Abstract
The interaction of Pseudomonas aeruginosa with surfaces has been described as a two-stage process requiring distinct signaling events and the reciprocal modulation of small RNAs (sRNAs). However, little is known regarding the relationship between sRNA-modulating pathways active under planktonic or surface-associated growth conditions. Here, we demonstrate that SagS (PA2824), the cognate sensor of HptB, links sRNA-modulating activities via the Gac/HptB/Rsm system postattachment to the signal transduction network BfiSR, previously demonstrated to be required for the development of P. aeruginosa. Consistent with the role of SagS in the GacA-dependent HtpB signaling pathway, inactivation of sagS resulted in hyperattachment, an HptB-dependent increase in rsmYZ, increased Psl polysaccharide production, and increased virulence. Moreover, sagS inactivation rescued attachment but abrogated biofilm formation by the ΔgacA and ΔhptB mutant strains. The ΔsagS strain was impaired in biofilm formation at a stage similar to that of the previously described two-component system BfiSR. Expression of bfiR but not bfiS restored ΔsagS biofilm formation independently of rsmYZ. We demonstrate that SagS interacts directly with BfiS and only indirectly with BfiR, with the direct and specific interaction between these two membrane-bound sensors resulting in the modulation of the phosphorylation state of BfiS in a growth-mode-dependent manner. SagS plays an important role in P. aeruginosa virulence in a manner opposite to that of BfiS. Our findings indicate that SagS acts as a switch by linking the GacA-dependent sensory system under planktonic conditions to the suppression of sRNAs postattachment and to BfiSR, required for the development of P. aeruginosa biofilms, in a sequential and stage-specific manner.
INTRODUCTION
Biofilms are complex communities of microorganisms attached to surfaces and embedded in a self-produced extracellular matrix (13), with biofilm formation initiated by bacteria attaching to a surface. The switch from the motile to the sessile mode of growth is an essential step in the formation of biofilms and the modulation of virulence. Several factors have been shown to impact the transition from the free-swimming to the surface-attached mode of growth, including appendages (type IV pili and flagella) (29, 39, 40, 51, 52, 61, 62) and the intracellular signaling molecule bis(3′-5′)-cyclic diguanylic monophosphate (cyclic di-GMP). First described to control extracellular cellulose biosynthesis in Acetobacter xylinum (48, 49), cyclic di-GMP has been demonstrated in several microorganisms, including Pseudomonas aeruginosa, to control the transition between a motile and a biofilm lifestyle via its concentration, with high levels fostering the sessile lifestyle and low cyclic di-GMP concentrations favoring motility (e.g., twitching and swarming) and the planktonic mode of growth (14, 27, 36, 47, 56, 60). Regulatory systems linking the modulation of cyclic di-GMP levels and attachment capabilities include the genetic pathway composed of BifA, SadB, and SadC, which regulates Pel and Psl exopolysaccharide production as P. aeruginosa transitions from a planktonic to a surface-associated lifestyle (11, 30, 34), and the Escherichia coli Csr system (28). CsrABCD, originally identified as a system to regulate glycogen biosynthesis, controls carbon and secondary metabolism, biofilm formation, motility, and quorum sensing via destabilization of respective mRNA targets. Regulation of the activity of the RNA binding protein CsrA is mediated in part by the actions of the two noncoding small RNAs (sRNAs) CsrB and CsrC (24, 25, 28, 46, 59, 64).
Similarly to the Csr system in E. coli, sRNAs in Pseudomonas aeruginosa have been shown to play an important role in attachment, polysaccharide production, virulence, and quorum sensing (6, 9, 23, 37, 42, 44). However, in contrast to the E. coli Csr system, the Rsm system in P. aeruginosa has not been linked to the modulation of cyclic di-GMP levels. sRNAs rsmZ and rsmY serve as antagonists of the translational regulator RsmA. The binding of RsmA to specific mRNA targets differentially affects their stability, turnover, and translation rates, thus controlling the expression of a significant number of virulence and attachment genes at the level of mRNA translation and/or stability (17, 25, 45, 63). Expression levels of the sRNAs are directly controlled by GacA/GacS, which are inversely controlled by the two-component hybrids RetS and LadS (18, 63). RetS negatively controls rsmY and rsmZ gene expression, while LadS positively controls sRNA levels. An additional component modulating sRNA levels was recently identified as the histidine phosphotransfer protein HptB. HptB is activated via three orphan sensor kinase hybrids (PA2824, PA1611, PA1976) and in turn relays the signal (a phosphor group) to the response regulator PA3346 and to RetS (6, 22). While HptB also intersects at the GacA response regulator, which directly controls sRNA production to reciprocally regulate the expression of exopolysaccharide components of the P. aeruginosa biofilm matrix and the organism's attachment and swarming motility, HptB appears to exclusively regulate rsmY expression (6–8, 17, 19, 22).
While attachment is enhanced by increased sRNA levels, subsequent surface-attached growth and biofilm formation by P. aeruginosa have recently been demonstrated to be hampered by elevated sRNA levels, in particular those of rsmZ (44). Modulation of rsmZ levels under biofilm growth conditions is dependent on the novel two-component system (TCS) BfiSR, which regulates the suppression of rsmZ via RNase G (CafA) (44). BfiSR was found to be required for biofilm development by P. aeruginosa, with inactivation of the biofilm-specific TCS arresting biofilm formation in a manner that coincided with its timing of phosphorylation (43). The interaction of P. aeruginosa with surfaces can thus be described as a two-stage process: initially, colonizing bacteria express and modulate genes and proteins required for the increase in sRNA levels (including LadS, GasAS, and HtpB), while the subsequent persistent biofilm developmental stages depend on the production and posttranslational modification of a different set of proteins (BfiSR, CafA) that promote biofilm formation by decreasing sRNAs. However, the coordinate regulation of these two contrasting sets of factors/proteins prior to and following the motile-sessile switch and subsequent biofilm formation remains to be elucidated.
Here, we report the identification of the probable sensor/response regulator hybrid PA2824, which we named SagS (surface attachment and growth sensor hybrid), as a sensor essential for biofilm formation that links, via interaction with and modulation of the phosphorylation state of BfiS, to the regulatory pathways that reciprocally modulate sRNA levels prior to and following attachment to enable biofilm formation. By regulating the motile-sessile switch, SagS links the Gac/HptB/Rsm and BfiSR signaling transduction systems into a multisensor signaling network.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and culture conditions.
All bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. P. aeruginosa strains PAO1 and PA14 were used as parental strains, the latter being required for the Arabidopsis virulence studies. All planktonic strains were grown in Lennox broth (LB) or Vogel-Bonner minimal medium (VBMM) (53) in shake flasks at 220 rpm.
Strain construction.
Isogenic mutants were constructed by allelic replacement using sucrose counterselection, as previously described (54), with the gene replacement vector pEX18Gm (21). Complementation and overexpression were accomplished by placing the respective genes under the control of an arabinose-inducible promoter in the pJN105 vector (38). C-terminal V5/6×His tagging was accomplished by subcloning into pET101D (Invitrogen), N-terminal 6×His tagging of BfiS was accomplished by subcloning into pQE30Xa (Qiagen), while a C-terminal hemagglutinin (HA) tag was introduced into SagS via PCR using the sequence AGCGTAGTCTGGGACGTCGTATGGGTA. The tagged constructs were introduced into pJN105 and pMJT1. Primers used for strain construction are listed in Table S2 in the supplemental material.
Biofilm formation.
Biofilms were grown in a once-through continuous-flow tube reactor system to obtain proteins and RNA or in flow cells to view the biofilm architecture, as previously described (1, 2, 43, 52, 57). Biofilms were grown at 22°C in 1/20-diluted LB or VBMM in the absence or presence of 0.1% arabinose. Quantitative analysis of confocal laser scanning microscopy (CSLM) images of flow cell-grown biofilms was performed using COMSTAT (20). Initial biofilm formation was measured by using the microtiter dish assay system with crystal violet (CV) staining (40) and by microscopy (12). The number of cells attached to the glass surface of a flow cell covering an area of 400 μm2 was quantified using ImageProPlus software.
Phosphoprotein detection and phosphotransfer analysis.
Detection of phosphoproteins via immunoblot analysis was carried out as previously described (43, 57). Briefly, 200 μg of total cell protein was subjected to separation by two-dimensional PAGE (2D-PAGE), followed by blotting onto polyvinylidene difluoride (PVDF) membranes and phosphoprotein detection using anti-phospho-(Ser/Thr)Phe antibodies (Cell Signaling Technologies, Danvers, MA). Analysis of phosphorylated BfiS-V5/6×His levels in protein extracts of P. aeruginosa strains in which sagS was inactivated or overexpressed was accomplished using phosphoprotein purification via metal oxide affinity chromatography (MOAC) as described previously (43), followed by the detection of BfiS-V5/5×His by immunoblotting with anti-V5 antibodies (Invitrogen Corp.). For phosphotransfer studies, V5/6×His-tagged BfiS and SagS, purified using Ni-nitrilotriacetic acid (NTA) spin columns (Qiagen), were combined and incubated in phosphorylation buffer (50 mM Tris-HCl [pH 7.5], 50 mM KCl, 5 mM MgCl2) for 60 min at room temperature, after which time the samples were subjected to MOAC phosphoprotein purification and anti-V5 immunoblot analysis. Purified BfiS and SagS alone were used as controls.
Psl polysaccharide dot blot analysis.
Psl polysaccharide was extracted from planktonic and 144-h-old biofilm cells essentially as described by Byrd et al. (10). Quantitation of Psl production was done by determining the anti-Psl dot blot spot volume (23) using the ImageMaster analysis software (GE Healthcare).
Motility assays.
