Abstract
The chvE-gguABC operon plays a critical role in both virulence and sugar utilization through the activities of the periplasmic ChvE protein, which binds to a variety of sugars. The roles of the GguA, GguB, and GguC are not known. While GguA and GguB are homologous to bacterial ABC transporters, earlier genetic analysis indicated that they were not necessary for utilization of sugars as the sole carbon source. To further examine this issue, in-frame deletions were constructed separately for each of the three genes. Our growth analysis clearly indicated that GguA and GguB play a role in sugar utilization and strongly suggests that GguAB constitute an ABC transporter with a wide range of substrates, including l-arabinose, d-fucose, d-galactose, d-glucose, and d-xylose. Site-directed mutagenesis showed that a Walker A motif was vital to the function of GguA. We therefore propose renaming gguAB as mmsAB, for multiple monosaccharide transport. A gguC deletion affected growth only on l-arabinose medium, suggesting that gguC encodes an enzyme specific to l-arabinose metabolism, and this gene was renamed araD1. Results from bioinformatics and experimental analyses indicate that Agrobacterium tumefaciens uses a pathway involving nonphosphorylated intermediates to catabolize l-arabinose via an l-arabinose dehydrogenase, AraAAt, encoded at the Atu1113 locus.
INTRODUCTION
The soilborne bacterium Agrobacterium tumefaciens is a Gram-negative plant pathogen that is found in a wide range of environments around the world (for a review, see reference 8). In its saprophytic state, Agrobacterium depends on—and competes for—a wide variety of sugars as well as other nutrients that may be available in the soil. In this report, we present experiments that help us understand the role played by the chvE-gguABC operon in such processes. Specifically, the activities of mutants deficient in gguA, gguB, and gguC support the hypothesis that the first two of these genes encode proteins involved in sugar uptake, while the GguC protein is involved in an arabinose metabolic pathway that also involves the activity of an arabinose dehydrogenase encoded at gene locus Atu1113.
Consistent with its occurrence in an environment that often results in a paucity of nutrients, A. tumefaciens appears to have evolved a variety of high-affinity uptake systems for the acquisition of nutrients. The genome sequence of A. tumefaciens indicates that it has 153 complete ATP-binding cassette (ABC) transport systems plus additional “orphan” subunits, which account for 60% of its total transporter complement (35). This compares with 57 ABC transporters in Escherichia coli (20). ABC transporters are used by organisms to transport substrates from the surrounding environment into cytoplasm, and they exist widely in organisms from bacteria to humans and have a broad range of substrates (for reviews, see references 15 and 26). In terms of structure, ABC transporters consist of two nucleotide-binding domains (NBDs) and two transmembrane domains (TMDs). These domains can be individual proteins or separate domains in one protein. The NBDs are involved in binding and hydrolysis of ATP, and they are highly conserved at the primary sequence level. Based on sequence and function, NBDs can be divided into three short sequence motifs: Walker A, Walker B, and the signature motif containing the sequence LSGGQ (26).
In A. tumefaciens the chvE-, gguA-, and gguB-encoded products are predicted to constitute a complete binding-protein-dependent ABC transporter. ChvE is a periplasmic sugar-binding protein homologous to the sugar-binding proteins involved in ABC transporter systems of numerous well-characterized sugar uptake systems (14). Besides its role in sugar utilization, ChvE also plays a role in the control of virulence (vir) gene expression from the tumor-inducing (Ti) plasmid. This occurs via a proposed interaction between ChvE and the periplasmic domain of the histidine kinase, VirA, resulting in increased sensitivity of that protein for inducing phenolics as well as causing an increase in maximal levels of vir gene expression at saturating levels of the phenolics (for a review, see reference 21). GguA and GguB are predicted to be an ATP-binding protein and a transmembrane protein, respectively. They belong to the carbohydrate uptake transporter 2 (CUT2) family, which transports only monosaccharides (28). Unlike classic ABC transporters, the integral membrane protein in the CUT2 family contains more transmembrane segments (usually at least 10), and the ATPase protein has two NBDs, in which arginine replaces lysine in the Walker A motif of the C-terminal NBD (28). Like AraG/RbsA family members (28), GguA contains two NBDs. Like most of the transmembrane components in the CUT2 family, GguB is predicted to contain 11 transmembrane domains (http://www.cbs.dtu.dk/services/TMHMM/). GguC, the last gene product encoded by the chvE-gguABC operon, has an as-yet-uncharacterized function.
Kemner et al. reported that insertion mutations in gguA, gguB, or gguC did not affect growth of A. tumefaciens in the presence of sugars such as galactose, glucose, and arabinose (17). However, our recent studies clearly indicate that ChvE can bind numerous different monosaccharides and is important in sugar utilization (14). Given that their amino acid sequences suggest that GguA and GguB are typical of sugar transporters, we sought to unravel this conflict and better understand the role of the chvE-gguABC operon in sugar utilization. Toward this end, we generated strains with deletions of gguA, gguB, or gguC and characterized their growth in medium containing various sugars. The data presented here indicate that GguA and GguB are, in fact, involved in the utilization of multiple sugars. Moreover, site-directed mutagenesis confirmed the importance of the putative ATP binding site of GguA. Therefore, we propose renaming gguAB as mmsAB, for multiple monosaccharide transport. Growth analysis also indicated that gguC is involved in arabinose utilization but not utilization of other sugars. Further investigation indicates that in A. tumefaciens, l-arabinose is catabolized by a pathway involving nonphosphorylated intermediates via an arabinose dehydrogenase encoded at gene locus Atu1113 on the chromosome of A. tumefaciens A348. In keeping with nomenclature of the gene in Azospirillum brasiliense (33), we name the gene at locus Atu1113 araAAt.
MATERIALS AND METHODS
Bacterial growth assays.
A. tumefaciens strains (Table 1) were first grown in Luria-Bertani (LB) medium (22) at 25°C. After 20 h of growth, the cultures were spun down, resuspended, and diluted to an optical density at 600 nm (OD600) of 0.1 in AB minimal medium (5) with 10 mM glycerol (Invitrogen) as the sole carbon source. The cultures were grown for another 20 h and then diluted to an OD600 of 0.06 in AB minimal medium in the presence of different sugars as the sole carbon source. The carbon sources, l-arabinose (Sigma), d-fucose (Sigma), d-galactose (Fisher Scientific), d-glucose (Fisher Scientific), glycerol, and d-xylose (Sigma), were used at 3 mM. Cultures were incubated with shaking at 25°C at 200 rpm. At intervals, OD600 measurements were taken with a spectrophotometer (Beckman DU 520). No growth was observed when the strains checked were incubated in minimal medium without a carbon source.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristics | Reference or source |
|---|---|---|
| E. coli XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac[F′ proAB lacIqZΔM15 Tn10 (Tcr)] | Stratagene |
| A. tumefaciens strains | ||
| A348 | C58 background carrying pTiA6 | 13 |
| AB510 | A348, gguA deletion | This study |
| AB520 | A348, gguB deletion | This study |
| AB530 | A348, gguC deletion | This study |
| AB540 | A348, gguAB deletion | This study |
| AB533 | A348, but with RGS-6His-tagged gguC | This study |
| AB600 | A348, with gene locus Atu1113 deletion | This study |
| AB601 | A348 but with RGS-6His tag at the N terminus of the Atu1113-encoded protein | This study |
| AB511 | A348, gguA and gbpR deletion | This study |
| Plasmids | ||
| pYW15b | Broad-host-range expression vector, PN25-MCS-STOP, IncW, and pBR322ori, Ampr | 30 |
| pBBR1MCS-5 | Broad-host-range plasmid cloning vector; Gmr | 18 |
| pK18mobsacB | Suicide plasmid in Xanthomonas campestris pv. campestris; Kmr | 27 |
| pJZ10 | gguA deletion plasmid in pK18mobsacB | This study |
| pJZ11 | gguB deletion plasmid in pK18mobsacB | This study |
| pJZ12 | gguC deletion plasmid in pK18mobsacB | This study |
| pJZ13 | gguAB deletion plasmid in pK18mobsacB | This study |
| pJZ14 | Gene locus Atu1113 deletion plasmid in pK18mobsacB | This study |
| pJZ15 | RGS-6His-tagged gguC recombination plasmid in pK18mobsacB | This study |
| pJZ16 | PN25-gguC in pYW15b | This study |
| pJZ17 | PN25-Atu1113 in pYW15b | This study |
| pJZ18 | pBBR1MCS-5+P2+gguA | This study |
| pJZ19 | pBBR1MCS-5+P2+gguB | This study |
| pJZ20 | pBBR1MCS-5+P2+gguA-His | This study |
| pJZ21 | pBBR1MCS-5+P2+gguA(K44A)-His | This study |
| pJZ22 | RGS-6His-tagged gene locus Atu1113 recombination plasmid in pK18mobsacB | This study |
Deletion strain construction.
