion integrated at the cotH locus (2; not shown). This observation, together with the total absence of β-galactosidase activity of strains carrying fusions Del1, Del2, and Del4 strongly suggested that the DNA region spanning from position −654 to −891 is essential for cotH gene expression.
The region upstream of the B. subtilis cotH gene is transcribed.
The long region required for cotH expression could be either the binding site for transcriptional activators or part of the transcription unit. We used an RT-PCR approach to discriminate between the two possibilities. For this purpose, total RNA was extracted from sporulating cells of a wild-type B. subtilis strain (PY79) 5 h after the initiation of sporulation and used as a template to produce cDNA with the synthetic oligonucleotide H (Fig. 1A; see Table S1 in the supplemental material) to prime the reaction. cDNA was then PCR amplified with oligonucleotide H and a collection of oligonucleotides annealing in the upstream region. We observed an amplification product of the expected size with oligonucleotide G22 and all oligonucleotides annealing downstream from it, while no PCR product was obtained with oligonucleotide G24 or all oligonucleotides annealing upstream of it. The amplifications performed with oligonucleotide pairs G22/H and G24/H are shown in Fig. 1B.
Therefore, these data indicate that the DNA region upstream of the cotH coding part is transcribed up to, at least, position −773 and that the cotH transcriptional start site is likely located downstream of position −845.
The analysis of the transcribed DNA region upstream of the cotH coding region revealed the presence of a long open reading frame (ORF), extending for 570 bp, and of several other short ORFs in the same orientation as cotH. However, all those ORFs do not have typical ribosome binding sites upstream of potential start codons. In addition, a translational fusion of the lacZ gene of E. coli and the longest ORF failed to produce β-galactosidase in both rich and sporulation-inducing media (not shown). Based on those results, we suggest that the long DNA region upstream of the cotH coding part is transcribed but not translated in the direction of cotH.
Identification and analysis of the cotH promoter.
To characterize the cotH promoter, we constructed a transcriptional gene fusion by using a 372-bp DNA region comprised of oligonucleotides Del3 and G27 (complementary to G22; Fig. 1A), containing the cotH promoter and the coding sequence of the Escherichia coli lacZ gene. The gene fusion was integrated at the amyE locus of the B. subtilis chromosome and the β-galactosidase activity measured at various times after the onset of sporulation in an otherwise wild-type strain and a collection of congenic sporulation mutants. As reported in Fig. 2A, we observed that β-galactosidase activity commenced 4 h after the onset of sporulation (T4), reached its maximum 1 h later (T5), and then decreased (Fig. 2A). Expression of cotH::lacZ was not detected in a mutant that fails to make σK (spoIVB::erm; data not shown), while the expression in a strain not producing GerE (gerE36; open symbols) was higher than that in an isogenic wild type (Fig. 2A, filled symbols). These results are in agreement with previous data obtained with a translational cotH::lacZ fusion (2) and confirm that cotH is transcribed by σK-containing RNA polymerase and is negatively regulated by GerE.
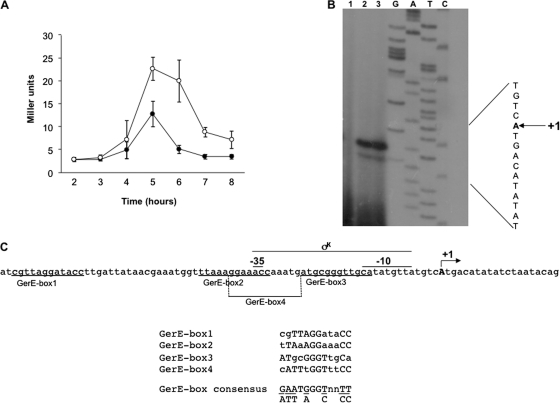
Fig. 2.
(A) Expression of a cotH::lacZ transcriptional fusion during sporulation in an otherwise wild-type (closed symbols) or gerE null mutant (open symbols) strain. Samples were collected at various times after the onset of sporulation. Enzyme activity is expressed in Miller units. Data are the means of three independent experiments, and error bars indicate the standard deviations. (B) Primer extension analysis of the cotH promoter region performed with total RNA extracted from sporulating cells 5 h after the onset of sporulation from a mutant that fails to make SigK (spoIVB::erm; lane 1), a gerE null mutant (gerE36; lane 3), and a wild-type strain (PY79; lane 2). Primer extension and sequencing reactions were primed with the synthetic oligonucleotide G25 (Table 1). (C) cotH promoter region. The transcription start site (“A”) is in bold and indicated as +1. The putative promoter sequences are indicated and the putative GerE boxes underlined. A comparison of the four GerE boxes with the GerE consensus is also reported.