Swimming, swarming, and twitching motilities were assessed in tryptone or LB medium containing 0.3%, 0.5%, and 1.0% agar, respectively, as previously described (39, 55, 57).
qRT-PCR.
Quantitative reverse transcriptase PCR (qRT-PCR) was used to determine the expression levels of pslA and rsmAYZ. Isolation of mRNA and cDNA synthesis were carried out as previously described (2, 3, 43, 57). qRT-PCR was performed using the Eppendorf Mastercycler ep realplex (Eppendorf AG, Germany) and the SYBR Fast qPCR kit (Kapa Biosystems, Woburn, MA) (see Table S2 in the supplemental material for a list of oligonucleotides). Relative transcript quantitation, with mreB used as a housekeeper control, was done as previously described (44).
Extraction and quantification of cyclic di-GMP from P. aeruginosa.
Cyclic di-GMP was extracted in triplicate from wild-type and mutant strains grown planktonically, essentially as described previously (4, 36, 56), using heat and ethanol precipitation followed by centrifugation. Supernatants were combined, dried using a SpeedVac, and resuspended in 10 mM ammonium bicarbonate buffer. Samples (10 μl or 20 μl) were analyzed using an Agilent 1100 high-performance liquid chromatography (HPLC) system equipped with an autosampler, degasser, and detector set to 253 nm and were separated using a reverse-phase C18 Targa column (2.1 by 40 mm by 5 μm) at a flow rate of 0.2 ml/min with the following gradients for each time range: for 0 to 9 min, 1% B; for 9 to 14 min, 15% B; for 14 to 19 min, 25% B; for 19 to 26 min, 90% B; and for 26 to 40 min, 1% B (where A is 10 mM ammonium acetate and B is methanol plus 10 mM ammonium acetate). Commercially available cyclic di-GMP was used as a reference for the identification and quantification of cyclic di-GMP in cell extracts. Moreover, the identity of HPLC-eluted cyclic di-GMP was confirmed by tandem mass spectrometry (MS/MS) using a QStar mass spectrometer (Applied Biosystems) by detecting the cyclic di-GMP fragments 691.1→151.9, 691.1→248.0, and 691.1→539.8 m/z, as described by Thormann et al. (60).
Determination of SagS protein interactions using pulldown assays.
Pulldown assays were used to determine whether SagS interacts with BfiS and BfiR. HA-tagged SagS was incubated with extracts containing 6×His-tagged BfiS or BfiR. Subsequently, BfiS or BfiR was immunoprecipitated using immobilized anti-6×His antibodies, and the immunoprecipitation eluates were separated by SDS-PAGE and assessed by immunoblot analysis for the presence of SagS using anti-HA antibodies (and vice versa). Pulldown assays were carried out using 200 μg protein from cellular extracts, with HA-tagged proteins used as bait and V5/6×His-tagged proteins used as prey (and vice versa). Mouse anti-HA and anti-His or anti-V5 antibodies (Invitrogen) were used for immunoprecipitation at 1 μg/ml and immunoblotting at 0.1 μg/ml. Two V5/6×His-tagged proteins, the inner membrane protein PA3343 and BdlA, were used as controls to determine the specificity of the SagS-BfiS complex formation in vitro.
Virulence testing.
The role of sagS in virulence was assessed using the Arabidopsis thaliana infection model. Plants were infected essentially as described by Starkey and Rahme (58), incubated for a period of 12 days in a 16-h-light (25°C)/8-h-dark (22°C) cycle, and analyzed as previously described (44).
Statistical analysis.
Student's t test was performed for pairwise comparisons of groups, and multivariant analyses were performed using a one-way analysis of variance (ANOVA) followed by an a posteriori test using Sigma Stat software. All experiments were carried out in triplicate.
RESULTS
The probable sensor/response regulator hybrid SagS.
The sagS gene encodes a 786-amino-acid polypeptide similar in sequence and domain organization to the sensor kinase/response regulator hybrid family of signal transduction proteins (Fig. 1A). This orphan sensor hybrid was previously identified to phosphorylate the GacA-dependent histidine phosphotransfer protein HptB in vitro and to interact with HptB in vivo and was thus implicated in playing a role in the GacAS-dependent control of sRNA levels (6, 22). Moreover, while sagS expression was found to be independent of the mode of growth (not shown), SagS was found to be phosphorylated only when P. aeruginosa was grown planktonically (43), suggesting that SagS might play a role in the motile-sessile transition, possibly via sRNA regulation.
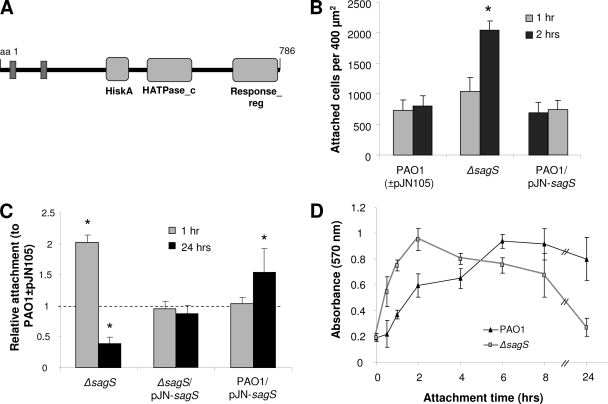
Fig. 1.
Attachment is dysregulated in the strain in which sagS, encoding a hybrid sensor kinase/response regulator, is inactivated, as determined by microscopy and crystal violet staining. (A) Domain organization of SagS. Domains are predicted by sequence homology using Pfam. HiskA, histidine kinase domain; HATPase_c, histidine ATPase domain C; Response_reg, response regulator domain; aa, amino acid. Predicted transmembrane-spanning segments are shown in dark gray. (B) Direct cell counts of wild-type and mutant strains attached to polystyrene 1 and 2 h after initial attachment. Numbers of cells attached per 400 μm2 were quantified using ImageProPlus software. *, significantly different from respective wild-type values (P < 0.05), as determined by ANOVA and Sigma Stat. (C) Evaluation of attachment to polystyrene by crystal violet staining 1 and 24 h after initial attachment, relative to that of P. aeruginosa PAO1 in the absence/presence of the empty plasmid (PAO1 ± pJN105). *, significantly different from respective wild-type values (P < 0.05), as determined by ANOVA and Sigma Stat. (D) Time course of attachment to polystyrene over the course of 24 h by P. aeruginosa PAO1 and the ΔsagS strain. Error bars indicate standard deviations.
SagS plays a temporal role in attachment.
To elucidate the role of SagS in the motile-sessile switch and, thus, the transition to a surface-associated lifestyle, we first determined whether inactivation or overexpression of sagS affected attachment. Inactivation of sagS resulted in significantly enhanced attachment, with the ΔsagS strain attaching twice as efficiently as the PAO1 strain within the first 2 h, as determined by microscopy and CV staining (Fig. 1B and C). The findings are consistent with previous reports by Goodman et al. demonstrating increased attachment capabilities for this mutant (17). However, increased attachment by the ΔsagS strain was only temporary, and the difference in attachment diminished with increasing attachment times. No difference in attachment was observed between PAO1 and the ΔsagS strain following 8 h of attachment, while continued incubation resulted in decreased attachment by the ΔsagS strain compared to that of the wild type (Fig. 1C and D). Complementation of the ΔsagS mutant with pJN-sagS restored the attachment phenotype to wild-type levels, while overexpression of sagS enhanced attachment after prolonged growth (Fig. 1B and C). Growth curves in liquid media suggested that the increase in attachment was not a result of a general increase in growth rate.
A ΔsagS mutant strain of P. aeruginosa is impaired in biofilm formation.
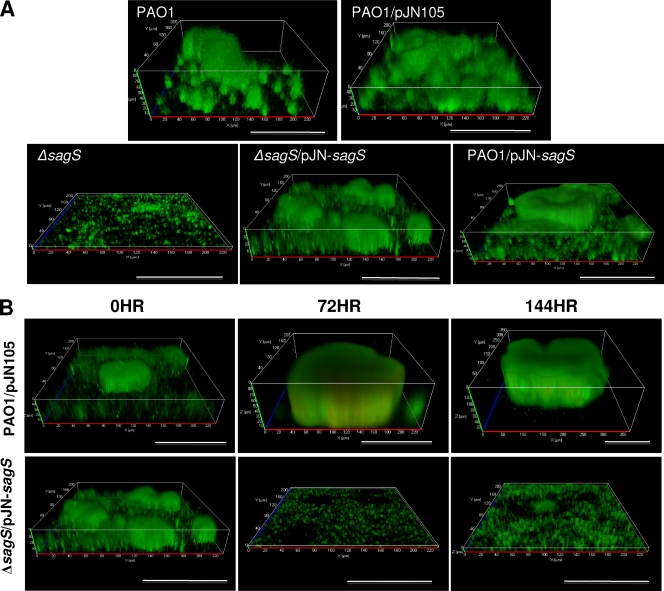
Since inactivation of sagS had only a temporary positive effect on attachment, with the difference in attachment diminishing upon continued surface exposure (Fig. 1), we asked whether inactivation or overexpression of this regulatory protein would alter or affect biofilm formation. We anticipated detecting no difference in biofilm formation between the ΔsagS strain and PAO1. However, the mutant strain failed to develop the biofilm architecture typically observed in PAO1 biofilms following 144 h under biofilm growth conditions (Fig. 2A; Table 1). Instead, the ΔsagS strain formed only thin biofilms that lacked large cellular aggregates and microcolonies and that were composed of ≥5-fold-less biomass than those of the wild type (Table 1). Overall, ΔsagS biofilms most closely resembled those of PAO1 following 24 h of growth. Daily monitoring of ΔsagS biofilms by confocal microscopy over a period of 24 to 144 h revealed that ΔsagS biofilms failed to accumulate the additional biomass typically seen in wild-type biofilms following continued growth under flowing conditions (after 48 h of growth) (Table 1; Fig. 2A) (43, 52), indicating that the observed biofilm architecture at the 144-h time point was not due to premature disaggregation. In contrast, overexpression of sagS resulted in biofilms that were similar in architecture to PAO1 biofilms but that displayed significantly increased biomass, as well as increased average and maximum thickness, when grown for 144 h (Fig. 2A; Table 1). Complementation restored the biofilm architecture to wild-type levels.