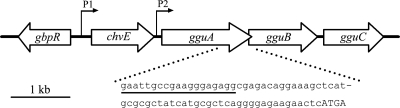
We used marker exchange eviction mutagenesis (27) to create strains carrying nonpolar deletions of gguA, gguB, gguAB, gguC, gguA plus gbpR, and the Atu1113 locus in the chromosome of A348 (13). Construction of these deletions used sequential-pair PCRs (16) and the suicide plasmid pK18mobsacB (27). For the gguA mutation, a 4-nucleotide (nt) overlap region between the stop codon of gguA and the start codon of gguB (Fig. 1) necessitated retaining the 3′ end of gguA in order to keep the start codon and ribosome binding site of gguB intact and allow expression of gguB. Primer P4, located 71 nt upstream from the start codon of gguB (within gguA), was used with P3 (located within gguB) to amplify a 675-bp fragment containing the downstream gguA-flanking sequence and the 3′ end of gguA. PCR to amplify the upstream gguA-flanking sequence used primers P1 and P2 to create a 600-bp product that included promoter P2. The 5′ ends of primers P4 and P1 had been designed to include complementary sequence so that the products from the first two PCRs could function as self-annealing templates in a third PCR with primers P3 and P4 to generate a 1.3-kb fragment carrying gguA-flanking sequences and a small segment at the 3′ end of gguA. This 1.3-kb PCR fragment was digested with HindIII and EcoRI and cloned into pK18mobsacB. The resulting construct pJZ10 was electroporated into A348. Colonies which underwent an initial recombination event were identified by their ability to grow on LB medium with kanamycin. Cells which underwent a second recombination, which is predicted to remove all vector sequences, were identified by growth on LB medium with 10% sucrose. A resulting isolate, containing the putative nonpolar gguA null mutation, was confirmed with PCR using primers that were (i) outside the region of exchange, which yielded a smaller PCR product than PCR with the wild type as the template, and (ii) internal to the gguA region, which yielded no PCR product. This gguA-null mutant was named AB510.
Fig. 1.
Gene map of the chvE-gguABC region of A. tumefaciens A348. The positions of promoters P1 and P2 are indicated. Arrows indicate the direction in which the genes are expressed. The enlarged region shows the 4-nt overlap sequence (uppercase) of start codon of gguB (ATG) and stop codon of gguA (TGA) and the position of gguA deletion primer P4 (underlined sequence).
Similar strategies were used to construct the gguB, gguC, gguAB, gguA plus gbpR, and gene locus Atu1113 nonpolar deletion strains AB520, AB530, AB540, AB511, and AB600, respectively. The primers used in these strain constructions are shown in Table 2. For the gguB deletion, downstream and upstream gguB-flanking sequences were amplified using primer pairs P7-P8 and P9-P10, respectively. Note that an additional TCA sequence was added before the gguA-gguB overlap sequence ACTCAT in primer P9 to keep the stop codon of gguA intact. The third PCR was conducted using primers P7 and P10 with the two PCR products from the first PCRs as the template. For the gguC deletion, primer pairs P15-P16 and P17-P18 were used to amplify downstream and upstream gguC-flanking sequences, respectively. Primers P15 and P18 were further used for the third PCR. For the gguAB deletion, downstream and upstream gguAB-flanking sequences were amplified using primer pairs P7-P40 and P41-P2, respectively. The third PCR was conducted using primers P7 and P2 with the two PCR products from the first PCRs as the template. For the deletion of the gene at locus Atu1113, primer pairs P27-P28 and P29-P30 were used to amplify downstream and upstream Atu1113-flanking sequences, respectively. In the third PCR, primers P27 and P30 were used. The gbpR deletion was created in the AB510 (ΔgguA) background. Primer pairs P36-P37, P38-P39 were used to amplify downstream and upstream gbpR-flanking sequences, respectively. Primers P36 and P39 were further used in the third PCR. The final PCR products were cloned into pK18mobsacB separately and transferred into Agrobacterium for homologous recombination. All mutant strains thus obtained were confirmed by the PCR strategy described above for AB510.
Table 2.
Primers used in this study
| Primer | Sequence (5′ to 3′)a | Description |
|---|---|---|
| P1 | TCGGCAATTCATCGCAATCCAGCGCCCG | Deletion of gguA |
| P2 | ACAGAATTCGACAGCGGCAAGCTGGTC | Deletion of gguA |
| P3 | GAGTACAAGCTTCCAGATAGAACAGCGCGAC | Deletion of gguA |
| P4 | GGATTGCGATGAATTGCCGAAGGGAGAGG | Deletion of gguA |
| P5 | ACTGTCTAGATCAAGGTTGTTCCGTCCTAC | Construction of gguA and gguB complementation |
| P6 | TCAGGTACCTCATGCCGAACTCATGAGTTCTTC | Construction of gguA complementation |
| P7 | GAGTACAAGCTTACGGAGAGGTGAGAGCCG | Deletion of gguB |
| P8 | CATGAGTTGATCGGCCCAACTCCTAT | Deletion of gguB |
| P9 | GGGCCGATCAACTCATGAGTTCTTCTCCC | Deletion of gguB |
| P10 | ACAGAATTCCAAGTCCTATGGCCACAGG | Deletion of gguB |
| P12 | TGATAGCGCGCATCGCCCGATTAATTCAGACG | Construction of gguB complementation |
| P13 | TCAGGTACCTTGACTGCGATAGGAGTTGG | Construction of gguB complementation |
| P14 | AATTAATCGGGCGATGCGCGCTATCATGCGCT | Construction of gguB complementation |
| P15 | ACTGGATCCCATCCGCCATTATGGAAACC | Deletion of gguC |
| P16 | GCAGGTGCATCTGACGGACTGCGTATGA | Deletion of gguC |
| P17 | TCCGTCAGATGCACCTGCATCTTATGAAAG | Deletion of gguC |
| P18 | ACACTGCAGCATCGTGCAGAATCTGCTGA | Deletion of gguC |
| P19 | ACTGAGCTCATGCACCGCGTAGAAGAGA | Construction of gguC complementation |
| P20 | TCAGAAGCTTTCAGATCTGCCGAACCACG | Construction of gguC complementation |
| P21 | ACTGTCTAGATCAAGGTTGTTCCGTCCTAC | Construction of gguA(K44A)-His complementation |
| P22 | GCGGGGGCCTCGACCTTGATGAAGGTCCT | Construction of gguA(K44A)-His complementation |
| P23 | CAAGGTCGAGGCCCCCGCGCCGTTCTCAC | Construction of gguA(K44A)-His complementation |
| P24b | TCAGGTACCTTAGTGATGGTGATGGTGATGTGAGTTCTTCTCCCCTGAGC | Construction of gguA(K44A)-His complementation |
| P25c | GGTGCAATGAGAGGATCGCATCACCATCACCATCACCGCGTAGAAGAGACAAG | RGS-6His-tagged gguC in situ replacement |
| P26 | GCGATCCTCTCATTGCACCTGCATCTTATGA | RGS-6His-tagged-gguC in situ replacement |
| P27 | GGAAGATCTCGGAGCGCTTCTGAGACTG | Deletion of gene locus Atu1113 |
| P28 | CCTCTCCATGTGAGAAGCCTCAACGCTTCC | Deletion of gene locus Atu1113 |
| P29 | AGGCTTCTCACATGGAGAGGGATCCGTTTG | Deletion of gene locus Atu1113 |
| P30 | GAGTACAAGCTTCCGGACGTCATCGAAAGC | Deletion of gene locus Atu1113 |
| P31 | ACTGAGCTCATGTCAGCCACCAAGCTTGC | Construction of gene locus Atu1113 complementation |