A primer extension experiment was then performed to map the cotH transcriptional start site. As shown in Fig. 2B, an extension product was obtained and appeared to be specific, since it was not observed when total RNA from a mutant that fails to make SigK (spoIVB::erm) was used for the extension reaction (Fig. 2B, lane 1) and was more intense with RNA from a strain not producing GerE (gerE36) than from a wild type (Fig. 2B, lanes 3 and 2, respectively). The 5′ terminus (indicated as +1) was mapped 812 bp upstream of the cotH first codon. The analysis of the nucleotide sequence upstream of the cotG 5′ terminus revealed the presence of a sequence perfectly matching all positions of the −10 and −35 consensus for a σK promoter (Fig. 2C) (15). As indicated in Fig. 2C, sequences resembling the proposed binding site for the GerE protein (18) are also present upstream of the promoter (GerE box 1), overlapping the −35 region (GerE box 2), between the −35 and −10 regions (GerE box 3), and on the opposite strand overlapping the −35 region (GerE box 4), consistent with a negative role of GerE on cotH transcription.
The 5′ part of cotH mRNA does not control CotH synthesis.
The 812-bp RNA sequence extending from the transcriptional start site (+1) to the first codon was analyzed using the RNAfold Web server (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) and showed the potentials for extensive RNA secondary structures (see Fig. S2 in the supplemental material). Although cis-acting regulatory RNA elements (riboswitches) in B. subtilis have an average length of 360 bp (19), we decided to analyze whether the long 5′ part of cotH mRNA had a regulatory role on the downstream coding region. With this aim, we first constructed two transcriptional gene fusions by using the coding sequence of the E. coli lacZ gene as a reporter gene and DNA regions containing the cotH promoter and either 53 or 812 bp downstream of the transcriptional start site. Therefore, the two fusions contained only the initial 53 bp or the entire 5′ untranslated region of cotH. Both gene fusions were checked by nucleotide sequence analysis and integrated at the amyE locus of the B. subtilis chromosome. The β-galactosidase activity was then measured at various times after the onset of sporulation in an otherwise wild-type strain and in an isogenic strain not producing GerE (gerE36). As reported in Fig. 3A, similar levels of β-galactosidase activity were observed with the short (open and closed squares in wild-type and gerE backgrounds, respectively) or the long (open and closed circles in wild-type and gerE backgrounds, respectively) fusion, suggesting that the 5′ region did not affect the transcription of the cotH coding part.
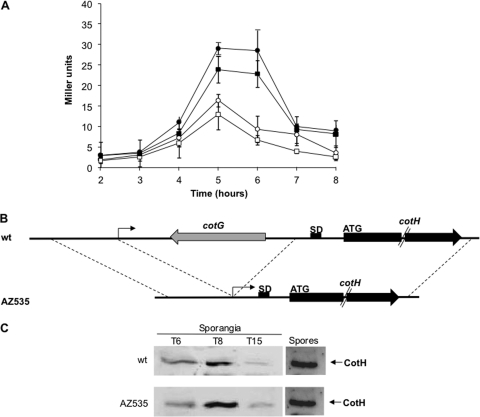
Fig. 3.
(A) β-Galactosidase activity of strains carrying the cotH promoter and either 53 (squares) or 812 (circles) bp downstream of the transcription start site fused to the lacZ gene of E. coli. Gene fusions were inserted in an otherwise wild-type (open symbols) or gerE null mutant (closed symbols) strain. Samples were collected at various times after the onset of sporulation, and enzyme activity is expressed in Miller units. Data are the means of three independent experiments, and error bars indicate the standard deviations. (B) Construction of the deletion mutant. (C) Western blot of proteins extracted from sporulating cells or from purified spores of a wild-type strain and of an isogenic mutant carrying the deletion indicated in panel B. Proteins were fractionated on 15% polyacrylamide gels, electrotransferred to membranes, and reacted with anti-CotH antibody.
In addition, we constructed a deletion mutant lacking 777 bp (from the transcriptional start site to 34 bp upstream of first codon) at the 5′ part of cotH (Fig. 3B). In this deletion mutant (AZ535), the cotH promoter was then positioned just upstream of the cotH ribosome binding site and the coding part. Sporulating cells of the wild type and the isogenic deletion mutant were harvested at various times during sporulation, lysed, and analyzed by Western blotting with anti-CotH antibody, as indicated in Materials and Methods. Similar amounts of CotH were found in the sporangia and on purified spores of both strains, indicating that, at least in these experimental conditions, the 5′ part of cotH mRNA does not affect production and assembly of CotH within the coat (Fig. 3C).
Purified spores of the wild type and the isogenic deletion mutant (AZ535) did not show any difference when analyzed for their heat resistance (10 and 30 min at 80°C), lysozyme (300 μg/ml) resistance, and Asn-induced germination (not shown), supporting the idea that the 5′ part of cotH mRNA does not affect CotH synthesis and spore coat formation.