Fig. 2.
SagS is required for the formation and maintenance of P. aeruginosa biofilms. (A) Biofilms of strains with sagS inactivated or overexpressed that were grown for 144 h were visualized by CSLM. P. aeruginosa PAO1 strains in the absence and presence of the empty plasmid were used as controls. (B) Inactivation of sagS expression in mature biofilms results in biofilm architectural collapse and biofilm dispersion. The P. aeruginosa ΔsagS strain complemented with sagS (the Δsag/pJN-sagS strain) and placed under the regulation of the arabinose-inducible PBAD promoter was grown under continuous-flow conditions in VBMM in the presence of 0.1% arabinose for 144 h, after which time the biofilms were visualized by confocal microscopy (0 h). Arabinose was subsequently eliminated from the growth medium, and the biofilm architecture was monitored following arabinose removal at the times indicated. The PAO1 strain harboring the empty pJN105 vector was used as a control. Biofilms were stained with the Live/Dead BacLight viability stain (Invitrogen Corp.). White bars = 100 μm.
Table 1.
COMSTAT analysis of P. aeruginosa wild-type and mutant biofilm structuresa
| P. aeruginosa strain and incubation time (h) | Total biomass (μm3/μm2) | Substratum coverage (%) | Avg thickness (μm) | Maximum thickness (μm) | Roughness coefficient (dimensionless) |
|---|---|---|---|---|---|
| PAO1 | |||||
| 24 | 0.95 ± 0.39* | 12.11 ± 5.08* | 0.78 ± 0.38* | 7.8 ± 1.14* | 1.69 ± 0.12 |
| 72 | 3.27 ± 1.29* | 32.43 ± 8.31 | 3.02 ± 1.52* | 18.29 ± 10.86* | 1.20 ± 0.31 |
| 144 | 7.99 ± 3.63 | 44.44 ± 22.58 | 7.17 ± 4.97 | 35.63 ± 26.73 | 1.24 ± 0.37 |
| Isogenic mutant after 144 h of growth | |||||
| PAO1 ΔsagS | 0.82 ± 0.63* | 7.07 ± 5.14* | 0.83 ± 0.62* | 9.00 ± 2.83* | 1.74 ± 0.20 |
| PAO1 ΔsagS/pJN-sagS | 11.89 ± 8.48 | 25.34 ± 11.36 | 11.90 ± 8.99 | 47.67 ± 29.96 | 1.38 ± 0.23 |
| PAO1/pJN-sagS | 14.04 ± 9.98* | 25.64 ± 11.26 | 16.97 ± 14.51* | 55.49 ± 28.84 | 1.09 ± 0.37 |
| PAO1 ΔbfiS/pJN-sagS | 2.10 ± 2.54* | 8.12 ± 6.98* | 1.89 ± 2.48* | 27.71 ± 16.19 | 1.82 ± 0.15 |
| PAO1 ΔsagS/pJN-bfiS | 1.33 ± 1.59* | 4.79 ± 2.00* | 1.24 ± 1.78* | 27.33 ± 13.08 | 1.89 ± 0.06 |
| PAO1 ΔsagS/pJN-bfiR | 27.76 ± 14.60* | 28.11 ± 12.21 | 29.40 ± 15.79* | 92.33 ± 24.98* | 1.12 ± 0.35 |
| PAO1 ΔgacA | 17.49 ± 10.77* | 16.14 ± 7.66 | 21.04 ± 14.45* | 94.22 ± 21.57* | 1.31 ± 0.40 |
| PAO1 ΔgacA ΔsagS | 0.65 ± 0.73* | 3.75 ± 3.18* | 0.67 ± 0.79* | 13.79 ± 8.61* | 1.90 ± 0.08 |
| PAO1 ΔrsmYZ | 19.00 ± 13.70* | 44.07 ± 27.85 | 20.28 ± 16.59* | 84.00 ± 31.59* | 1.09 ± 0.40 |
| PAO1 ΔrsmYZ ΔsagS | 0.29 ± 0.64* | 2.51 ± 2.74* | 0.25 ± 0.59* | 8.33 ± 9.44* | 1.95 ± 0.06 |
| PAO1 ΔhptB | 1.64 ± 1.98* | 17.44 ± 24.20 | 1.61 ± 1.88* | 8.22 ± 3.99* | 1.51 ± 0.56 |
| PAO1 ΔhptB ΔsagS | 0.64 ± 0.69* | 7.19 ± 6.34* | 0.57 ± 0.69* | 6.89 ± 2.93* | 1.82 ± 0.17* |
The COMSTAT analysis was carried out with biofilms grown in triplicate, using at least 6 images per replicate.
, significantly different from the value for 144-h-old PAO1. P was ≤0.05, as determined by ANOVA and SigmaStat.
SagS expression is required for the maintenance of mature biofilm architecture.
Our observations indicated that SagS was essential for the stage-specific development of P. aeruginosa biofilm formation, as the ΔsagS strain was arrested in biofilm formation, while sagS overexpression had no effect on continued biofilm development or on biofilm architecture (Fig. 2A). To determine whether sagS is also required for the maintenance of biofilm architecture, we made use of a complemented mutant strain (the ΔsagS/pJN-sagS strain), which harbored sagS under the control of the arabinose-inducible PBAD promoter. This allowed mutant biofilms to form wild-type-like biofilms within 144 h of growth in the presence of arabinose (Fig. 2B, 0 h), after which time arabinose was removed from the growth medium to stop the transcription of sagS. The resulting biofilm architecture was viewed over a period of 144 h after arabinose removal using confocal microscopy. Wild-type P. aeruginosa harboring an empty pJN105 vector was used as a control.
Loss of sagS expression due to arabinose removal resulted in the collapse of the mutant biofilm architecture within 3 days (Fig. 2B, 72 h; Table 2). While a similar architectural collapse was observed following inactivation of bfiS, bfmR, and mifR expression (43), the collapse observed following sagS inactivation was distinct, coinciding with biofilm dispersion, as evidenced by the formation of large voids (Fig. 2B) indicative of dispersion events. The voids were detectable as soon as 1 day following arabinose removal (not shown). Cells that remained attached at the surface showed increased motility, as determined by microscopy (not shown). The collapse was furthermore apparent by significant reductions in biofilm variables, including biofilm biomass and thickness, which further decreased upon continued incubation (Table 2). Under the conditions tested, no reduction in the biomass and thickness of the wild-type biofilm architecture was observed (Fig. 2B; Table 2). These findings indicate that the effect of SagS is reversible with respect to biofilm architecture and biofilm development.
Table 2.
COMSTAT analysis of P. aeruginosa wild-type and complemented mutant biofilm structure following removal of arabinose, which results in a lack of sagS expressiona
| P. aeruginosa strain | Time (h)b | Total biomass (μm3/μm2) | Substratum coverage (%) | Avg thickness (μm) | Maximum thickness (μm) | Roughness coefficient (dimensionless) |
|---|---|---|---|---|---|---|
| PAO1/pJN105 | 0 | 11.92 ± 9.28 | 28.85 ± 13.29 | 11.97 ± 11.48 | 37.17 ± 26.78 | 1.25 ± 0.28 |
| 72 | 31.66 ± 10.56 | 29.37 ± 11.36 | 30.99 ± 10.58 | 66.33 ± 13.90 | 1.22 ± 0.30 | |
| 144 | 36.92 ± 18.00 | 45.01 ± 19.18 | 37.91 ± 18.76 | 111.33 ± 19.06 | 0.92 ± 0.44 | |
| PAO1 ΔsagS/pJN-sagS | 0 | 11.89 ± 8.48 | 25.34 ± 11.36 | 11.90 ± 8.99 | 47.67 ± 29.96 | 1.38 ± 0.23 |
| 72 | 1.88 ± 0.84* | 16.28 ± 9.72* | 1.83 ± 0.99* | 14.50 ± 12.00* | 1.55 ± 0.30 | |
| 144 | 1.40 ± 0.84* | 15.55 ± 7.77* | 1.21 ± 0.85* | 6.33 ± 2.53* | 1.61 ± 0.20* |
The COMSTAT analysis was carried out with biofilms grown in triplicate, using at least 6 images per replicate.
, significantly different from the value for PAO1/pJN105 at the respective time point. P was ≤0.05, as determined by ANOVA and SigmaStat.
Time zero for biofilms was 144 h. Time corresponds to the time following the switch to an arabinose-free medium.
Inactivation of sagS arrests biofilm development prior to the irreversible attachment stage.
Quantitative analysis of confocal microscope images of ΔsagS biofilms indicated that the ΔsagS strain was arrested in biofilm formation. This prompted us to determine the biofilm developmental stage at which the ΔsagS mutant was arrested. We have previously demonstrated that wild-type P. aeruginosa biofilms exhibit distinct, stage-specific protein phosphorylation patterns over the course of development and that mutant strains impaired in biofilm formation demonstrate phosphorylation profiles that are indicative/predictive of the developmental stage at which developmental arrest occurs (43). A comparison of the phosphorylation patterns of ΔsagS biofilms grown for 144 h to those of P. aeruginosa wild-type biofilms grown for 8, 24, 72, and 144 h using anti-phospho-Ser/Thr antibodies indicated that ΔsagS biofilms failed to exhibit phosphorylation events typically observed during normal biofilm development following 144 h of growth (Table 3; see also Table S3 in the supplemental material). For instance, ΔsagS biofilms lacked all of the phosphorylated proteins typically found in mature, 144-h-old biofilms and instead exhibited stage-specific phosphorylation events typically detected in 8- and 24-h-old wild-type biofilms; the phosphoproteome contained 26 out of 38 and 12 out of 33 phosphorylated proteins that are specific to 8-h-old and 24-h-old wild-type biofilms, respectively (Table 3; see also Table S3 in the supplemental material). Furthermore, ΔsagS biofilms lacked evidence of BfiS phosphorylation (see Table S3 in the supplemental material), which was previously detected following 8 h of biofilm growth (43), indicating that ΔsagS biofilms are probably arrested prior to ΔbfiS biofilms at the transition from the planktonic to the initial attachment stage.
