Abstract
Vibrio cholerae secretes the Zn-dependent metalloprotease hemagglutinin (HA)/protease (mucinase), which is encoded by hapA and displays a broad range of potential pathogenic activities. Expression of HA/protease has a stringent requirement for the quorum-sensing regulator HapR and the general stress response regulator RpoS. Here we report that the second messenger cyclic diguanylic acid (c-di-GMP) regulates the production of HA/protease in a negative manner. Overexpression of a diguanylate cyclase to increase the cellular c-di-GMP pool resulted in diminished expression of HA/protease and its positive regulator, HapR. The effect of c-di-GMP on HapR was independent of LuxO but was abolished by deletion of the c-di-GMP binding protein VpsT, the LuxR-type regulator VqmA, or a single-base mutation in the hapR promoter that prevents autorepression. Though expression of HapR had a positive effect on RpoS biosynthesis, direct manipulation of the c-di-GMP pool at a high cell density did not significantly impact RpoS expression in the wild-type genetic background. In contrast, increasing the c-di-GMP pool severely inhibited RpoS expression in a ΔhapR mutant that is locked in a regulatory state mimicking low cell density. Based on the above findings, we propose a model for the interplay between HapR, RpoS, and c-di-GMP in the regulation of HA/protease expression.
INTRODUCTION
Cholera is an acute waterborne diarrheal disease caused by Vibrio cholerae of serogroups O1 and O139. Infecting V. cholerae cells adhere to the intestinal mucosa, where they express major virulence factors such as the toxin coregulated pilus (TCP) (21) and cholera toxin (CT), which is largely responsible for the profuse rice-watery diarrhea typical of this illness (12, 26). Cholera patients shed V. cholerae in their stools, which usually contain about 108 hyperinfective V. cholerae cells per ml (38). The V. cholerae cells can then survive and persist in fresh water and estuarine aquatic ecosystems to eventually gain entrance to a new host.
The Zn-dependent metalloprotease hemagglutinin (HA)/protease (18) has been proposed to facilitate V. cholerae detachment from the intestinal mucosa when infecting V. cholerae cells reach a high density (4, 14, 43, 44). Inactivation of hapA, which encodes HA/protease, has been shown to enhance adherence to mucin-coated polystyrene plates (43), adherence to mucin-secreting differentiated HT29-18N2 cultured cells (4), and colonization of the suckling mouse intestine (41, 43). The mucinase activity of HA/protease (13), together with its capacity to cleave the mucin-binding adhesin GbpA at a high cell density, provides a mechanism that supports the “detachase” function attributed to this protein (25). In addition, HA/protease has been reported to perturb the paracellular barrier of cultured intestinal epithelial cells (37, 55) by acting on tight junction-associated proteins (56). In agreement with this finding, we have shown that HA/protease enhances enterotoxicity in the rabbit ileal loop model (43).
The expression of flagellar motility is required for intestinal colonization (28, 43). In addition, experiments conducted with the rabbit ileal-loop model suggest that an RpoS-dependent upregulation of motility in the stationary phase is responsible for the detachment of V. cholerae cells from the intestinal mucosa and their escape into the luminal fluid (40). We have shown that motility also enhances enterotoxicity (fluid accumulation) in rabbit ileal loops in which the deletion of both hapA and motY had the most prominent attenuating effect (43). The above evidences suggest that production of HA/protease and motility could occur at one or more stages of infection to jointly promote pathogenesis and/or detachment.
The expression of HA/protease and motility are positively regulated by the cyclic AMP receptor protein (CRP) and the quorum-sensing regulator HapR (2, 22, 42). We have shown that CRP acts upstream of HapR by inducing the biosynthesis of cholera autoinducer 1 (CAI-1) to activate quorum sensing (30, 31, 42). At high cell density, HapR binds to the promoter of hapA to enhance protease production (52). Gene expression profiling has shown that HapR also positively affects the expression of numerous genes required for the expression of flagellar motility and chemotaxis (40, 58). Expression of HapR has clearly been shown to diminish the intracellular concentration of the second messenger cyclic diguanylic acid (c-di-GMP), an inhibitor of flagellar motility (17, 54).
HA/protease production also requires the general stress response regulator RpoS (42, 57). Although RpoS has also been shown to enhance flagellar motility and chemotaxis (40), its role in c-di-GMP signaling in V. cholerae has not been established.