The cotG-cotH chromosomal region.
The unusual size and the lack of a regulatory function of the 5′ portion of cotH mRNA, together with the presence of the divergent cotG gene between the promoter and the beginning of the coding region of cotH (Fig. 1A), prompted us to investigate in more detail the cotG-cotH chromosomal region. We used the amino acid sequence of the B. subtilis subsp. subtilis strain 168 CotG protein (accession number NP_391488.1) as a query in a BLAST (1) analysis to probe all translated ORFs of all completely sequenced prokaryotic genomes. The scanning of the 1,006 genomes (917 from bacteria and 89 from archaea) using the default parameters with no filters retrieved only six sequences with an E value below the default parameters used, four from bacilli and two from geobacilli (Table 1). The six CotG sequences exhibited different lengths, ranging from 77 (Geobacillus sp. WCH70) to 198 (B. subtilis subsp. spizizeni strain W23) amino acid residues (Table 1). Since our BLAST analysis included 76 entire genomes of Bacillales, results of Table 1 confirmed the very narrow phylogenetic distribution of cotG and indicated, as a consequence, that the cotG-cotH gene organization found in B. subtilis is not prevalent among spore-forming bacilli. Next, we analyzed the cotG-cotH chromosome organization in the six cotG-containing organisms. The cotG ortholog is adjacent to a divergently transcribed cotH ortholog in all four Bacillus sequences and in one of the two Geobacillus sequences, Geobacillus sp. Y4.1MC1 (see Fig. S3 in the supplemental material). In Geobacillus sp. strain WCH70, cotG and cotH orthologs are divergently transcribed but are separated by an ORF coding for a putative transposase of the IS116/IS110/IS902 protein family (Fig. S3). RT-PCR experiments indicate that, as in B. subtilis, also in B. amyloliquefaciens FZB42 and B. atrophaeus 1942, cotG lies within the cotH transcript (see Fig. S4 in the supplemental material). Additional experiments will be needed to clarify if a similar situation also occurs in geobacilli. Those results indicate that while the cotG-cotH gene organization is not prevalent among spore-forming bacilli, it is conserved in all organisms that contain a cotG ortholog, suggesting its common evolutionary origin. This hypothesis is also supported by the analysis of a phylogenetic tree based on CotH amino acid sequence, showing that the six CotG-containing strains joined the same cluster (see Fig. S5 in the supplemental material).
Table 1.
List of cotG genes retrieved from completely sequenced genomes
| Organism | Accession no. | E value | Protein length (aa) |
|---|---|---|---|
| B. subtilis subsp. subtilis strain 168 | NP_391488.1 | 4e−92 | 195 |
| B. subtilis subsp. spizizenii strain W23 | YP_003867887.1 | 1e−61 | 198 |
| B. amyloliquefaciens FZB42 | YP_001422884.1 | 6e−31 | 176 |
| B. atrophaeus 1942 | YP_003975035.1 | 2e−28 | 174 |
| Geobacillus sp. strain WCH70 | YP_002950034.1 | 6e−05 | 77 |
| Geobacillus sp. strain Y4.1MC1 | YP_003988838.1 | 5e−04 | 183 |
Structure, origin and evolution of the cotG gene.
A Clustal W analysis (38) performed on the six CotG sequences of Table 1 revealed that conserved amino acids were more abundant at the N-terminal region (amino acids 1 to 45) than at the C terminus and not apparent in the internal part of the proteins (see Fig. S6 in the supplemental material). When the same analysis was limited to CotG of bacilli, conserved amino acids were found also in the central parts of the proteins and were more apparent in the C-terminal regions (see Fig. S7 in the supplemental material).