| P32 | ACGCTGTCGACTCAGTCGTAGAACGAATCCAC | Construction of gene locus Atu1113 complementation |
| P33d | CGCGGCTCGCATCACCATCACCATCACTCAGCCACCAAGCTTGCCA | RGS-6His-tagged gene locus Atu1113 in situ replacement |
| P34 | ATGGTGATGCGAGCCGCGCATGGAGAGGGATCCGTTTG | RGS-6His-tagged gene locus Atu1113 in situ replacement |
| P35 | CGGCTCGAGCCGGACGTCATCGAAAGC | RGS-6His-tagged gene locus Atu1113 in situ replacement |
| P36 | GAGTACAAGCTTGATCTGGGACAGCTGGTTC | Deletion of gbpR |
| P37 | TTGGTCAGCTCATGCGCAGGTGGCTCAT | Deletion of gbpR |
| P38 | CTGCGCATGAGCTGACCAACACCTGACG | Deletion of gbpR |
| P39 | ACAGAATTCCGTTTTCAGCGGAAATGCT | Deletion of gbpR |
| P40 | GGATTGCGATTCGGCCCAACTCCTATCG | Deletion of gguAB |
| P41 | TTGGGCCGAATCGCAATCCAGCGCCCG | Deletion of gguAB |
Boldface type indicates engineered restriction sites.
Underlining indicates the added 6-His tag before the stop codon of gguA.
Underlining indicates the added RGS-6His tag after the start codon of gguC.
Underlining indicates the added RGS-6His tag after the start codon of the protein encoded by the gene locus Atu1113.
Constructions of plasmids used in complementation of the gguA, gguB, gguC, and gene locus Atu1113 deletions.
Besides the inducible promoter P1 upstream of chvE, a second promoter, P2, which drives transcription of gguABC, is located upstream of gguA (11, 17). For gguA complementation, primers P5 and P6 were used to amplify the gguA open reading frame with its promoter, P2. The 1.7-kb PCR fragment was digested with XbaI and KpnI and cloned into pBBR1MCS-5 (18), yielding pJZ18. For gguB complementation, primers P5 and P12 were used to amplify a 0.2-kb fragment carrying promoter P2. Primers P13 and P14 were used to amplify the 1.2-kb gguB open reading frame. The two PCR products described above were mixed and used as a template for a third PCR with primers P5 and P13 to obtain the gguB open reading frame driven by promoter P2. This 1.4-kb product was digested with XbaI and KpnI and cloned into pBBR1MCS-5, yielding pJZ19. For gguC complementation, primers P19 and P20 were used to amplify the 1.0-kb gguC open reading frame. The PCR product was digested with SacI and HindIII and then cloned into pYW15b, yielding pJZ16, in which gguC is driven by the constitutively expressed PN25 promoter (30). For gene locus Atu1113 complementation, primers P34 and P35 were used to amplify the 930-bp gene locus Atu1113. The PCR product was digested with SacI and SalI and cloned into pYW15b, yielding pJZ17.
RGS-6His-tagged gguC and RGS-6His-tagged gene locus Atu1113 in situ replacement.
In order to monitor the expression of GguC, we fused an RGS-6His tag to the N terminus of gguC and placed it at the original gguC location in the chromosome (RGS is arginine-glycine-serine). Two initial PCRs used primer pairs P15-P25 and P18-P26. Subsequently, the products from the first two PCRs were used as templates to create a fragment carrying the RGS-6His-tagged gguC gene in addition to 600 bp of upstream and 600 bp of downstream sequences. The latter PCR used primers P15 and P18. The final PCR fragment was digested with BamHI and PstI and was cloned into pK18mobsacB to create plasmid pJZ15. The plasmid was introduced into AB530, and transformants were screened as described above. A resulting isolate, containing N-terminal RGS-6His-tagged GguC, was confirmed with PCR using primers that were outside the region of mutagenesis, which yielded a larger PCR product than PCR with AB530 as the template. The sequence authenticity of the N-terminal RGS-6His-tagged GguC of this isolate was further verified by sequencing. The strain was named AB533.
The same method was also used to create an RGS-6His-tagged gene at the Atu1113 chromosomal locus using primers shown in Table 2. The first PCRs were done using primer pairs P27-P28 and P29-P30. Subsequently, primers P27 and P30 were used for overlap PCR with the first two PCR products as the template. The final PCR fragment was cloned into pK18mobsacB and subsequently introduced into AB600 for crossover events. The strain thus obtained with the RGS-6His at the N terminus of the protein product encoded by gene locus Atu1113 was named AB601, and it was confirmed by PCR and verified by sequencing as described for AB533.
Site-directed mutation in the ATP-binding site of GguA.
Overlap extension PCR was used to generate a mutation (K44A) in the GguA Walker A ATP-binding site. Primers P5 and P22 were used to amplify the promoter P2 of gguA and the first 44 codons of gguA. Primers P23 and P24 were used to amplify the remainder of gguA with the addition of six histidines at the C terminus immediately preceding the stop codon. These two PCR products were mixed and used as the template for the third PCR with primers P5 and P24. The third PCR product was digested by KpnI and XbaI and cloned into pBBR1MCS-5 to make pJZ21. As a control, the wild-type gguA was also amplified with primers P5 and P24 and cloned into pBBR1MCS-5 to make pJZ20.
A. tumefaciens CFE preparation.
A single colony of the tested strain was inoculated into LB medium and incubated with aeration at 25°C overnight. The cultures were then inoculated (OD600 = 0.1) into AB minimal medium with 10 mM l-arabinose. The cultures were grown approximately 20 h and collected for cell-free extract (CFE) preparation. Cell pellets were resuspended in 20 mM phosphate-buffered saline (PBS) (pH 7.0) buffer and subjected to sonication. After centrifugation at 13,000 × g for 15 min, the supernatant (CFE) was collected.
l-Arabinose dehydrogenase and l-arabinose isomerase assay.
l-Arabinose 1-dehydrogenase activity was assayed routinely in the direction of l-arabinose oxidation by measuring the production of NAD(P)H by monitoring absorbance at 340 nm at 25°C as previously described (31). The standard assay mixture contained 10 mM l-arabinose in 100 mM Tris-HCl (pH 9.0) buffer. The reaction was started by the addition of 10 mM NAD(P)+ solution (100 μl) with a final reaction volume of 1 ml. CFE protein concentrations were determined with the BCA protein assay kit (Pierce), and approximately 100 μg CFE was added to the 1-ml reaction mixture.
l-Arabinose isomerase activity was measured as described by Cribbs and Englesberg (7). CFE (40 μl) was incubated with 60 μl of assay mix (100 mM Tris-HCl [pH 7.5], 100 mM l-arabinose, 1 mM MnCl2) at 25°C for 30 min. The reaction was terminated with the addition of 0.9 ml of 100 mM HCl, and the cysteine-carbazole test was used to measure production of ribulose (10).