Cyclic di-GMP has been shown to regulate the transition between the sessile and motile lifestyles by activating biofilm formation and inhibiting motility in a broad spectrum of bacterial species (8, 19, 46, 49). The V. cholerae genome contains 31 genes encoding GGDEF domain family proteins, 10 genes encoding proteins with GGDEF and EAL domains, 12 genes encoding proteins with only EAL domains, and 9 genes encoding proteins with HD-GYP domains (15). The GGDEF domain family proteins have been shown to exhibit a diguanylate cyclase (DGC) activity that makes c-di-GMP from GTP. The EAL and HD-GYP domain families have been reported to exhibit a phosphodiesterase (PDE) activity that degrades c-di-GMP to GMP (15). In V. cholerae, overexpression of the DGC VCA0956 abolishes swimming, whereas expression of the PDE VieA enhances it (51). Transcriptional profiling has revealed that genes involved in flagellum biosynthesis, motility, and chemotaxis are repressed in response to an increase in intracellular c-di-GMP content (5).
In this study, we examined the role of c-di-GMP in the regulation of HA/protease expression. We report that c-di-GMP negatively regulates the production of HA/protease and propose a model for the interplay between c-di-GMP, HapR, and RpoS in this regulation.
MATERIALS AND METHODS
Strains, media, plasmids, and oligonucleotide primers.
Strains, plasmids, and primers used in the study are listed and described briefly in Tables 1 and 2. All V. cholerae strains were derived from C7258 (El Tor biotype, serotype Ogawa). For HA/protease production, strains were grown in tryptic soy broth (TSB) at 37°C with agitation (225 rpm) to stationary phase. Culture media were supplemented with ampicillin (Amp; 100 μg/ml), polymixin B (PolB; 100 units/ml), chloramphenicol (Cm; 5 μg/ml), or kanamycin (Km; 50 μg/ml) as required.
Table 1.
V. cholerae strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| C7258 | Wild type O1; El Tor biotype | Peru, 1991 |
| N16961 | Wild type O1; El Tor biotype; hapR negative | Bangladesh, 1975 |
| AJB50 | C7258 ΔrpoS | 30 |
| AJB51 | C7258 ΔhapR | 30 |
| AJB52 | C7258 ΔhapRΔrpoS | This study |
| AJB501 | C7258 ΔluxO | This study |
| AJB502 | C7258 rpoS::pCVDRpoS-FLAG | This study |
| AJB503 | AJB51 rpoS::pCVDRpoS-FLAG | This study |
| AJB504 | AJB501 rpoS::pCVDRpoS-FLAG | This study |
| N16961-FLAG | N16961 rpoS::pCVDRpoS-FLAG | This study |
| SZS007 | C7258 luxC-lacZ reporter | 47 |
| SZS012 | SZS007 ΔluxO | 47 |
| HX01 | SZS007 hapRp* | This study |
| HX02 | SZS007 ΔvpsT | This study |
| HX03 | AJB51 ΔvpsT | This study |
| AJB505 | SZS007 ΔvqmA | This study |
| Plasmids | ||
| pAT1568 | PDE VieA in pBAD33 | 51 |
| pAT1615 | Inactive VieA(E170A) in pBAD33 | 51 |
| pAT1662 | DGC VCA0956 in pBAD33 | 51 |
| pCMW75 | DGC QrgB expression vector | 54 |
| pTT3 | rrnB T1T2 transcription terminator in pUC19 | 44 |
| pCVDΔRpoS2 | DNA fragment carrying rpoS deletion in pCVD442 | 30 |
| pUCΔvqmA | SacI-BamHI and BamHI-SphI DNA fragments flanking vqmA sequentially cloned in pUC19 | This study |
| pCVDΔvqmA | SacI-SphI fragment harboring vqmA deletion in pCVD442 | This study |
| pUCΔvpsT | SacI-BamHI and BamHI-SphI DNA fragments flanking vpsT sequentially cloned in pUC19 | This study |
| pΔvpSTKm | 1.2-kb BamHI fragment encoding a Km resistance cassette from pUC4Ka in pUCΔvpsT | This study |
| pCVDΔvpsTKm | SacI-SphI fragment from pΔvpsTKm in pCVD442 | This study |
| pTT3RpoS-FLAG | SphI-PstI fragment containing rpoS ORF in pTT3 | This study |
| pCVDRpoS-FLAG | SphI-XbaI rpoS-FLAG and rrnB T1T2 fragment in pCVD442 | This study |
| pCVDhapRp* | Mutant hapR promoter in pCVD442 | This study |
The GenBank accession number of pUC4K is X06404.1.