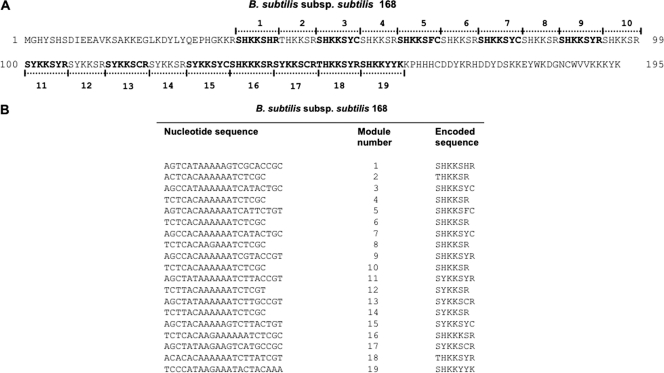
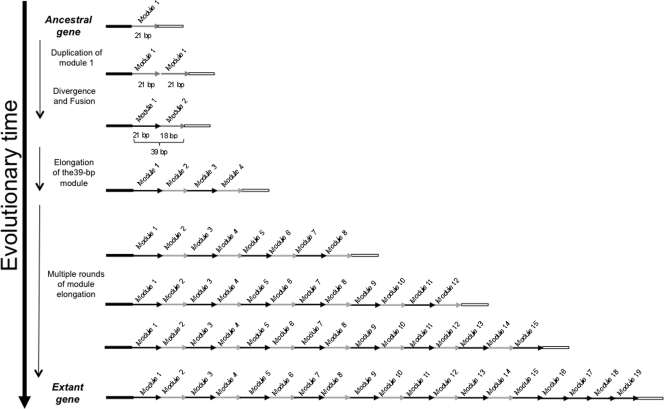
It has been previously reported that the central part of CotG of B. subtilis 168 is characterized by the presence of 9 repeats of 13 amino acids (34). We now suggest for that part of CotG a more complex organization with 7 tandem repeats of 7 and 6 amino acids followed by 5 repeats of 7 amino acids (Fig. 4A). At the DNA level, this part of cotG consists of 19 paralogous regions of two different sizes: 12 21-bp-length copies and 7 18-bp-length copies (Fig. 4B). The intriguing modular structure of the internal part of this gene suggests that it is the outcome of several rounds of gene elongation events of an ancestral module, as postulated for other bacterial genes (9, 10, 11, 12). A plethora of different molecular pathways might have occurred to originate the extant 19 modules, starting from a single ancestral one. Useful hints on the most probable pathway might be inferred by the analysis of the number of mismatches existing between the different modules (Tables 2, 3, and 4). As shown in these tables, most of the mutations fell in the 21-bp modules (modules 1, 3, 5, 7, 9, 11, 13, 15, 16, 17, 18, and 19), whereas the seven 18-bp modules (modules 2, 4, 6, 8, 10, 12, and 14) remained almost identical. In addition, comparing the tandem organized module pairs (1-2 versus 3-4 and 3-4 versus 5-6, etc.), the sequence divergence between each pair of 39-bp modules is very low.
Fig. 4.
(A) Amino acid sequence of CotG of B. subtilis subsp. subtilis 168. The 19 modules present in the central part of the protein are indicated. The 7-amino-acid modules are in bold. (B) The 19 paralogous regions, 12 of 21 bp and 7 of 18 bp, encoding the protein modules.
Table 2.
Numbers of mutations between the different 21-bp modules of the B. subtilis 168 cotG gene
| Module | No. of mutations in modulea: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3 | 5 | 7 | 9 | 11 | 13 | 15 | 16 | 17 | 18 | 19 | |
| 1 | — | 5 | 7 | 7 | 5 | 6 | 7 | 5 | 11 | 6 | 8 | 11 |
| 3 | — | 4 | 2 | 2 | 3 | 2 | 4 | 10 | 5 | 7 | 8 | |
| 5 | — | 2 | 2 | 3 | 2 | 4 | 9 | 8 | 6 | 11 | ||
| 7 | — | 1 | 1 | 2 | 3 | 10 | 6 | 7 | 9 | |||
| 9 | — | 3 | 2 | 4 | 12 | 7 | 5 | 9 | ||||
| 11 | — | 0 | 2 | 12 | 5 | 5 | 9 | |||||
| 13 | — | 2 | 13 | 4 | 8 | 10 | ||||||
| 15 | — | 13 | 6 | 6 | 11 | |||||||
| 16 | — | 10 | 8 | 9 | ||||||||
| 17 | — | 10 | 10 | |||||||||
| 18 | — | 10 | ||||||||||
| 19 | — | |||||||||||
Dashes are shown to indicate the same module in the row and column.
Table 3.
Numbers of mutations between the different 18-bp modules of the B. subtilis 168 cotG gene
| Module | No. of mutations in modulea: |
||||||
|---|---|---|---|---|---|---|---|
| 2 | 4 | 6 | 8 | 10 | 12 | 14 | |
| 2 | — | 1 | 0 | 0 | 1 | 0 | 0 |
| 4 | — | 0 | 0 | 1 | 0 | 0 | |
| 6 | — | 0 | 1 | 0 | 0 | ||
| 8 | — | 1 | 0 | 0 | |||
| 10 | — | 1 | 1 | ||||
| 12 | — | 0 | |||||
| 14 | — | ||||||
Dashes are shown to indicate the same module in the row and column.
Table 4.
Numbers of mutations between the different tandem modules of the B. subtilis 168 cotG gene
| Module pair | No. of mutations in module paira: |
|||||
|---|---|---|---|---|---|---|
| 3–4 | 5–6 | 7–8 | 9–10 | 11–12 | 13–14 | |
| 1–2 | 6 | 7 | 7 | 6 | 6 | 7 |
| 3–4 | — | 4 | 2 | 3 | 3 | 2 |
| 5–6 | — | 2 | 3 | 3 | 2 | |
| 7–8 | — | 2 | 1 | 2 | ||
| 9–10 | — | 4 | 3 | |||
| 11–12 | — | 0 | ||||
| 13–14 | — | |||||
Dashes are shown to indicate the same module in the row and column.