Table 3.
Comparison of phosphorylation patterns between ΔsagS and ΔbfiS biofilms
| Biofilm developmental stage (h) | No. of detected phosphorylation events in wild-type P. aeruginosa PAO1 | No. of detected phosphorylation events in 144-h-old biofilm of: |
||
|---|---|---|---|---|
| ΔbfiS mutant | ΔsagS mutant | ΔbfiS mutant and ΔsagS mutant | ||
| Planktonic | 22 | 3 | 2 | 1 |
| Motile-sessile switcha | 8 | 6 | 6 | 5 |
| Reversible attachment (8) | 38 | 25 | 26 | 18 |
| Irreversible attachment (24) | 33 | 11 | 12 | 7 |
| Maturation (144) | 39 | 0 | 0 | 0 |
| Dispersion (216) | 19 | 0 | 0 | 0 |
| Biofilm-specificb | 7 | 6 | 7 | 6 |
| Constitutive | 26 | 26 | 26 | 26 |
Comparison of the phosphorylation patterns of both the ΔsagS and ΔbfiS strains indicated that while both mutant biofilms exhibited stage-specific phosphorylation events typically detected in 8-h-old and 24-h-old wild-type biofilms, inactivation of sagS and bfiS affected distinct sets of phosphorylated proteins under biofilm growth conditions (Table 3).
SagS controls Psl production.
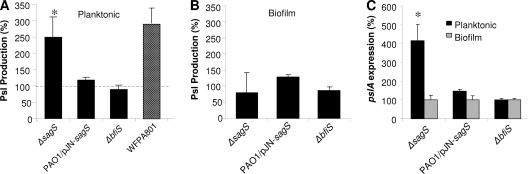
To begin elucidating the mechanisms by which SagS affects transitioning between the motile and sessile states, mutants in which sagS was inactivated or overexpressed were tested for properties known to affect initial attachment, including cellular motility, cyclic di-GMP levels, and polysaccharide production, which have been shown to play a role in promoting biofilm formation in diverse P. aeruginosa strains (15, 16, 26, 50). Increased Psl production by the ΔsagS strain in a planktonic-stage-specific manner was detected by immunoblot analysis using anti-Psl antibodies (Fig. 3A and B). Under planktonic growth conditions, Psl abundance in the ΔsagS strain was comparable to that observed in the Psl-overproducing strain WFPA801 (Fig. 3A). Moreover, qRT-PCR demonstrated increased expression of pslA involved in Psl polysaccharide biosynthesis in the ΔsagS strain under free-swimming growth conditions but not in biofilms (Fig. 3C). While cellular motility has been shown to affect initial attachment, no differences with respect to twitching, swimming, and swarming were detected for strains in which sagS was inactivated or overexpressed (not shown). Mutant strains demonstrating increased attachment and matrix polymer production have been previously linked to increased cyclic di-GMP levels (5, 56). However, no difference in cyclic di-GMP levels between ΔsagS mutant cells and wild-type cells grown planktonically was detected, indicating that increased adhesiveness and Psl production are independent of cyclic di-GMP levels in the ΔsagS strain.
Fig. 3.
Inactivation of sagS correlates with increased Psl production under planktonic growth conditions. Psl production by P. aeruginosa strains grown planktonically (A) and as biofilms (B), relative to that of P. aeruginosa PAO1, as determined using dot blot analysis of anti-Psl dot blot spot volumes. The Psl-overproducing strain WFPA801 was grown in the presence of 2% arabinose. (C) pslA expression, as determined by qRT-PCR, in P. aeruginosa strains grown planktonically and as biofilms, relative to that of P. aeruginosa PAO1. Error bars indicate standard deviations. *, significantly different from values for respective wild-type cells grown under identical conditions/modes of growth (P < 0.05) as determined by ANOVA and Sigma Stat.
SagS controls rsmY and rsmZ expression under planktonic growth conditions.
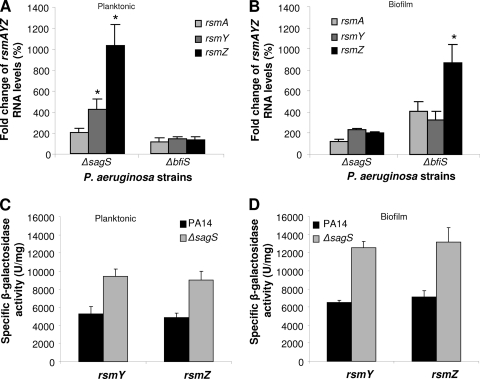
The Gac/Rsm network directly modulates the levels of sRNAs rsmY and rsmZ to regulate the expression of exopolysaccharide components of the P. aeruginosa biofilm matrix and the organism's attachment and swarming motility (6–8, 17, 19). SagS has been previously linked to this network via the histidine phosphotransfer protein HptB, with HptB having been shown to be dependent on the GacA response regulator (6, 22), which directly controls rsmY and rsmZ transcription (6–8, 17, 19). This prompted us to ask whether SagS is involved in the regulation of rsmYZ levels. Inactivation of sagS correlated with elevated rsmA levels and significantly increased rsmYZ levels compared to those of the wild type under planktonic growth conditions (Fig. 4A). Under biofilm growth conditions, rsmAYZ levels were increased 2-fold in ΔsagS biofilms (Fig. 4B). No difference in rsmAYZ gene expression was detected when transcript levels of PAO1/pJN-sagS, ΔsagS/pJN-sagS, and the wild type were compared. In contrast, analysis of the bfiS mutant strain, which is arrested in biofilm formation at a stage similar to that of the ΔsagS strain (43, 44), demonstrated that the RNA levels of rsmA, rsmY, and rsmZ were unaltered in the ΔbfiS strain under planktonic growth conditions but that they were significantly increased compared to wild-type levels upon biofilm growth (Fig. 4A and B). We also measured the activities of rsmY-lacZ and rsmZ-lacZ chromosomal transcriptional fusions in planktonic, 24-h-old, and 144-h-old biofilm cells. In agreement with the expression levels, transcription from rsmY and rsmZ promoters was found to be repressed by SagS regardless of growth conditions (Fig. 4C and D). This is in contrast to what was observed with the ΔbfiS strain, for which rsmYZ transcription levels were comparable to those of the wild type regardless of growth conditions, while rsmYZ RNA levels were significantly increased under biofilm growth conditions (44). These findings strongly suggest that SagS and BfiS reciprocally modulate rsmAYZ abundance at distinct levels in a growth-mode-dependent manner. The mutants further differed with respect to pslA expression and Psl production (Fig. 3). While increased rsmYZ levels in the ΔsagS strain correlated with increased pslA expression and Psl production under planktonic growth conditions, no difference in Psl production was detected in the ΔbfiS strain regardless of the growth conditions (Fig. 3).
Fig. 4.
SagS controls rsmY and rsmZ expression under planktonic but not biofilm growth conditions. (A and B) rsmAYZ RNA levels in P. aeruginosa strains grown planktonically (A) and as biofilms (B), relative to those in PAO1, as determined by qRT-PCR. (C and D) Transcriptional reporter fusion assays for rsmY and rsmZ expression in P. aeruginosa and ΔsagS mutant strains grown planktonically (C) and as biofilms (144 h) (D). Error bars indicate standard deviations. *, significantly different from respective wild-type cells grown under identical conditions/modes of growth (P < 0.05) as determined by ANOVA and Sigma Stat.
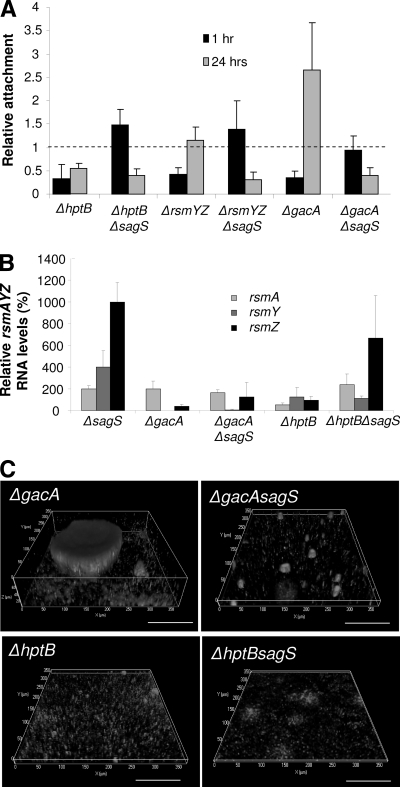
Deletion of sagS enhances early attachment but abrogates biofilm formation by the gacA and hptB mutant strains.