Immunoblot analysis of the expression of ChvE, GguC, GguA, GguA(K44A), and the l-arabinose dehydrogenase encoded at gene locus Atu1113.
Bacteria were grown in AB medium with various sugars as the sole carbon source. They were collected and adjusted to an OD600 of 20. After addition of sample buffer (19), they were boiled for 10 min. Samples (10 μl each) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and subsequently transferred to a polyvinylidene difluoride membrane (Millipore). For immunoblot analysis, penta-His mouse monoclonal antibody (Qiagen) was used to detect 6His-tagged GguA and GguA(K44A), while RGS-His mouse monoclonal antibody (Qiagen) was used to detect the RGS-6His-tagged protein encoded at gene locus Atu1113 and RGS-6His-tagged GguC. Proteins were visualized with horseradish peroxidase-conjugated anti-mouse immunoglobulin secondary antibody (Amersham) and ECL Plus (Amersham). To check the ChvE expression level, anti-ChvE rabbit antibody was used as the primary antibody (14).
RESULTS
gguA, gguB, and gguC mutants are differentially deficient in carbon source utilization.
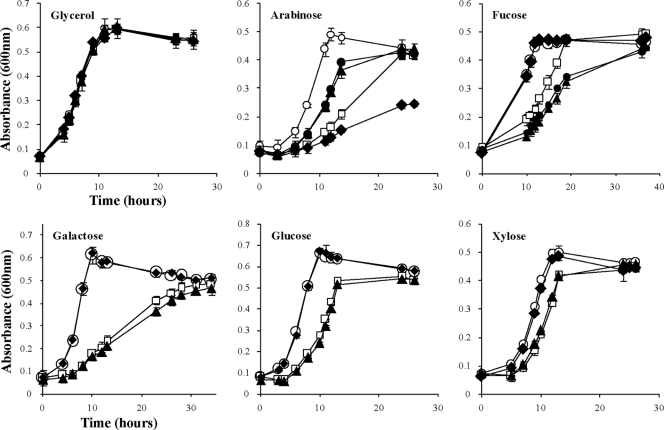
In-frame deletions in genes downstream from chvE (gguA, gguB, and gguC) were used to characterize sugar utilization in A. tumefaciens. For this purpose, mutants AB510 (ΔgguA), AB520 (ΔgguB), and AB530 (ΔgguC) were derived from wild-type strain A348. Because there is a 4-nt overlap region between the stop codon of gguA and the start codon of gguB (17) (Fig. 1), the deletion of gguA was constructed so that the start codon and ribosome-binding site of gguB remained intact (see Materials and Methods for details). The strains carrying the gguA, gguB, and gguC deletions were tested for growth on various sugars as the sole carbon source. For this purpose, mutant and wild-type (A348) strains were grown in AB medium supplied with either glycerol or one of several sugars (l-arabinose, d-fucose, d-galactose, d-glucose, or d-xylose). All of the mutant strains were able to utilize glycerol and grow as well as the wild-type strain (Fig. 2). Compared to A348, AB510 (ΔgguA) and AB520 (ΔgguB) grew slowly in each of the different sugar (3 mM)-containing media but reached the same OD600 as A348 after extended cultivation. These data are consistent with the idea that GguA and GguB constitute one transporter which can take up multiple monosaccharides, including l-arabinose, d-fucose, d-galactose, d-glucose, and d-xylose. The capacity of the mutant strains to grow on these sugars (albeit slowly) suggests that there is at least one other pathway for transporting these sugars. One unexpected observation was that while the strains carrying the gguA and gguB deletions exhibited virtually identical growth curves when provided galactose, glucose, or xylose as the sole carbon source, they exhibited different results when grown with arabinose or fucose. Intriguingly, when the double mutant (ΔgguAB) strain AB540 was tested for growth with arabinose or fucose, it exhibited the same growth phenotype as AB520 (ΔgguB) (Fig. 2). In contrast to the strains with gguA and gguB deletions, strain AB530 (ΔgguC) grew normally with all sugars tested with the sole exception of arabinose (Fig. 2). Not only did AB530 grow more slowly in arabinose, but it was unable to reach the same cell density as wild-type strain A348 even after cultivation was extended to 150 h (data not shown).
Fig. 2.
Growth curves of A. tumefaciens wild type and gguABC mutants in minimal medium containing different sugars. Bacteria were grown at 25°C in AB minimal medium with 3 mM glycerol or various monosaccharides as the sole carbon source. At intervals, the optical density at 600 nm of the cultures was determined. Strains used were the wild-type A348 (○), the gguA mutant AB510 (□), the gguB mutant AB520 (▴), the gguC mutant AB530 (♦), and the gguAB mutant AB540 (•). Note that AB540 was tested only in minimal medium with arabinose and fucose. Data are the averages of triplicate values with standard deviations.
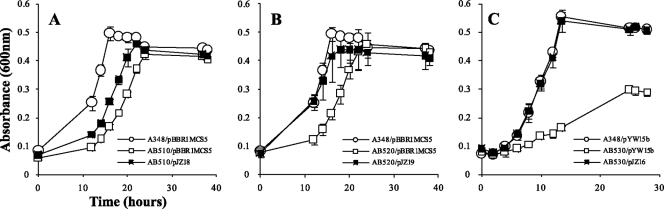
Experiments were carried out to determine whether mutant phenotypes conferred by the gguA, gguB, and gguC deletions could be complemented by adding back an intact version of the deleted gene. Interestingly, when the constitutively expressed promoter PN25 was used to drive expression of gguA or gguB, the strains grew very poorly (data not shown). Given that these constructs were on multicopy plasmids, it appears that their overexpression may be toxic. We therefore used the strategy employed in the analysis of gguA and gguB in Brucella suis, which used the P2 promoter of the operon (1). The data indicate that the gguA mutant (AB510) was partially complemented in trans with plasmid-borne gguA (pJZ18), whereas plasmid-borne gguB (pJZ19) fully complemented AB520 (ΔgguB) (Fig. 3). The partial complementation of the gguA deletion on strain AB510 by pJZ18 may be due to the lack of translational coupling between gguA and gguB that is suggested by the overlapping stop and start codons in the wild-type strain (Fig. 1). Such coupling is a common feature in operons encoding ABC transporters (3, 4, 25). Finally, plasmid pJZ16, in which gguC is driven by the constitutively expressed promoter PN25, fully restored growth of the gguC deletion mutant strain AB530 in minimal medium containing arabinose (Fig. 3).
Fig. 3.
Growth curves of complementation of gguABC mutants in AB minimal medium with 3 mM arabinose. (A) AB510 (gguA mutant) and (B) AB520 (gguB mutant) were complemented with pBBR1MCS-5 derived plasmids pJZ18 and pJZ19, respectively. (C) AB530 (gguC mutant) was complemented with pYW15b based plasmid pJZ16. Data are the averages of triplicate values with standard deviations.
K44 is necessary for GguA function.