Assuming that the evolution of this gene has followed the most parsimonious pathway, data from Tables 2 to 4 suggest two alternative possibilities: either (i) the ancestral module was 21 bp long (module 1 in Fig. 4A) and elongated, giving rise to another module which underwent an evolutionary divergence consisting of the deletion of the last triplet and additional base pairs substitutions, resulting in a 18-bp module (module 2 of Fig. 4A), or (ii) the ancestral module was 18 bp long (module 2 in Fig. 4A) and elongated, giving rise to a new copy that diverged by acquiring a triplet (module 1 in Fig. 4A). Either way, modules 1 and 2 would have formed the first tandem module (1-2) of 39 bp, which in turn elongated, originating module 3-4, and the two copies diverged, differing by only 6 bp (5 base substitutions between modules 1 and 3 and 1 between modules 2 and 4). This evolutionary pathway predicts that the newly generated 39-bp module elongated until the construction of the first 14 modules. Then, the last part of the repeated region (consisting of five 21-bp repeats) would have originated via duplication of one or more 21-bp modules. Figure 5 shows a model of such an evolutionary process, considering the 21-bp module as the ancestral one. The last five modules (15, 16, 17, 18, and 19) are the most divergent ones; however, the traces of their origin from the other paralogous 21-bp long modules are evident.
Fig. 5.
Model for the origin and evolution of the cotG gene of Bacillus subtilis, considering the 21-bp module as the ancestral one.
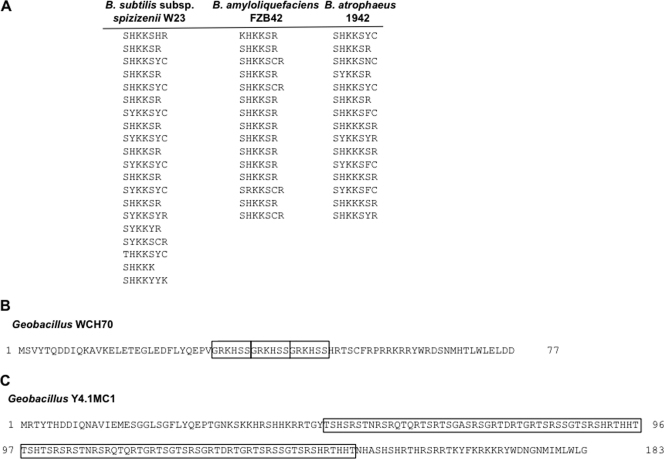
In all CotG-containing bacilli, CotG has a modular structure, although the numbers and the lengths of the repeats differ in the four microorganisms (Fig. 6A). Indeed, 20 repeats were found in the CotG of B. subtilis subsp. spizizeni W23, and there were 15 in B. amyloliquefaciens FZB42 and B. atrophaeus 1942. This finding suggests that, starting from the same ancestral gene, a different number of elongation events in the four different bacteria originated the extant cotG genes. A similar scenario can be depicted for CotG of the two CotG-containing Geobacillus strains, where the central part of CotG has a modular structure but the various modules do not share a significant degree of sequence identity/similarity with those of B. subtilis. In Geobacillus sp. strain WCH70, CotG is only 77 amino acids long (Table 1) and contains three repeats (Fig. 6B), while in Geobacillus sp. strain Y4.1MC1, CotG is 183 amino acids long and contains two almost identical repeats of 49 amino acids (Fig. 6C). Although those modules have a primary structure different from that present in CotG-containing bacilli, it is likely that also in this case, gene elongation events occurred during evolution to originate the extant cotG orthologs.
Fig. 6.
(A) Amino acid modules of CotG of B. subtilis subsp. spizizenii W23, B. amyloliquefaciens FZB42, and B. atrophaeus 1942. (B) Amino acid sequence of CotG of Geobacillus WCH70. The three identical modules of 6 amino acids present in the central part of the protein are boxed. (C) Amino acid sequence of CotG of Geobacillus Y4.1MC1. The two 49 repeats present in the central part of the protein are boxed.
DISCUSSION
A first result of this work is the identification of the cotH transcriptional start site 812 bp upstream of the first codon. A transcriptional start site for the cotH gene had been previously mapped 99 bp upstream of the first codon (28). However, we believe that the −99 site is not an internal promoter and that its erroneous identification was due either to RNA processing events or to the inhibition of cDNA synthesis during primer extension experiments (27). This hypothesis is supported by the following evidence: (i) the in silico analysis of the untranslated region showed the potentials for extensive secondary RNA structures (see Fig. S2 in the supplemental material), (ii) DNA fragments containing the upstream promoter, but not the putative internal promoter, were able to drive transcription (Fig. 1A and B, 2A, and 3B), and (iii) RT-PCR experiments failed to detect mRNA synthesis in fragments containing only the putative internal promoter (not shown).