Given that our findings implicated SagS in the regulation of rsmYZ levels (Fig. 4), we next determined whether SagS regulatory function and its associated phenotypes are dependent on the components of the Gac/Rsm system. Components of this network have been demonstrated to play a role in attachment and biofilm formation, with inactivation of gacA and deletion of its small regulatory RNA targets, rsmYZ, resulting in reduced attachment compared to that of the parental strain, P. aeruginosa PAK (8, 41). Inactivation of hptB in P. aeruginosa PAK resulted in hyperattachment, while the same mutation in P. aeruginosa PAO1 affected attachment only in the presence of glucose and Casamino Acids (6, 33). Here, we demonstrate that in P. aeruginosa PAO1, inactivation of rsmYZ, gacA, and hptB results in reduced attachment following 1 h of incubation under static conditions (Fig. 5A). Continued incubation, however, resulted in increased attachment or hyperattachment by the ΔgacA and ΔrsmYZ strains (Fig. 5A), indicating that, similarly to the ΔsagS mutation, mutations in gacA and rsmYZ have only a temporary effect on attachment. In contrast, the ΔhptB strain demonstrated reduced attachment regardless of attachment time (Fig. 5A). A second-site mutation in sagS rescued the attachment defect of the ΔgacA, ΔhptB, and ΔrsmYZ strains following 1 h of attachment while also suppressing attachment upon continued incubation (Fig. 5A).
Fig. 5.
Second-site mutations in the ΔsagS strain implicate three SagS-dependent phenotypes as HptB dependent, linking SagS to the GacS/GacA/RsmZ regulatory pathway. (A) Evaluation of attachment to polystyrene by crystal violet staining 1 and 24 h after initial attachment in the ΔsagS strain harboring various second-site mutations, relative to that in P. aeruginosa PAO1. (B) rsmAYZ RNA levels in various ΔsagS strains grown planktonically, relative to those of PAO1, as determined by qRT-PCR. (C) Biofilm architecture of the ΔsagS strain harboring various second-site mutations. Biofilms were visualized by CSLM after 144 h. White bars = 100 μm.
In agreement with the attachment phenotype following 24 h of incubation, the ΔgacA strain formed hyperbiofilms characterized by significantly increased biofilm biomass and height compared to those of wild-type biofilms. Deletion of sagS in the ΔgacA strain significantly reduced biofilm biomass accumulation by more than 20-fold, with mutant biofilms resembling those of ΔsagS biofilms (Fig. 5C; Table 1). The ΔhptB strain formed only thin biofilms that lacked microcolonies and were, on average, only 2 μm thick, with a secondary-site mutation in sagS resulting in a further 2- to 3-fold reduction in the biofilm biomass (Fig. 5C; Table 1). Thus, introducing a sagS deletion into the gacA and hptB mutants abrogates biofilm formation of the parent strain, underscoring the importance of SagS in biofilm formation. Our findings indicate that SagS functions upstream of GacA to regulate attachment and biofilm formation. Moreover, the similarities between the attachment (after 24 h) and biofilm phenotypes (Fig. 5A and C) of the ΔsagS and ΔhptB strains suggested that HptB and SagS play similar or convergent regulatory roles, with inactivation of both resulting in comparable impairments of biofilm formation.
In order to establish the epistatic relationships between SagS, HptB, and GacA in regulating rsmAYZ, respective RNA levels in the ΔgacA and ΔhptB strains and the strains harboring a second-site mutation in sagS were determined under planktonic conditions. In agreement with previous results, inactivation of gacA resulted in significantly reduced rsmYZ levels under planktonic growth conditions. Deletion of sagS in the ΔgacA strain did not significantly alter rsmAYZ levels. The observation that rsmAYZ RNA levels are similarly reduced in ΔgacA and ΔgacA ΔsagS strain backgrounds indicated that SagS is not directly required for GacA function and instead functions independently. HptB has been shown to exclusively regulate rsmY expression in P. aeruginosa PAK (6, 22). While we were able to demonstrate reduced rsmY expression in an ΔhptB PAK strain (not shown), inactivation of hptB in P. aeruginosa PAO1 was correlated with 2-fold-reduced rsmA levels, and rsmYZ levels remained comparable to those of the wild type (Fig. 5B). A second-site sagS mutation in the ΔhptB strain resulted in significantly increased rsmA (4-fold) and rsmZ (7-fold) levels without affecting rsmY (Fig. 5B). While our findings confirm the dependency of rsmYZ levels on GacA, rsmY levels in ΔsagS mutant cells grown planktonically appeared to be dependent primarily on HptB, previously shown to be GacA dependent (6).
BfiR is required for SagS to enable biofilm formation.
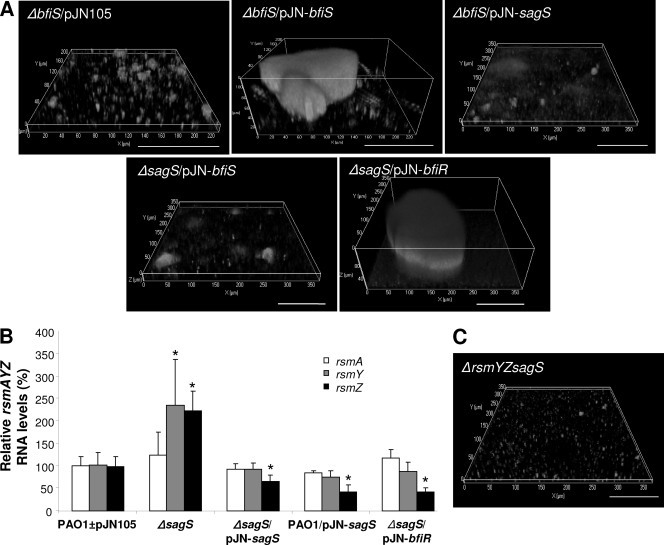
Phosphoproteome analysis suggested ΔsagS biofilms to be arrested prior to ΔbfiS biofilms at the transition from the planktonic to the initial attachment stage, indicating a possible link between these two sensory proteins. We reasoned that if SagS and BfiSR play distinct roles in biofilm formation, overexpression of bfiS or bfiR in the ΔsagS strain would not result in the restoration of wild-type biofilm formation (and vice versa). However, if SagS and BfiSR have overlapping or sequential functions during biofilm development, overexpression of bfiS or bfiR is anticipated to restore biofilm formation by the ΔsagS strain to wild-type levels.
Overexpression of sagS in the ΔbfiS strain had no effect on biofilm formation. Similarly, overexpression of bfiS, encoding the sensor kinase of the BfiSR TCS, in the ΔsagS strain did not affect the overall biofilm architecture (Fig. 6A; Table 1). The findings indicate the lack of a signal(s) required for the activation of either sensor kinase. In contrast, overexpression of bfiR, encoding the cognate response regulator of BfiS, in the ΔsagS strain resulted in the restoration of the biofilm architecture (Fig. 6A). Instead of forming thin biofilms lacking large cellular aggregates and microcolonies, ΔsagS/pJN-bfiR biofilms were, on average, 30 μm thick, mostly due to the presence of large microcolonies. Overexpression of bfiR furthermore resulted in a 25-fold increase in biofilm biomass compared to those of ΔsagS biofilms and a 3- to 4-fold increase compared to those of PAO1 biofilms of comparable age (Table 1).
Fig. 6.
Biofilm formation of the ΔsagS strain is restored by BfiR and correlates with reduced rsmYZ levels. (A) Expression of bfiR, but not bfiS, restores biofilm formation by the ΔsagS strain. Confocal images showing the biofilm architecture of ΔbfiS mutants harboring an empty plasmid or overexpressing bfiS and sagS and the biofilm architecture of ΔsagS mutants overexpressing bfiS and bfiR. (B) rsmAYZ RNA levels in ΔsagS biofilms complemented with or overexpressing sagS or bfiR, relative to the levels in P. aeruginosa PAO1 and PAO1/pJN105 (PAO1±pJN105), as determined by qRT-PCR. (C) The biofilm architecture of the ΔsagS strain is not restored to wild-type levels upon inactivation of rsmYZ, as revealed by confocal microscopy. White bars = 100 μm.
Restoration of the ΔsagS biofilm architecture to wild-type levels by complementation or overexpression of bfiR correlated with rsmYZ levels that were comparable to those of wild-type and vector-only controls (Fig. 6B). Compared to ΔsagS biofilms, complemented biofilms had 3.37-fold-reduced rsmZ levels, while overexpression of sagS or bfiR coincided with an ∼5.2-fold reduction in rsmZ abundance. It should be noted that rsmA levels were unaltered (Fig. 6B). These findings underscored previous observations of a negative correlation between rsmZ levels and biofilm formation (44). However, while the findings suggested that the biofilm formation defect observed for the ΔsagS strain results, at least in part, from elevated rsmYZ abundance, deletion of rsmYZ in the ΔsagS strain did not restore biofilm formation to wild-type levels. Instead, ΔrsmYZ ΔsagS biofilms were comparable in biofilm architecture to ΔsagS biofilms (Fig. 6C; Table 1), suggesting that while SagS is involved in modulating sRNA levels, the ΔsagS biofilm phenotype is independent of rsmYZ.
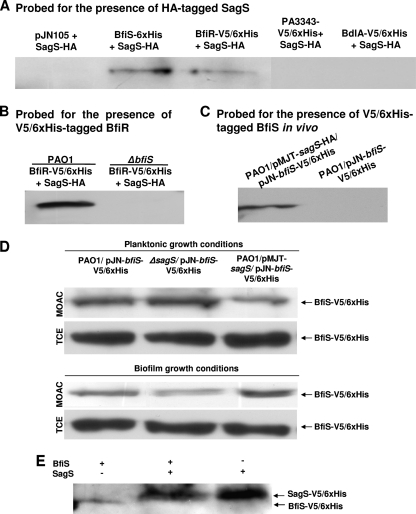
SagS interacts directly with BfiS but only indirectly with BfiR.