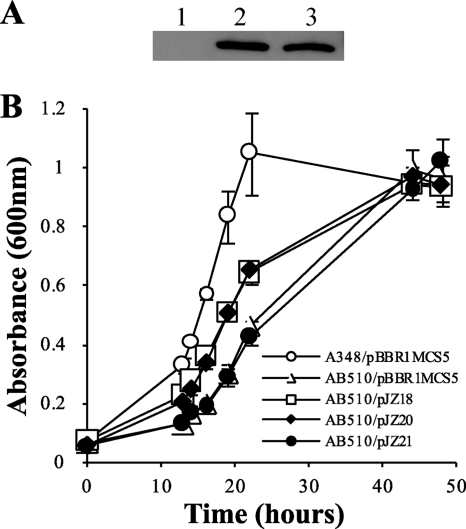
A typical Walker A motif (G38ENGAGKST46) is found in the N-terminal region of GguA, and the conserved lysine residue (codon 44) is important for binding ATP/ADP in other homologs (9). To further investigate the function of GguA, site-directed mutagenesis was employed to change lysine 44 to alanine. In order to compare the expression levels of GguA(K44A) and wild-type GguA, a 6His tag was added to the C terminus of both GguA(K44A) and wild-type GguA. Immunoblot analysis showed that GguA-His and GguA(K44A)-His expressed from the genes driven by the P2 promoter, were equally abundant (Fig. 4A). Growth assays showed that AB510/pJZ20 (6His-tagged wild-type gguA) grew similarly to AB510/pJZ18 (wild-type gguA) in arabinose medium, suggesting that the 6His tag did not affect the function of GguA. In contrast, strain AB510/pJZ21 [gguA(K44A)-His] demonstrated delayed growth in arabinose, similar to that of AB510 carrying the empty vector (Fig. 4B). These results strongly suggest that the K44 residue is vital for GguA function and that GguA is most likely an ATP-binding protein.
Fig. 4.
Effect of the K44A mutation of GguA on protein expression and strain growth ability. (A) Expression level of GguA in the gguA deletion strain AB510 carrying 6His-tagged wild-type gguA (pJZ20, lane 2), 6His-tagged K44A gguA mutant (pJZ21, lane 3), or vector control (pBBR1MCS-5, lane 1). (B) Growth curves of gguA mutant AB510 carrying wild-type gguA (pJZ18), 6His-tagged wild-type gguA (pJZ20), 6His-tagged K44A gguA mutant (pJZ21), or vector control (pBBR1MCS-5). The strains were grown in minimal medium containing 10 mM arabinose. Data are the averages of triplicate values with standard deviations.
Expression from the P2 promoter is induced by some sugars and regulated by GbpR.
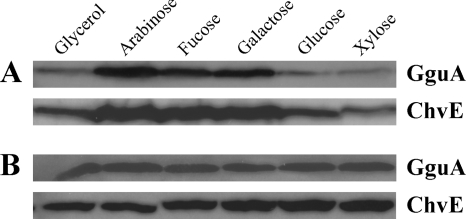
The chvE-gguABC operon carries two promoters. Expression from the P1 promoter located just 5′ to chvE was shown to be inducible by arabinose and galactose, while expression from the P2 promoter between chvE and gguA was previously suggested to be constitutively expressed and not induced by arabinose or other sugars tested (17). Plasmid pJZ20, carrying gguA-His controlled by promoter P2, was used to analyze P2 activity based on the expression of GguA. Immunoblot analysis was used to examine GguA abundance in response to different sugars. Surprisingly, GguA-His was upregulated in the presence of arabinose, fucose, and galactose (Fig. 5A). Doty et al. demonstrated that the arabinose-inducible promoter P1 is regulated by GbpR, a LysR-type regulator (11). The gbpR locus is adjacent to chvE but divergently transcribed. To determine whether expression from P2 is also regulated by GbpR, we constructed the gbpR and gguA double-deletion strain AB511. Plasmid pJZ20, harboring gguA-His, was introduced into this strain, and immunoblot analysis was used to detect GguA. In the absence of GbpR, GguA, like ChvE, was not subject to regulation by carbon source (Fig. 5B). These results strongly suggest that GbpR regulates promoter P2 as well as promoter P1.
Fig. 5.
GguA and ChvE expression in minimal medium containing 10 mM glycerol, arabinose, fucose, galactose, glucose, or xylose as the sole carbon source. The gguA deletion strain AB510 (A) or the gguA and gbpR deletion strain AB511 (B) carrying 6His-tagged wild-type gguA in pJZ21 was grown in minimal medium with the indicated sugars. After GguA was detected with mouse anti-penta-His antibody, the membrane was stripped and subsequently used to determine the ChvE level with rabbit anti-ChvE antibody.
As the growth assay presented in Fig. 2 shows, AB530 demonstrated defective growth only in media containing l-arabinose. We wished to determine whether gguC expression is also only induced by l-arabinose. To do this, we added an RGS-6His tag to the N terminus of gguC via double crossover as described in Materials and Methods. GguC was upregulated in minimal medium not only with arabinose but also with fucose and galactose (Fig. 6). This is consistent with the result obtained by Kemner et al. showing that P1 is the primary promoter of chvE-gguABC operon and is upregulated in the presence of arabinose, fucose, and galactose. It is also consistent with the result described above showing that P2 is also induced by these sugars.
Fig. 6.
GguC expression is induced by arabinose, galactose, and fucose. Strain AB533 with RGS-6His-tagged gguC was grown in minimal medium containing 10 mM glycerol, arabinose, fucose, galactose, glucose, or xylose as the sole carbon source. After 24 h cultivation, cells were spun down and adjusted to an OD of 20 in PBS buffer. Ten-microliter samples were loaded in each lane. GguC was detected with mouse anti-RGS-His antibody.
l-Arabinose degradation in A. tumefaciens occurs via a pathway lacking phosphorylated intermediates.
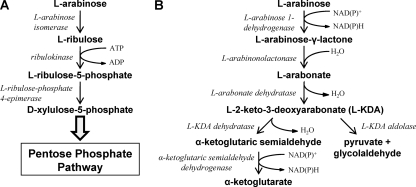
The sequence of GguC indicates that it has a fumarylacetoacetase (FAA) hydrolase domain at the C terminus (Pfam [http://pfam.sanger.ac.uk/]). A BLAST search also indicated that GguC is homologous to KdaD (2-keto-3-deoxy-d-arabinonate dehydratase), an enzyme involved in d-arabinose metabolism in Sulfolobus solfataricus (see Discussion). This, together with the observed growth defect of AB530 on l-arabinose medium, suggests that GguC is an enzyme involved in l-arabinose metabolism. In bacteria, there are two known pathways for l-arabinose metabolism. One pathway, consisting of phosphorylated intermediates, has been extensively studied. In many bacteria, including E. coli, l-arabinose isomerase, ribulokinase, and l-ribulose-phosphate 4-epimerase convert l-arabinose into d-xylulose-5-phosphate (23) (Fig. 7A). The second pathway (Fig. 7B), involving nonphosphorylated intermediates, utilizes l-arabinose 1-dehydrogenase, l-arabinonolactonase, and l-arabonate dehydratase to convert l-arabinose to L-2-keto-3-deoxyarabonate (l-KDA). Metabolism of l-KDA can follow two possible pathways. First, it may be converted into α-ketoglutarate via l-KDA dehydratase and α-ketoglutaric semialdehyde dehydrogenase, as in Azospirillum brasiliense (31–33). Alternatively, l-KDA aldolase can convert l-KDA into pyruvate and glycolaldehyde. A BLAST search did not identify homologs to E. coli genes involved in l-arabinose metabolism in the A. tumefaciens genome. However, several genes homologous to these involved in the alternative pathway found in A. brasiliense were found in the genome of A. tumefaciens (Table 3). Significantly, the product encoded by the gene locus Atu1113 has 58% identity to AraA (l-arabinose 1-dehydrogenase) in A. brasiliense (31). Furthermore, the residues D168 and N172, which are important for catalytic function in l-arabinose 1-dehydrogenase in A. brasiliense, were also found in the protein encoded by gene locus Atu1113. This suggests that A. tumefaciens degrades l-arabinose in a pathway involving nonphosphorylated intermediates.