While the correct position and sequence of the cotH promoter have been identified in this study, our results confirm previous data based on the analysis of a translational gene fusion (2) indicating that cotH transcription is driven by the sporulation-specific σ factor σK of the RNA polymerase and that the transcriptional regulator GerE acts as a repressor of cotH expression.
The long 5′ part of cotH mRNA (812 bp from +1 to the first codon) is most likely not translated in the direction of cotH. It contains several open reading frames (ORFs), but none of them has typical ribosome binding sites upstream of potential start codons. Additional experiments would be needed to definitely conclude that none of those ORFs is translated.
Long 5′ untranslated regions are not common in bacteria unless they are cis-acting regulatory RNA elements (riboswitches) that control the expression of the downstream coding regions. An average length of 360 bp has been determined for such regulatory elements in B. subtilis (19). The 5′ untranslated part of cotH mRNA is unusually long (812 bp) and can form secondary structures potentially involving the cotH ribosome binding site (see Fig. S2 in the supplemental material) but does not seem to have a regulatory role on the downstream coding region. Although we cannot rule out the possibility that the 5′ untranslated region of cotH mRNA may have a regulatory role in particular environmental conditions, data reported in Fig. 3 do not show any effect on CotH synthesis in standard laboratory conditions (Materials and Methods).
The unusual size of the 5′ end of cotH and the lack of a regulatory role in the analyzed conditions were puzzling and induced us to analyze in more detail the presence of the divergently transcribed gene cotG, entirely contained between the cotH transcriptional and translational start sites (Fig. 1A). The analysis of the structure, organization, and phylogenetic distribution of cotG revealed that it is most likely the outcome of gene elongation events involving an ancestral module. Gene elongation increases the size of genes by duplication of internal motifs and represents one of the most important mechanisms in the evolution of complex genes from simple ones (8, 11). A gene elongation event can be the outcome of an in-tandem duplication of a DNA sequence. When the elongation event involves an entire gene, a deletion of the intervening sequence between the two copies occurs, and this is followed by a mutation converting the stop codon of the first copy into a sense codon and resulting in the fusion of the two gene copies (7). When the elongation event involves only an internal region of a gene, as in the case of cotG, a gene rearrangement occurs to position the duplicated sequences in the same frame. Examples of tandem arrays of multiple short repeats within a gene in pathogenic bacteria have been described (27). In those cases, low-complexity regions have been hypothesized to originate by mutational processes, such as slipped-strand mispairing and unequal crossing-over taking place during DNA replication (27). Two or more paralogous moieties (modules) that constitute or are contained in the new gene may diverge and undergo further duplication events, leading to a gene constituted by or containing more repetitions. The biological significance of gene elongation might rely on (i) the improvement of the function of a protein by increasing the number of active sites and/or (ii) the acquisition of an additional function by modifying a redundant segment. Examples of genes sharing internal sequence repetitions have been described in both prokaryotes and eukaryotes (see reference 14 and references therein). The most extensively documented example in prokaryotes is represented by the pair of genes hisA and hisF, showing an evident split into two modules half the size of the entire gene (8, 9, 12).
All six CotG-containing strains have significantly different GC contents in their cotG genes with respect to their entire genomes (Table 5). In particular, in the four bacilli, the GC content of cotG is lower than that in their entire genomes, while in the two geobacilli, the GC content of the gene is higher than that in the genome (Table 5). However, this difference is mainly due to the repeated part of the cotG genes of geobacilli that have GC contents significantly higher than that of their entire cotG genes (Table 5).
Table 5.
GC contents of cotG genes compared to those in the corresponding entire genomes
| Organism | % GC |
||
|---|---|---|---|
| Entire cotG | Repeated region of cotG | Entire genome | |
| Bacillus subtilis subsp. subtilis strain 168 | 38.3 | 36.7 | 43.5 |
| Bacillus subtilis subsp. spizizenii strain W23 | 36.3 | 36.4 | 44.0 |
| Bacillus amyloliquefaciens FZB42 | 41.4 | 42.3 | 46.5 |
| Bacillus atrophaeus 1942 | 36.3 | 37.0 | 43.0 |
| Geobacillus sp. strain WCH70 | 47.9 | 59.2 | 42.8 |
| Geobacillus sp. strain Y4.1MC1 | 51.9 | 60.4 | 44.0 |
At the protein level, homologies are found in the N- and C-terminal regions of all analyzed CotG proteins (see Fig. S6 in the supplemental material), while the central part shows homologies only excluding the Geobacillus strains from the analysis (see Fig. S7 in the supplemental material). Based on this, on the narrow phylogenetic distribution of cotG, and on the different GC contents of cotG genes with respect to their entire genomes, we hypothesize that an ancestral cotG gene was constituted by the 5′ and 3′ regions and by an internal low-complexity region acting as an ancestral module. This ancestral minigene was either acquired by bacilli and geobacilli through independent horizontal gene transfer events from a still-unknown microorganism or acquired by a common ancestor of all CotG-containing bacteria. The latter hypothesis is supported by the chromosomal organization of cotG-cotH locus, similar in all six CotG-containing microorganisms. The internal low-complexity region of cotG genes would have then evolved independently through elongation events.