The observation that ΔsagS biofilm formation was restored to wild-type levels by bfiR overexpression suggested either that SagS and BfiSR belong to distinct pathways that are short-circuited by bfiR overexpression or instead that these proteins may function through direct interaction to enable biofilm formation and to receive an as-yet-unknown signal from SagS that is absent in ΔsagS biofilms. To test these hypotheses, we assessed whether SagS forms a complex with BfiSR in vitro. A pulldown of a complex of 6×His-tagged BfiS and hemagglutinin-tagged SagS (SagS-HA), as well as a complex of BfiS-V5/6×His and SagS-HA, could be clearly demonstrated when pulldown assays were probed for HA-tagged SagS (Fig. 7A). Two proteins were used as controls to determine the specificity of the SagS-BfiS interaction, namely, the inner membrane protein PA3343, characterized by 5 predicted transmembrane helices harboring a CheY-like receiver domain and a GGDEF domain having 64% similarity to the sensory histidine kinase of Synechocystis spp. (65), and BdlA (36). Neither BdlA nor PA3343 was detected in a complex with SagS-HA (Fig. 7A). Complex formation was also detectable when SagS-HA was used as bait to detect 6×His-tagged BfiS (not shown) and BfiR (Fig. 7B). However, the interaction of SagS with BfiR was found to be indirect and to occur via BfiS, as no interaction between BfiR and SagS was detected in a ΔbfiS mutant background (Fig. 7B). Complex formation of SagS and BfiS was confirmed by in vivo pulldown assays (Fig. 7C) and by using purified enzymes (not shown), thus indicating that the two proteins work in concert.
Fig. 7.
SagS directly interacts with and modulates the phosphorylation status of BfiS in a growth-mode-dependent manner. (A) Extracts of PAO1 bearing HA-tagged SagS were incubated with extracts containing 6×His-tagged BfiS or BfiR or a vector control (pJN105), and incubation was followed by anti-6×His immunoprecipitation. The immunoprecipitation eluates were subsequently assessed by immunoblot analysis for the presence of SagS-HA using anti-HA antibodies. Tagged PA3343 and BdlA were used as negative controls. (B) SagS/BfiR interactions in the wild-type (PAO1/pJN-bfiR-V5/6×His, PAO1/pJN-sagS-HA) and ΔbifS mutant (ΔbfiS/pJN-bfiR-V5/6×His, ΔbfiS/pJN-sagS-HA) backgrounds were assessed via anti-HA pulldown assays followed by immunoblot analysis using anti-V5 antibodies for the presence of BfiR-V5/6×His. Cell extracts obtained from PAO1/pJN105 were used as controls. (C) SagS and BfiS form a complex in vivo as demonstrated by pulldown assays using PAO1/pMJT-sagS-HA/pJN-bfiS-V5/6×His. PAO1/pJN-bfiS-V5/6×His was used as a negative control. (D) Phosphorylation of BfiS is modulated by the absence/presence of SagS in a growth-mode-dependent manner, as revealed by anti-V5 immunoblotting for BfiS-V5 in phosphoproteins purified by MOAC and in total cell protein extracts (TCE). (E) Under planktonic conditions, SagS dephosphorylates BfiS.
BfiS phosphorylation is SagS dependent.
The observations of SagS forming a complex with BfiS to enable biofilm formation (Fig. 7A to C) raised the possibility of SagS being involved in the transfer of a phosphoryl group to BfiS. This was supported by the finding of ΔsagS biofilms lacking evidence of BfiS Ser/Thr phosphorylation (see Table S3 in the supplemental material). We therefore determined the consequences of SagS/BfiS interaction on the phosphorylation status of BfiS under planktonic and biofilm growth conditions in vivo by quantifying the amount of phosphorylated BfiS present following the purification of total phosphoproteins via metal oxide affinity chromatography (MOAC) in PAO1, ΔsagS, and PAO1/pMJT-sagS biofilm and planktonic cells overexpressing bfiS-V5/6×His. Under biofilm growth conditions, overexpression of sagS resulted in increased detection of phosphorylated BfiS via immunoblotting using anti-V5 antibodies compared to the level detected in the wild type (Fig. 7D). In contrast, deletion of sagS correlated with reduced levels of phosphorylated BfiS (Fig. 7D) compared with those found in the wild type. The findings are in strong support of BfiS phosphorylation being SagS dependent under biofilm growth conditions. Interestingly, differential expression of sagS had the opposite effect on BfiS phosphorylation under planktonic growth conditions, with overexpression of sagS correlating with reduced BfiS phosphorylation levels and a lack of sagS resulting in increased levels of phosphorylated BfiS (Fig. 7D). The findings suggested the phosphorylation of BfiS to be SagS dependent, with SagS reducing or interfering with BfiS phosphorylation under planktonic conditions.
To demonstrate the transfer of a phosphoryl group between SagS and BfiS in vitro, we made use of the finding that both V5/6×His-tagged proteins are phosphorylated when overexpressed in E. coli (Fig. 7E). Purified SagS and BfiS were coincubated and subjected to phosphoprotein enrichment via MOAC, with the resulting protein eluates being subjected to SDS-PAGE and immunoblot analysis using anti-V5 antibodies. Coincubation of SagS with BfiS led to a significant decrease in the phosphorylation of BfiS in vitro (Fig. 7E), confirming phosphotransfer between SagS and BfiS, with SagS acting as a phosphatase under planktonic growth conditions.
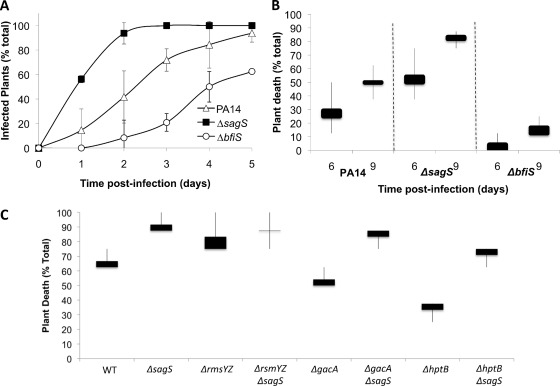
Inactivation of sagS renders P. aeruginosa hypervirulent.
Transitions between free-swimming and surface-associated modes of growth have been previously linked to global shifts in the virulence mechanisms of P. aeruginosa (17, 63). In P. aeruginosa, this switch has been shown to depend on sRNA levels and the GacA/Rsm system orchestrating the reciprocal expression of virulence factors required for acute infection and the coordinate repression of genes promoting adaptation to chronic persistence (17, 31, 32, 66, 67). Furthermore, BfiSR, which regulates biofilm formation postattachment via modulation of rmsYZ levels, has also been shown to be essential for pathogenesis. As the present findings implicated SagS in the regulation of rsmYZ levels, as well as in the modulation of the phosphorylation state of BfiS, we asked whether the function of this hybrid regulator impacts P. aeruginosa virulence properties. Using the A. thaliana infection model, we observed that the ΔsagS strain elicits an earlier onset of initial signs of infection than the wild type, as indicated by lesions and discoloration (Fig. 8A). Moreover, the ΔsagS strain was hypervirulent and resulted in A. thaliana plant death 3 days earlier than did the wild type (Fig. 8B). These results suggested a contribution of SagS signaling to the virulence of P. aeruginosa. In contrast, ΔbfiS mutants were avirulent (44) and showed significantly delayed onsets of initial signs of infection and significantly reduced virulence compared to the wild type and the ΔsagS mutant (Fig. 8A and B). Considering that the main difference observed between the two strains is the growth mode dependence of rsmYZ levels, we determined the virulence properties of the ΔsagS strain in the absence of rsmYZ (Fig. 8C). The triple mutant was found to exhibit a hypervirulent phenotype comparable to that of the ΔsagS single mutant. Inactivation of sagS in the ΔgacA or ΔhptB strain rendered these strains hypervirulent, significantly increasing their virulence. These results indicated that the SagS virulence phenotype and the difference in virulence between the ΔbfiS and ΔsagS strains are not dependent on rsmYZ levels.
Fig. 8.
Effect of sagS inactivation on P. aeruginosa virulence. A. thaliana plants, grown with the indicated PA14 strains, were monitored for signs of infections, indicated by discoloration of plants (A), and for death, manifesting as wilting and necrosis spread throughout the plant (B). (C) Inactivation of sagS renders the ΔgacA, ΔhptB, and ΔrsmYZ mutant strains more virulent. The graphs represent the averages of results from three experiments, with 8 plants used per replicate per strain. Lines indicate the differences between the highest and lowest survival rates observed, while bars represent distributions between the means and medians of the replicates. WT, wild type.
DISCUSSION
The opportunistic pathogen P. aeruginosa is capable of causing a wide variety of human diseases, ranging from bacteremia caused primarily by free-swimming cells to biofilm-related diseases due to colonization of medical devices and chronic infections of immunocompromised patients and those suffering from cystic fibrosis. The ability of P. aeruginosa to persist as free-swimming or biofilm cells is dependent on posttranslational modification and/or the differential expression of genes required for the maintenance of either mode of growth, resulting in the modulation of small regulatory RNA (sRNA) levels (7, 8, 17, 18, 44, 63). Here, we demonstrate that SagS (PA2824) is essential for the transition from the free-swimming to the sessile mode of growth. The transition from the free-swimming to the sessile mode of growth regulated by SagS correlated with the modulation of sRNA levels, with elevated levels of rsmYZ being observed in planktonically growing cells. The planktonic-growth-specific modulation of rsmYZ in the ΔsagS strain was found to be upstream of GacA but dependent on HptB (Fig. 5B). Indeed, SagS has been demonstrated to be one of three sensor kinase hybrids that undergo autophosphorylation and transfer a phosphoryl group specifically to the histidine phosphotransfer protein HptB, which in turn relays the signal to the response regulator PA3346 (6, 22). HptB is also capable of phosphorylating RetS, and both HptB and RetS intersect at the GacA response regulator, which directly controls sRNA production.