Fig. 7.
Pathways of l-arabinose metabolism in bacteria. (A) The well-characterized and long-established phosphorylative pathway in E. coli and many other bacteria. The product d-xylulose-5-phosphate is further catabolized via the pentose phosphate pathway. (B) The nonphosphorylative pathway operative in A. brasiliense (first pathway) and in several other bacteria (second pathway).
Table 3.
Genes in A. tumefaciens homologous to those involved in l-arabinose metabolism in A. brasiliense
| ORF in A. brasiliense (enzyme name; lengtha) | Homolog in A. tumefaciensb | Lengtha | Expect value | % Identity/ranged | Annotation |
|---|---|---|---|---|---|
| araA (l-arabinose 1-dehydrogenase; 309) | Atu1113 | 308 | 2e-93 | 58/303 | Dehydrogenase |
| Atu5435 | 390 | 1e-07 | 33/132 | Oxidoreductase | |
| Atu3347 | 377 | 2e-07 | 24/251 | Hypothetical protein | |
| araB (l-arabinolactonase; 300) | Atu5464 | 560 | 6e-29 | 35/283 | IclR family transcriptional regulator |
| Atu0702 | 295 | 9e-26 | 34/274 | Calcium-binding protein, regucalcin | |
| Atu4190 | 293 | 1e-20 | 30/256 | Calcium-binding protein, regucalcin | |
| Atu3962 | 269 | 1e-20 | 29/272 | Calcium-binding protein, regucalcin | |
| Atu4029 | 306 | 3e-08 | 31/145 | Gluconolactonase precursor | |
| araC (l-arabonate dehydratase; 593) | Atu3971 | 574 | 3e-136 | 47/568 | Dihydroxy-acid dehydratase |
| Atu3219 | 603 | 1e-114 | 46/543 | Dihydroxy-acid dehydratase | |
| Atu2736 | 594 | 3e-103 | 41/593 | Dihydroxy-acid dehydratase | |
| Atu1918 | 611 | 8e-56 | 33/584 | Dihydroxy-acid dehydratase | |
| Atu0598 | 606 | 1e-33 | 27/518 | Phosphogluconate dehydratase | |
| araD (l-2-keto-3-deoxyarabonate dehydratase; 309) | Atu5457 | 302 | 7e-20 | 31/286 | Dihydrodipicolinate synthase |
| Atu1024 | 294 | 4e-11 | 27/137 | Dihydrodipicolinate synthase | |
| araE (α-ketoglutaric semialdehyde dehydrogenase; 481)c | Atu3498 | 478 | 2e-175 | 63/476 | Aldehyde dehydrogenase |
| Atu4247 | 484 | 2e-90 | 41/466 | Aldehyde dehydrogenase | |
| Atu3403 | 485 | 2e-86 | 41/467 | Succinate semialdehyde dehydrogenase | |
| Atu5137 | 484 | 6e-84 | 39/477 | NAD-dependent succinyl-semialdehyde dehydrogenase | |
| Atu4762 | 492 | 6e-78 | 39/466 | Succinate semialdehyde dehydrogenase |
Number of amino acids.
Accession number obtained from a BLAST search.
The top five significant hits from the BLAST search are shown.
Number of amino acids over which the identity exists.
To test the hypothesis that A. tumefaciens uses such a pathway to metabolize l-arabinose, we first looked for evidence of the enzyme activities that represent the first steps of these two different pathways, l-arabinose isomerase and l-arabinose 1-dehydrogenase. NAD+- and NADP+-dependent l-arabinose 1-dehydrogenase enzyme activity (138.50 ± 4.02 and 80.64 ± 3.31 nmol min−1 mg−1, respectively) was found in cell extracts of A. tumefaciens cells grown in AB medium with l-arabinose as the sole carbon source. In contrast, no l-arabinose isomerase activity was observed in A. tumefaciens, though the activity was readily observed in cell extracts from E. coli (data not shown). This result indicates that A. tumefaciens uses a pathway including nonphosphorylated intermediates to metabolize l-arabinose, starting with the enzyme l-arabinose dehydrogenase.
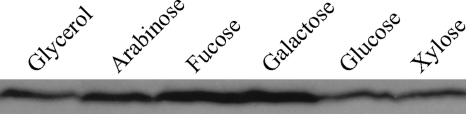
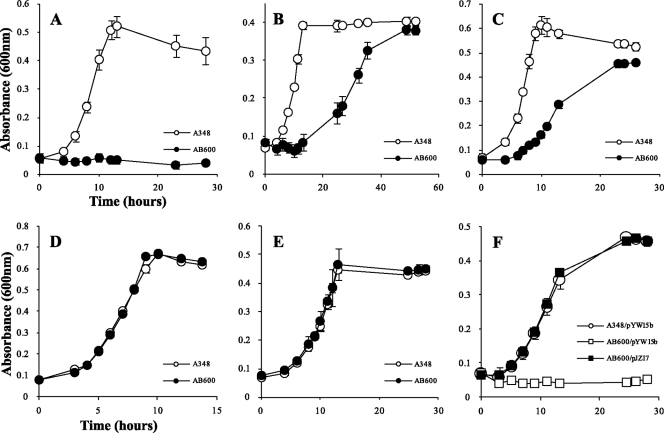
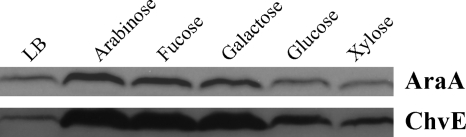
As noted above, the Atu1113 locus is homologous to araA, the gene encoding l-arabinose dehydrogenase, in A. brasiliense. We constructed AB600, which carries a deletion of the Atu1113 locus, and measured its growth in minimal medium with different sugars. AB600 did not grow with l-arabinose as the sole carbon source (Fig. 8), further supporting the suggestion that the gene at this locus is involved in l-arabinose degradation and that A. tumefaciens uses the nonphosphorylation pathway. For consistency and clarification, the Atu1113 locus was named araAAt. Unexpectedly, AB600 also grew poorly on d-galactose and very poorly on d-fucose (Fig. 8). However, the strain grew normally on galactose, glucose, and xylose. This indicates that AraAAt is also involved in d-fucose and d-galactose metabolism. The growth defect of AB600 was successfully complemented with plasmid pJZ17 (Fig. 8). Enzyme assays with overexpressed and purified AraAAt further confirmed that AraAAt is an arabinose, fucose, and galactose dehydrogenase (data not shown). To monitor the expression of AraAAt, an RGS-6His tag was added to the N terminus of AraAAt, and the construct was reintroduced to its original site on the chromosome to make strain AB601. This strain was used in assays to detect the expression of AraAAt in response to different carbon sources. Western blotting showed that AraAAt is induced by the metabolic substrates arabinose, fucose, and galactose (Fig. 9).
Fig. 8.
Growth curves of A. tumefaciens wild-type strain A348 and gene locus Atu1113 deletion strain AB600 (A, B, C, D, and E) and complementation strains (F) in minimal medium containing different sugars. Bacteria were grown at 25°C in AB minimal medium with 3 mM concentrations of the monosaccharides arabinose (A and F), fucose (B), galactose (C), glucose (D), and xylose (E) as the sole carbon sources. At intervals, the optical density at 600 nm of the cultures was determined. Data are the averages of triplicate values with standard deviations.