In B. subtilis, cotG is entirely contained between the promoter and coding part of cotH, and a similar situation has been observed also in B. amyloliquefaciens and B. atrophaeus. It is not clear whether this chromosomal organization has given a selective advantage to the cell; however, this event has been fixed by evolution. It is instead clear that, soon after the insertion of cotG, the cell has adjusted the regulation of the cotG-cotH expression in order to avoid the collision of RNA polymerase molecules during transcription of cotG and cotH. Although the genome organization of the cotH-cotG cluster is conserved and is likely to have occurred only once during evolution, we still do not know whether in the geobacilli the presence of cotG has caused the detachment of the promoter from the coding part of cotH, and further studies will be needed to address this point.
Supplementary Material
ACKNOWLEDGMENTS
We thank L. Di Iorio for technical assistance.
This work was supported by a grant (KBBE-2007-207948) from the EU 7th Framework to E.R.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 7 October 2011.
REFERENCES
- 1. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baccigalupi L., et al. 2004. GerE-independent expression of cotH leads to CotC accumulation in the mother cell compartment during Bacillus subtilis sporulation. Microbiology 150:3441–3449 [DOI] [PubMed] [Google Scholar]
- 3. Cangiano G., et al. 2010. Direct and indirect control of late sporulation genes by GerR of Bacillus subtilis. J. Bacteriol. 192:3406–3413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cutting S., Vander Horn P. B. 1990. Genetic analysis, p. 27–74 In Harwood C., Cutting S. (ed.), Molecular biological methods for Bacillus. John Wiley and Sons, Chichester, United Kingdom [Google Scholar]
- 5. Driks A., Roels S., Beall B., Moran C. P., Jr., Losick R. 1994. Subcellular localization of proteins involved in the assembly of the spore coat of Bacillus subtilis. Genes Dev. 8:234–244 [DOI] [PubMed] [Google Scholar]
- 6. Eichenberger P., et al. 2004. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2(10):e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fani R. 2004. Gene duplication and gene loading, p. 67–81 In Miller R. V., Day M. J. (ed.), Microbial evolution: gene establishment, survival, and exchange. ASM Press, Washington, DC [Google Scholar]
- 8. Fani R., Fondi M. 2009. Origin and evolution of metabolic pathways. Phys. Life Rev. 6:23–52 [DOI] [PubMed] [Google Scholar]
- 9. Fani R., Liò P., Chiarelli I., Bazzicalupo M. 1994. The evolution of the histidine biosynthetic genes in prokaryotes: a common ancestor for the hisA and hisF genes. J. Mol. Evol. 38:489–495 [DOI] [PubMed] [Google Scholar]
- 10. Fani R., Brilli M., Fondi M., Liò P. 2007. The role of gene fusions in the evolution of metabolic pathways: the histidine biosynthesis case. BMC Evol. Biol. 7(Suppl. 2):S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fondi M., Emiliani G., Fani R. 2009. Origin and evolution of operons and metabolic pathways. Res. Microbiol. 160:502–512 [DOI] [PubMed] [Google Scholar]
- 12. Fondi M., Emiliani G., Lio P., Gribaldo S., Fani R. 2009. The evolution of histidine biosynthesis in archaea: insights into the his genes structure and organization in LUCA. J. Mol. Evol. 69:512–526 [DOI] [PubMed] [Google Scholar]
- 13. Fritze D. 2004. Taxonomy and systematics of the aerobic endospore forming bacteria: Bacillus and related genera, p. 17–34 In Ricca E., Henriques A. O., Cutting S. M. (ed.), Bacterial spore formers. Horizon Bioscience, Norfolk, United Kingdom [Google Scholar]
- 14. Gao L., Lynch M. 2009. Ubiquitous internal gene duplication and intron creation in eukaryotes. Proc. Natl. Acad. Sci. U. S. A. 106:20818–20823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Halberg R., Kroos L. 1994. Sporulation regulatory protein SpoIIID from Bacillus subtilis activates and represses transcription by both mother-cell-specific forms of RNA polymerase. J. Mol. Biol. 243:425–436 [DOI] [PubMed] [Google Scholar]
- 16. Henriques A. O., Moran C. P., Jr 2007. Structure, assembly and function of the spore surface layers. Annu. Rev. Microbiol. 61:555–588 [DOI] [PubMed] [Google Scholar]
- 17. Horton R. M., Hunt H. D., Ho. S. N., Pullen, Pease J. K. R. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61–68 [DOI] [PubMed] [Google Scholar]