Similarly to RetS, SagS is a member of the sensor kinase/response regulator hybrid family of signal transduction proteins (Fig. 1) and is encoded by a single gene operon, with no genes encoding response regulators in the close proximity of sagS (17, 33, 65). BLAST and BLINK analysis revealed the presence of orthologs primarily in the genomes of proteobacteria, including Pseudomonas putida, Pseudomonas fluorescens, Pseudomonas syringae, Geobacter spp., Shewanella spp., and Vibrio spp. In addition to sensor kinase/response regulator domain structure and genomic context, the two proteins share other similarities. For instance, inactivation of sagS and retS coincided with increased Psl production and increased attachment in a microtiter plate assay (Fig. 1 and 3) (17), which is consistent with previous reports indicating that increased levels of rsmYZ enhance attachment (17, 18, 63). Modulation of these properties in the ΔsagS strain was independent of motility or cyclic di-GMP levels. A similar disconnect between attachment capabilities, Psl production, and motility was recently demonstrated by Merritt et al. (35), indicating that these characteristics are not necessarily linked. The effect of sagS inactivation on attachment, however, was only temporary, with the reduction of attached biomass being noticeable as soon as 8 h following initial attachment (Fig. 1 and 2; Table 1) (43). Moreover, a ΔsagS mutant was impaired at early stages of biofilm development in a manner similar to that observed for the ΔretS and ΔbfiS strains (Fig. 2) (17, 43, 44). Similar analysis performed with the ΔbfiS strain showed a strictly antagonistic control of rsmYZ, with elevated levels of sRNAs in biofilm cells but no variation in the sRNAs of cells in the planktonic growth mode (Fig. 4). The findings identified the probable sensor/response regulator hybrid SagS as part of the regulatory system linking the opposing factors/proteins that reciprocally modulate sRNA levels prior to and after attachment to enable biofilm formation (17, 18, 63). However, while the rsmYZ levels in the ΔsagS strain appeared to be dependent on HptB, the postattachment phenotypes of the two mutants were very similar, suggesting similar or convergent (but not necessarily dependent) regulatory roles for the two proteins in biofilm formation. Moreover, the ΔsagS biofilm phenotype was independent of rsmYZ levels, as indicated by the finding that while restoration of ΔsagS biofilm formation by BfiR correlated with a reduction in rsmYZ levels, inactivation of rsmYZ in the ΔsagS strain did not restore biofilm formation to wild-type levels (Fig. 6). The finding suggested that SagS, the potential cognate sensor for HptB, serves an additional function besides its involvement in controlling the levels of sRNAs to enable biofilm formation (Fig. 9). This function, which will be the subject of future studies, appears to be required (in a Gac/Rsm system-independent manner) for the activation of the two-component system BfiSR, which is essential for biofilm formation.
Fig. 9.
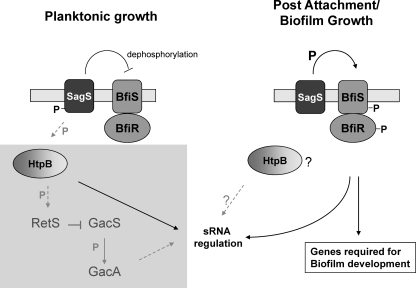
Model for the role of SagS in linking the Gac/HptB/Rsm and BfiSR signaling systems prior to and following the motile-sessile switch. The proposed model of differential SagS activity prior to and following the motile-sessile switch integrates present findings (indicated by black arrows) and previously published data (gray arrows with dashed lines). Under planktonic conditions, SagS is phosphorylated and directly interferes with BfiSR activity, probably via dephosphorylation of BfiSR. Moreover, SagS modulates sRNA levels under planktonic conditions in an HptB-dependent manner. GacAS and HptB contribute to the regulation of the small regulatory RNAs (sRNAs) rsmY and rsmZ, which act through RsmA sequestration. RetS participates in a phosphotransfer with HptB and directly interferes with GacS activity (18, 22). Transition to the surface (postattachment and under biofilm growth conditions) coincides with SagS dephosphorylation, presumably via phosphoryl group transfer from SagS to BfiS, resulting in BfiSR activation (43), suppression of sRNA levels (in particular, rsmZ levels), and activation of genes and gene products required for biofilm development by P. aeruginosa (43, 44). While both SagS and HptB play a role in biofilm formation, SagS function appears to be independent of HptB (and GacA). The lightly shaded square area indicates components belonging to the Gac/HptB/Rsm signaling system (without indicating proper location in the cell). P, phosphoryl group or phosphoryl group transfer; ?, unknown contribution to biofilm formation or sRNA regulation under biofilm growth conditions; IM, inner membrane.
Pulldown assays demonstrated that SagS interacts directly with BfiS and only indirectly with BfiR via BfiS (Fig. 7). However, overexpression of bfiR but not bfiS restored ΔsagS biofilm formation to wild-type levels, suggesting that in order for biofilm formation to occur, SagS is required to interact with BfiS, resulting in the relay of a signal which is lacking in ΔsagS biofilms (Fig. 6 and 7). Considering that SagS is not phosphorylated under biofilm growth conditions after 8 h of attachment, a time at which BfiSR phosphorylation becomes detectable by anti-phospho-Ser/Thr immunoblot analysis (43), the signal likely consists of a phosphoryl group relayed from the hybrid sensor SagS to BfiS. The experiments described here are consistent with a model in which SagS enhances the phosphorylation of BfiS under biofilm growth conditions while blocking phosphorylation or dephosphorylating BfiS under planktonic conditions (Fig. 9).
The signals that trigger the opposing activities of SagS with respect to BfiS phosphorylation are unclear. However, SagS-BfiS complex formation and the phosphotransfer between these two sensor proteins are reminiscent of complex formation and the phosphotransfer of the RetS and GacS heterodimer described previously (18), with complex formation blocking GacS autophosphorylation (and subsequent phosphotransfer to the response regulator GacA), leading to a reduction in rsmYZ expression. The finding suggested that heterologous sensor kinases can interact directly to form multisensor signaling networks, with RetS interaction acting as a switch in modulating the downstream activity of the GacAS system (18). The observation furthermore suggested that by modulating signal transduction at the level of sensor kinase phosphorylation, multiple inputs can be integrated without the need for cross-phosphorylation between sensors and noncognate response regulators (18). In a manner similar to that of RetS, SagS acts as a switch to regulate the transition from the planktonic to the surface-attached mode of growth by directly interacting with and modulating the phosphorylation status of BfiS. Present findings are consistent with a model in which SagS suppresses sRNA accumulation and sequestration of the mRNA-binding protein RsmA under planktonic growth conditions, probably by acting in concert with HptB (6, 22) while blocking BfiS phosphorylation (Fig. 7D and E). Upon transition to surface-associated growth, however, SagS interacts with BfiSR to promote BfiS phosphorylation (Fig. 7D) and, likely, BfiSR activation, with this TCS playing an essential role in suppressing sRNA levels and enabling P. aeruginosa to transition from reversible to irreversible attachment (43, 44). Our findings are summarized in a model shown in Fig. 9. Additional support for SagS acting as a switch is given by the observation that SagS and BfiS modulate rsmYZ levels in opposing manners, which are dependent on the mode of growth (Fig. 4). Moreover, SagS and BfiSR differ in their contributions to the virulence of P. aeruginosa when tested in a nonmammalian virulence model. While the ΔbfiS mutant was avirulent, a ΔsagS mutant strain was hypervirulent (Fig. 8) (44). Taking into account that both the ΔsagS and ΔbfiS biofilms are arrested in biofilm development (with ΔsagS biofilms being arrested prior to ΔbfiS biofilms) (Fig. 2; see also Table S3 in the supplemental material), the findings suggest possible roles for SagS and BfiSR in contributing to the switch from acute infection to chronic persistence.
In conclusion, our data suggest that the sensor kinase hybrid SagS regulates the transition between the motile and sessile modes of growth by modulating sRNA levels in a growth-mode-dependent manner (Fig. 9). Moreover, SagS interacts with BfiS in a manner reminiscent of the RetS-GacS heterodimer interaction. To our knowledge, this is the first description of a P. aeruginosa protein enabling the motile-sessile switch by linking the reciprocal modulation of sRNA levels via the Gac/HptB/Rsm system to the signal transduction network composed of BfiSR, BfmSR, and MifSR, previously demonstrated to be required for the development of P. aeruginosa biofilms, in a sequential and stage-specific manner, into multisensor signaling networks.
Supplementary Material
ACKNOWLEDGMENTS
We thank D. Wozniak for kindly providing the anti-Psl antibody and D. Haas, A. Filloux, and H.-L. Peng for kindly providing the ΔhptB, ΔrsmYZ, and ΔgacA mutant strains and the respective parental strains.
This work was supported by grants from the Army Research Office (W911NF0710604) and the National Institutes of Health (2R15 HL073835-02).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 23 September 2011.