Fig. 9.
Comparison of AraAAt and ChvE expression. Strain AB601 with the RGS-His-tagged Atu1113 gene locus was grown in LB or in minimal medium containing 10 mM arabinose, fucose, galactose, glucose, or xylose as the sole carbon source. After AraA was detected with mouse anti-RGS-His antibody, the membrane was stripped and subsequently used to determine the ChvE level with rabbit anti-ChvE antibody.
DISCUSSION
A. tumefaciens is a plant pathogen routinely found as a free-living saprophyte in the soil, where it is likely to encounter both deficiency of and competition for nutrients. To adapt to this environment and compete with other soilborne microorganisms, the evolution of multiple transport systems for the uptake of nutrients appears to be important as a means to acquire these nutrients, thereby obtaining energy for growth and pathogenesis. In this study, we revisited the previously characterized operon chvE-gguABC. Our results indicate that (i) gguAB is involved in transport of several different monosaccharides and (ii) A. tumefaciens uses a pathway to degrade l-arabinose that does not involve phosphorylated intermediates, while gguC encodes an enzyme in this metabolic pathway.
Tests of several different sugars—l-arabinose, d-fucose, d-galactose, d-glucose, and d-xylose—as sole carbon sources by the gguA and gguB deletion strains (AB510 and AB520, respectively) (Fig. 2) strongly supports the hypothesis that gguAB is involved in the import of multiple substrates. These results contrast with those of Kemner et al. (17), who reported no growth differences between the wild type and strains carrying disruptions in gguA, gguB, or gguC. However, our liquid growth assay is more sensitive than the colony size assay used by Kemner et al. Furthermore, Kemner et al. (17) used a sugar concentration of 1 mM, whereas our studies employed a concentration of 3 mM. The K44A mutation in GguA eliminated that protein's capacity to complement the gguA deletion (Fig. 4B). Given that this lysine is conserved and essential for activity in all ATP-binding proteins that are components of ABC transporters as well as the growth studies referred to above, we suggest that the current name gguAB be changed to mmsAB for multiple monosaccharide transport. The first gene in chvE-mmsAB-gguC operon, chvE, was found to be a sugar-binding protein and can bind l-arabinose, d-fucose, d-galactose, d-glucose, and d-xylose (14). Because it was first identified as a protein involved in sugar-mediated control of virulence in A. tumefaciens, we suggest that the chvE name be retained. Based on gene location and these experimental results, ChvE-MmsAB apparently constitute a complete binding-protein-dependent ABC transporter involved in the uptake of several different sugars.
The finding that the mmsAB deletion strains still grow, albeit slowly, in medium supplied with various sugars suggests the existence of an additional, less efficient uptake system(s) for these sugars. Many substrates are transported into the cell by multiple transport systems. For example, in E. coli galactose is transported by at least five systems (34). In Agrobacterium radiobacter, two high-affinity glucose-binding proteins (GBP1 and GBP2) were also found to bind d-galactose and d-xylose, respectively, and were involved in the transport of these sugars (6). On the basis of the amino-terminal sequence, the GBP1 and GBP2 are encoded by the gene loci Arad_3358 and Arad_3364 in the A. radiobacter genome (29), respectively. A BLAST search using GBP1 and GBP2 sequences against the A. tumefaciens genome showed that GBP1 is homologous to ChvE, while GBP2 is homologous to the protein encoded by the gene locus Atu3576. This gene locus is a part of a larger gene locus encoding transmembrane and ATP-binding proteins. We can infer that A. tumefaciens can also use the ABC transporter encoded by the Atu3576-related locus to transport glucose and xylose.
In our studies, AB530 (ΔgguC) exhibited ligand-specific effects—growth in arabinose was vastly reduced, whereas growth in the other tested sugars was unaffected. Additionally, sequence analysis suggested that GguC has some homology to an enzyme involved in d-arabinose metabolism (see below). Two pathways for l-arabinose degradation by bacteria have been described. The first converts arabinose into a phosphorylated intermediate, while the second forms a reduced, nonphosphorylated intermediate that is subsequently metabolized. Our data indicate that A. tumefaciens uses the nonphosphorylation pathway to degrade l-arabinose. This pathway was recently characterized in A. brasiliense, and the corresponding genes have been cloned (31–33).
The enzyme activity assays with cell extracts and the growth assays with the araAAt mutant provide strong evidence that AraAAt is an l-arabinose dehydrogenase. Furthermore, the growth assay of AB600 (araAAt deletion strain) indicates that AraAAt is also involved in d-fucose and d-galactose metabolism. Consistent with the growth assays, accumulation of AraAAt was also shown by immunoblot analysis to be stimulated by these three metabolic sugars. In A. brasiliense, l-arabinose 1-dehydrogenase, encoded by araA, was also shown to oxidize other sugars, such as d-galactose and d-xylose, in vitro. In contrast to araAAt in A. tumefaciens, the expression of araA in A. brasiliense was found to be induced only by l-arabinose, and a strain with an araA disruption exhibited a growth defect only in a minimal medium with l-arabinose as the sole carbon source. Thus, it appears that araAAt has novel and substantial roles in the metabolism of at least arabinose, galactose, and fucose.
The enzymatic activity encoded by gguC remains unresolved. One clue concerning its possible activity comes from the characterization of the metabolic pathway for d-arabinose, the C4 epimer of l-arabinose in Sulfolobus solfataricus, a member of the Archaea (2). KdaD (2-keto-3-deoxy-d-arabinonate dehydratase) catalyzes the dehydration step from 2-keto-3-deoxy-d-arabinonate (d-KDA). Interestingly, GguC has some homology to KdaD (expect value, 2e-17; 34% identity over 184 residues), and both proteins have a fumarylacetoacetate hydrolase domain in their C termini. Based on sequence alignment, most of the active sites for metal and substrate binding were found in GguC (data not shown). We propose that GguC is functionally homologous to KdaD but uses l-KDA as the substrate (Fig. 7). An important observation, however, is that while a deletion of araAAt completely eliminates growth in l-arabinose, the deletion of gguC only slows growth in this sugar, suggesting redundancy in the pathway. One candidate for a second KdaD was revealed by a BLAST search. We found that the protein encoded at gene locus Atu5457 in A. tumefaciens is a homolog to AraD (expect value, 2e-20; 31% identity over 286 residues) (Table 3), the gene product 2-keto-3-deoxy-l-arabinonate dehydratase involved in catabolism of l-arabinose in A. brasiliense (33), and may have a similar function. Given the growth-defective phenotype of the gguC mutant (AB530) in arabinose medium, we hypothesize that A. tumefaciens uses two distinct but functionally similar enzymes to break down 2-keto-3-deoxy-l-arabinonate. To follow the nomenclature for 2-keto-3-deoxy-l-arabinonate dehydratase in A. brasiliense, we propose renaming gguC as araD1 and gene locus Atu5457 as araD2.
Biochemical studies have revealed that Sinorhizobium meliloti uses the same pathway as A. brasiliense to metabolize l-arabinose (12). Recently, by mutagenesis, the gene cluster araABCDEF was found to be necessary for l-arabinose utilization (24). Among this gene cluster, araABCD were homologous to chvE-mmsAB-araD1. While l-arabinose dehydrogenase activity was found in this strain, the gene for that enzyme has not yet been identified. Based on our BLAST search, we found that the gene product of SMc00588 has significant homology to AraAAt (expect value, 5e-140; 58% identity over 308 residues), making it a good candidate for l-arabinose dehydrogenase.