- 18. Ichikawa H., Halberg R., Kroos L. 1999. Negative regulation by the Bacillus subtilis GerE protein. J. Biol. Chem. 274:8322–8327 [DOI] [PubMed] [Google Scholar]
- 19. Irnov I., Sharma C. M., Vogel J., Winkler W. C. 2010. Identification of regulatory RNAs in Bacillus subtilis. Nucleic Acids Res. 38:6637–6651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Isticato R., Pelosi A., De Felice M., Ricca E. 2010. CotE binds to CotC and CotU and mediates their interaction during spore coat formation in Bacillus subtilis. J. Bacteriol. 192:949–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim H., et al. 2006. The Bacillus subtilis spore coat protein interaction network. Mol. Microbiol. 59:487–502 [DOI] [PubMed] [Google Scholar]
- 22. Kimura M. 1980. Simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111–120 [DOI] [PubMed] [Google Scholar]
- 23. Krajcíková D., Lukácová M., Müllerová D., Cutting S. M., Barák I. 2009. Searching for protein-protein interactions within the Bacillus subtilis spore coat. J. Bacteriol. 191:3212–3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Levin P. A., et al. 1993. An unusually small gene required for sporulation by Bacillus subtilis. Mol. Microbiol. 9:761–771 [DOI] [PubMed] [Google Scholar]
- 25. McKenney P. T., et al. 2010. A distance-weighted interaction map reveals a previously uncharacterized layer of the Bacillus subtilis spore coat. Curr. Biol. 20:934–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mota L. J., Tavares P., Sá-Nogueira I. 1999. Mode of action of AraR, the key regulator of L-arabinose metabolism in Bacillus subtilis. Mol. Microbiol. 33:476–489 [DOI] [PubMed] [Google Scholar]
- 27. Moxon E. R. 1999. Whole-genome analysis of pathogens, p. 191–204 In Stearns S. C. (ed.), Evolution in health and disease. Oxford University Press, New York, NY [Google Scholar]
- 28. Naclerio G., Baccigalupi L., Zilhao R., De Felice M., Ricca E. 1996. Bacillus subtilis spore coat assembly requires cotH gene expression. J. Bacteriol. 178:4375–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nicholson W. L., Setlow P. 1990. Sporulation, germination and outgrowth, p. 391–450 In Harwood C., Cutting S. (ed.), Molecular biological methods for Bacillus. John Wiley and Sons, Chichester, United Kingdom [Google Scholar]
- 30. Ramamurthi K. S., Clapham K. S., Losick R. 2006. Peptide anchoring spore coat assembly to the outer forespore membrane in Bacillus subtilis. Mol. Microbiol. 62:1547–1557 [DOI] [PubMed] [Google Scholar]
- 31. Ramamurthi K. S., Losick R. 2008. ATP-driven self-assembly of a morphogenetic protein in Bacillus subtilis. Mol. Cell 31:406–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ricca E., Cutting S., Losick R. 1992. Characterization of bofA, a gene involved in inter-compartmental regulation of pro-sK processing during sporulation in Bacillus subtilis. J. Bacteriol. 174:3177–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roels S., Driks A., Losick R. 1992. Characterization of spoIVA, a sporulation gene involved in coat morphogenesis in Bacillus subtilis. J. Bacteriol. 174:575–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sacco M., Ricca E., Losick R., Cutting S. 1995. An additional GerE-controlled gene encoding an abundant spore coat protein from Bacillus subtilis. J. Bacteriol. 177:372–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 36. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning, a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 37. Tamura K., et al. 2011. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 doi:10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Youngman P., Perkins J. B., Losick R. 1984. A novel method for the rapid cloning in Escherichia coli of Bacillus subtilis chromosomal DNA adjacent to Tn917 insertion. Mol. Gen. Genet. 195:424–433 [DOI] [PubMed] [Google Scholar]
- 40. Zheng L., Donovan W. P., Fitz-James P. C., Losick R. 1988. Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilis endospore. Genes Dev. 2:1047–1054 [DOI] [PubMed] [Google Scholar]
- 41. Zilhão R., et al. 1999. Assembly requirements and role of CotH during spore coat formation in Bacillus subtilis. J. Bacteriol. 181:2631–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zilhão R., et al. 2004. Interactions among CotB, CotG, and CotH during assembly of the Bacillus subtilis spore coat. J. Bacteriol. 186:1110–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.