REFERENCES
- 1. Allegrucci M., et al. 2006. Phenotypic characterization of Streptococcus pneumoniae biofilm development. J. Bacteriol. 188:2325–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allegrucci M., Sauer K. 2007. Characterization of colony morphology variants isolated from Streptococcus pneumoniae biofilms. J. Bacteriol. 189:2030–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Allegrucci M., Sauer K. 2008. Formation of Streptococcus pneumoniae non-phase-variable colony variants is due to increased mutation frequency present under biofilm growth conditions. J. Bacteriol. 190:6330–6339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Amikam D., Steinberger O., Shkolnik T., Ben-Ishai Z. 1995. The novel cyclic dinucleotide 3′-5′ cyclic diguanylic acid binds to p21ras and enhances DNA synthesis but not cell replication in the Molt 4 cell line. Biochem. J. 311:921–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barraud N., et al. 2009. Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di-GMP levels, and enhanced dispersal. J. Bacteriol. 191:7333–7342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bordi C., et al. 2010. Regulatory RNAs and the HptB/RetS signalling pathways fine-tune Pseudomonas aeruginosa pathogenesis. Mol. Microbiol. 76:1427–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brencic A., Lory S. 2009. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol. Microbiol. 72:612–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brencic A., et al. 2009. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol. Microbiol. 73:434–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burrowes E., Baysse C., Adams C., O'Gara F. 2006. Influence of the regulatory protein RsmA on cellular functions in Pseudomonas aeruginosa PAO1, as revealed by transcriptome analysis. Microbiology 152:405–418 [DOI] [PubMed] [Google Scholar]
- 10. Byrd M. S., et al. 2009. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol. Microbiol. 73:622–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Caiazza N. C., Merritt J. H., Brothers K. M., O'Toole G. A. 2007. Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 189:3603–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Caiazza N. C., O'Toole G. A. 2004. SadB is required for the transition from reversible to irreversible attachment during biofilm formation by Pseudomonas aeruginosa PA14. J. Bacteriol. 186:4476–4485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Costerton J. W., Lewandowski Z., Caldwell D. E., Korber D. R., Lappin-Scott H. M. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711–745 [DOI] [PubMed] [Google Scholar]
- 14. D'Argenio D. A., Miller S. I. 2004. Cyclic di-GMP as a bacterial second messenger. Microbiology 150:2497–2502 [DOI] [PubMed] [Google Scholar]
- 15. Flemming H.-C., Neu T. R., Wozniak D. J. 2007. The EPS matrix: the “house of biofilm cells.” J. Bacteriol. 189:7945–7947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Friedman L., Kolter R. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51:675–690 [DOI] [PubMed] [Google Scholar]
- 17. Goodman A. L., et al. 2004. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 7:745–754 [DOI] [PubMed] [Google Scholar]
- 18. Goodman A. L., et al. 2009. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 23:249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Heurlier K., et al. 2004. Positive control of swarming, rhamnolipid synthesis, and lipase production by the posttranscriptional RsmA/RsmZ system in Pseudomonas aeruginosa PAO1. J. Bacteriol. 186:2936–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Heydorn A., et al. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395–2407 [DOI] [PubMed] [Google Scholar]
- 21. Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J., Schweizer H. P. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86 [DOI] [PubMed] [Google Scholar]
- 22. Hsu J. L., Chen H. C., Peng H. L., Chang H. Y. 2008. Characterization of the histidine-containing phosphotransfer protein B-mediated multistep phosphorelay system in Pseudomonas aeruginosa PAO1. J. Biol. Chem. 283:9933–9944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Irie Y., et al. 2010. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol. Microbiol. 78:158–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jackson D. W., Simecka J. W., Romeo T. 2002. Catabolite repression of Escherichia coli biofilm formation. J. Bacteriol. 184:3406–3410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jackson D. W., et al. 2002. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J. Bacteriol. 184:290–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jackson K. D., Starkey M., Kremer S., Parsek M. R., Wozniak D. J. 2004. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J. Bacteriol. 186:4466–4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jenal U. 2004. Cyclic di-guanosine-monophosphate comes of age: a novel secondary messenger involved in modulating cell surface structures in bacteria? Curr. Opin. Microbiol. 7:185–191 [DOI] [PubMed] [Google Scholar]
- 28. Jonas K., et al. 2008. The RNA binding protein CsrA controls cyclic di-GMP metabolism by directly regulating the expression of GGDEF proteins. Mol. Microbiol. 70:236–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Klausen M., et al. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 48:1511–1524 [DOI] [PubMed] [Google Scholar]
- 30. Kuchma S. L., et al. 2007. BifA, a cyclic-di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 189:8165–8178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Laskowski M. A., Kazmierczak B. I. 2006. Mutational analysis of RetS, an unusual sensor kinase-response regulator hybrid required for Pseudomonas aeruginosa virulence. Infect. Immun. 74:4462–4473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Laskowski M. A., Osborn E., Kazmierczak B. I. 2004. A novel sensor kinase-response regulator hybrid regulates type III secretion and is required for virulence in Pseudomonas aeruginosa. Mol. Microbiol. 54:1090–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lin C.-T., et al. 2006. Identification of an HptB-mediated multi-step phosphorelay in Pseudomonas aeruginosa PAO1. Res. Microbiol. 157:169–175 [DOI] [PubMed] [Google Scholar]
- 34. Merritt J. H., Brothers K. M., Kuchma S. L., O'Toole G. A. 2007. SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J. Bacteriol. 189:8154–8164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Merritt J. H., et al. 2010. Specific control of Pseudomonas aeruginosa surface-associated behaviors by two c-di-GMP diguanylate cyclases. mBio 1(4):e00183–10 doi:10.1128/mBio.00183-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morgan R., Kohn S., Hwang S.-H., Hassett D. J., Sauer K. 2006. BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. J. Bacteriol. 188:7335–7343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mulcahy H., et al. 2008. Pseudomonas aeruginosa RsmA plays an important role during murine infection by influencing colonization, virulence, persistence, and pulmonary inflammation. Infect. Immun. 76:632–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Newman J. R., Fuqua C. 1999. Broad-host-range expression vectors that carry the arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227:197–203 [DOI] [PubMed] [Google Scholar]
- 39. O'Toole G. A., Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295–304 [DOI] [PubMed] [Google Scholar]
- 40. O'Toole G. A., Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449–461 [DOI] [PubMed] [Google Scholar]
- 41. Parkins M. D., Ceri H., Storey D. G. 2001. Pseudomonas aeruginosa GacA, a factor in multihost virulence, is also essential for biofilm formation. Mol. Microbiol. 40:1215–1226 [DOI] [PubMed] [Google Scholar]
- 42. Pessi G., et al. 2001. The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J. Bacteriol. 183:6676–6683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Petrova O. E., Sauer K. 2009. A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog. 5:e1000668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Petrova O. E., Sauer K. 2010. The novel two-component regulatory system BfiSR regulates biofilm development by controlling the small RNA rsmZ through CafA. J. Bacteriol. 192:5275–5288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reimmann C., Valverde C., Kay E., Haas D. 2005. Posttranscriptional repression of GacS/GacA-controlled genes by the RNA-binding protein RsmE acting together with RsmA in the biocontrol strain Pseudomonas fluorescens CHA0. J. Bacteriol. 187:276–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Romeo T. 1998. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol. Microbiol. 29:1321–1330 [DOI] [PubMed] [Google Scholar]
- 47. Romling U., Amikam D. 2006. Cyclic di-GMP as a second messenger. Curr. Opin. Microbiol. 9:218–228 [DOI] [PubMed] [Google Scholar]
- 48. Ross P., et al. 1990. The cyclic diguanylic acid regulatory system of cellulose synthesis in Acetobacter xylinum. Chemical synthesis and biological activity of cyclic nucleotide dimer, trimer, and phosphothioate derivatives. J. Biol. Chem. 265:18933–18943 [PubMed] [Google Scholar]
- 49. Ross P., et al. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325:279–281 [DOI] [PubMed] [Google Scholar]
- 50. Ryder C., Byrd M., Wozniak D. J. 2007. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr. Opin. Microbiol. 10:644–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sauer K., Camper A. K. 2001. Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J. Bacteriol. 183:6579–6589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sauer K., Camper A. K., Ehrlich G. D., Costerton J. W., Davies D. G. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schweizer H. P. 1991. The agmR gene, an environmentally responsive gene, complements defective glpR, which encodes the putative activator for glycerol metabolism in Pseudomonas aeruginosa. J. Bacteriol. 173:6798–6806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schweizer H. P., Hoang T. T. 1995. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 158:15–22 [DOI] [PubMed] [Google Scholar]
- 55. Shrout J., et al. 2006. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 62:1264–1277 [DOI] [PubMed] [Google Scholar]
- 56. Simm R., Morr M., Kader A., Nimtz M., Romling U. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53:1123–1134 [DOI] [PubMed] [Google Scholar]
- 57. Southey-Pillig C. J., Davies D. G., Sauer K. 2005. Characterization of temporal protein production in Pseudomonas aeruginosa biofilms. J. Bacteriol. 187:8114–8126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Starkey M., Rahme L. G. 2009. Modeling Pseudomonas aeruginosa pathogenesis in plant hosts. Nat. Protoc. 4:117–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Suzuki K., Babitzke P., Kushner S. R., Romeo T. 2006. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev. 20:2605–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Thormann K. M., et al. 2006. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J. Bacteriol. 188:2681–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Toutain C. M., Zegans M. E., O'Toole G. A. 2005. Evidence for two flagellar stators and their role in the motility of Pseudomonas aeruginosa. J. Bacteriol. 187:771–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Vallet I., Olson J. W., Lory S., Lazdunski A. E., Filloux A. 2001. The chaperone/usher pathways of Pseudomonas aeruginosa: identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc. Natl. Acad. Sci. U. S. A. 98:6911–6916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ventre I., et al. 2006. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc. Natl. Acad. Sci. U. S. A. 103:171–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Weilbacher T., et al. 2003. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol. Microbiol. 48:657–670 [DOI] [PubMed] [Google Scholar]
- 65. Winsor G. L., et al. 2009. Pseudomonas genome database: facilitating user-friendly, comprehensive comparisons of microbial genomes. Nucleic Acids Res. 37:D483–D488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zolfaghar I., et al. 2005. Mutation of retS, encoding a putative hybrid two-component regulatory protein in Pseudomonas aeruginosa, attenuates multiple virulence mechanisms. Microbes Infect. 7:1305–1316 [DOI] [PubMed] [Google Scholar]
- 67. Zolfaghar I., Evans D. J., Ronaghi R., Fleiszig S. M. 2006. Type III secretion-dependent modulation of innate immunity as one of multiple factors regulated by Pseudomonas aeruginosa RetS. Infect. Immun. 74:3880–3889 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.