In conclusion, this study provides the first evidence for a pathway lacking phosphorylated intermediates to degrade l-arabinose in A. tumefaciens. Further study is still needed to elucidate the entire pathway for l-arabinose metabolism. Finally, our results suggest that l-arabinose is one of the substrates transported by the binding-protein-dependent ABC transport system ChvEMmsAB, which also transports d-fucose, d-galactose, d-glucose, and d-xylose.
ACKNOWLEDGMENTS
We thank Arlene A. Wise for critically reading an earlier version of the manuscript. We thank Erik Ronzone for the initial BLAST analysis indicating that GguC is homologous to KdaD of Sulfolobus solfataricus.
This work was partially funded by grant 0818613 from the National Science Foundation.
Footnotes
Published ahead of print on 7 October 2011.
REFERENCES
- 1. Alvarez-Martinez M. T., et al. 2001. The Brucella suis homologue of the Agrobacterium tumefaciens chromosomal virulence operon chvE is essential for sugar utilization but not for survival in macrophages. J. Bacteriol. 183:5343–5351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brouns S. J., et al. 2006. Identification of the missing links in prokaryotic pentose oxidation pathways: evidence for enzyme recruitment. J. Biol. Chem. 281:27378–27388 [DOI] [PubMed] [Google Scholar]
- 3. Camacho L. R., Ensergueix D., Perez E., Gicquel B., Guilhot C. 1999. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol. Microbiol. 34:257–267 [DOI] [PubMed] [Google Scholar]
- 4. Cheng J., Guffanti A. A., Krulwich T. A. 1997. A two-gene ABC-type transport system that extrudes Na+ in Bacillus subtilis is induced by ethanol or protonophore. Mol. Microbiol. 23:1107–1120 [DOI] [PubMed] [Google Scholar]
- 5. Chilton M. D., et al. 1974. Agrobacterium tumefaciens DNA and PS8 bacteriophage DNA not detected in crown gall tumors. Proc. Natl. Acad. Sci. U. S. A. 71:3672–3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cornish A., Greenwood J. A., Jones C. W. 1989. Binding-protein-dependent sugar transport by Agrobacterium radiobacter and A. tumefaciens grown in continuous culture. J. Gen. Microbiol. 135:3001–3013 [DOI] [PubMed] [Google Scholar]
- 7. Cribbs R., Englesberg E. 1964. l-arabinose negative mutants of the L-ribulokinase structural gene affecting the levels of l-arabinose isomerase in Escherichia coli. Genetics 49:95–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Decleene M., Deley J. 1976. Host range of crown gall. Bot. Rev. 42:389–466 [Google Scholar]
- 9. Deyrup A. T., Krishnan S., Cockburn B. N., Schwartz N. B. 1998. Deletion and site-directed mutagenesis of the ATP-binding motif (P-loop) in the bifunctional murine ATP-sulfurylase/adenosine 5′-phosphosulfate kinase enzyme. J. Biol. Chem. 273:9450–9456 [DOI] [PubMed] [Google Scholar]
- 10. Dische Z., Borenfreund E. 1951. A new spectrophotometric method for the detection and determination of keto sugars and trioses. J. Biol. Chem. 192:583–587 [PubMed] [Google Scholar]
- 11. Doty S. L., Chang M., Nester E. W. 1993. The chromosomal virulence gene, chvE, of Agrobacterium tumefaciens is regulated by a LysR family member. J. Bacteriol. 175:7880–7886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Duncan M. J. 1979. l-arabinose metabolism in rhizobia. J. Gen. Microbiol. 113:177–179 [Google Scholar]
- 13. Garfinkel D. J., Nester E. W. 1980. Agrobacterium tumefaciens mutants affected in crown gall tumorigenesis and octopine catabolism. J. Bacteriol. 144:732–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. He F., et al. 2009. Molecular basis of ChvE function in sugar binding, sugar utilization, and virulence in Agrobacterium tumefaciens. J. Bacteriol. 191:5802–5813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Higgins C. F. 1992. ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol. 8:67–113 [DOI] [PubMed] [Google Scholar]
- 16. Higuchi R., Krummel B., Saiki R. K. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351–7367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kemner J. M., Liang X., Nester E. W. 1997. The Agrobacterium tumefaciens virulence gene chvE is part of a putative ABC-type sugar transport operon. J. Bacteriol. 179:2452–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kovach M. E., et al. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176 [DOI] [PubMed] [Google Scholar]
- 19. Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 20. Linton K. J., Higgins C. F. 1998. The Escherichia coli ATP-binding cassette (ABC) proteins. Mol. Microbiol. 28:5–13 [DOI] [PubMed] [Google Scholar]
- 21. McCullen C. A., Binns A. N. 2006. Agrobacterium tumefaciens and plant cell interactions and activities required for interkingdom macromolecular transfer. Annu. Rev. Cell Dev. Biol. 22:101–127 [DOI] [PubMed] [Google Scholar]
- 22. Miller J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 23. Neidhardt F. C. 1987. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, DC [Google Scholar]
- 24. Poysti N. J., Loewen E. D., Wang Z., Oresnik I. J. 2007. Sinorhizobium meliloti pSymB carries genes necessary for arabinose transport and catabolism. Microbiology 153:727–736 [DOI] [PubMed] [Google Scholar]
- 25. Pradhan P., Li W., Kaur P. 2009. Translational coupling controls expression and function of the DrrAB drug efflux pump. J. Mol. Biol. 385:831–842 [DOI] [PubMed] [Google Scholar]
- 26. Rees D. C., Johnson E., Lewinson O. 2009. ABC transporters: the power to change. Nat. Rev. Mol. Cell Biol. 10:218–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schafer A., et al. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73 [DOI] [PubMed] [Google Scholar]
- 28. Schneider E. 2001. ABC transporters catalyzing carbohydrate uptake. Res. Microbiol. 152:303–310 [DOI] [PubMed] [Google Scholar]
- 29. Slater S. C., et al. 2009. Genome sequences of three Agrobacterium biovars help elucidate the evolution of multichromosome genomes in bacteria. J. Bacteriol. 191:2501–2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang Y., Mukhopadhyay A., Howitz V. R., Binns A. N., Lynn D. G. 2000. Construction of an efficient expression system for Agrobacterium tumefaciens based on the coliphage T5 promoter. Gene 242:105–114 [DOI] [PubMed] [Google Scholar]
- 31. Watanabe S., Kodaki T., Makino K. 2006. Cloning, expression, and characterization of bacterial l-arabinose 1-dehydrogenase involved in an alternative pathway of l-arabinose metabolism. J. Biol. Chem. 281:2612–2623 [DOI] [PubMed] [Google Scholar]
- 32. Watanabe S., Kodaki T., Makino K. 2006. A novel alpha-ketoglutaric semialdehyde dehydrogenase: evolutionary insight into an alternative pathway of bacterial l-arabinose metabolism. J. Biol. Chem. 281:28876–28888 [DOI] [PubMed] [Google Scholar]
- 33. Watanabe S., Shimada N., Tajima K., Kodaki T., Makino K. 2006. Identification and characterization of L-arabonate dehydratase, L-2-keto-3-deoxyarabonate dehydratase, and L-arabinolactonase involved in an alternative pathway of l-arabinose metabolism. Novel evolutionary insight into sugar metabolism. J. Biol. Chem. 281:33521–33536 [DOI] [PubMed] [Google Scholar]
- 34. Wilson D. B. 1978. Cellular transport mechanisms. Annu. Rev. Biochem. 47:933–965 [DOI] [PubMed] [Google Scholar]
- 35. Wood D. W., et al. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317–2323 [DOI] [PubMed] [Google Scholar]