Table 2.
Oligonucleotide primers
| Primer | Sequence (5′→3′)a |
|---|---|
| HapA1010 | GCACGGCGTTCAGTTATGCTTGTA |
| HapA1551 | AGGTAAAACGCGCGGTTAAACACG |
| HapR-F1 | GCCTCTAGATGAATTTGACGAGCAAGATC |
| HapR-R1 | GATGCATGCCTCAAACCAGACTTTGAG |
| HapRmut-F | CTTTAAGTAGCAAATAGCAAAATAATCATTAG |
| HapRmut-R | CTAATGATTATTTTGCTATTTGCTACTTAAAG |
| HapR589 | CGCCTCAAAAACGCAAACTACAACT |
| HapR1046 | ATGGAGTAGAAGATGCCGTGGAAC |
| RecA578 | GTGCTGTGGATGTCATCGTTGTTG |
| RecA863 | CCACCACTTCTTCGCCTTCTTTGA |
| RpoS3 | GAAGCATGCCAATACCGTAACCAAA |
| RpoS13 | GCCGAATTCGAGTGTCAGCAATAC |
| RpoS576 | CGATTTTGAAGATGAAGCACTGGAAG |
| RpoS994 | GCCAGATCTGTATTCGACGTTAAAC |
| RpoS1003 | TGTTTGGTTCATCAGCGCACGTTC |
| RpoS1020 | CTTCTGCAGTTACTTGTCGTCATCG |
| RpoSP189 | GGCTAGCAAAGGAAGGACAAACTGT |
| VpsT56 | CCAGATTGTTGAAAGAGGCGTTAG |
| VpsT252 | TGCGGACAGTTTATGATGACCTCT |
| VpsT690 | CTGCAAACACATATCGAGCTCTTCT |
| VpsT1220 | GCAGGATCCAAGCATTCTAACGTTTA |
| VpsT1395 | GAAGGATCCTGATGATGTTTTGACC |
| VpsT2093 | GGGATTATGAAACGTTTGGAAAAGCC |
| VqmA645 | GTAGAGCTCCATTCGGGTTAACTTG |
| VqmA1519 | GAAGGATCCGAACACACAGAGGTAT |
| VqmA2455 | GAAGGATCCCAGTAAGCAACAACGT |
| VqmA3334 | CAAGCATGCATGAGTGGTTCCTGTT |
Restriction sites used for directional cloning are underlined.
Construction of mutants.
To construct a vqmA deletion mutant, we amplified DNA fragments flanking the vqmA locus (VCA1078) from C7258 genomic DNA using primer pairs VqmA645/VqmA1519 and VqmA2455/VqmA3334. The PCR products were sequentially cloned in pUC19 to yield pUCΔvqmA and confirmed by DNA sequencing. Then the chromosomal DNA fragment harboring the vqmA deletion was transferred to the suicide vector pCVD442 (11) to yield pCVDΔvqmA and transferred by conjugation to strain SZS007 (Table 1). Exconjugants were selected in LB agar containing Amp and PolB. Finally, the segregant AJB505, harboring the vqmA deletion, was isolated by sucrose selection as described previously (30). An identical strategy was used to construct ΔvpsT mutants, except that primer pairs VpsT690/VpsT1220 and VpsT1395/VpsT2093 were used and a Km resistance gene was inserted in place of the deleted vpsT DNA. The vpsT deletion/insertion was introduced into strains SZS007 and AJB51 (ΔhapR) by conjugation as described above. The ΔluxO strain AJB501 was constructed by allelic exchange as previously described (47) but using strain C7258 as the conjugal receptor. Similarly, a mutant with deletions of rpoS and hapR (AJB52) was constructed by conjugal transfer of the suicide vector pCVDΔRpoS2 (30) to the hapR mutant AJB51 followed by sucrose selection. To construct a strain defective for HapR autorepression, we amplified the hapR promoter from strain C7258 using primers HapR-F1 and HapR-R1. The PCR product was cloned in pUC19 and confirmed by DNA sequencing. Then we used a QuikChange site-directed mutagenesis kit (Stratagene) to introduce an A→G mutation in the hapR promoter using primers HapRmut-F and HapRmut-R. The mutant promoter was subcloned in pCVD442 and transferred by conjugation to strain SZS007, and segregant HX01, which retained the mutant promoter designated hapRp*, was isolated by sucrose selection as above. The A→G (originally described as A18G) mutation has been shown to eliminate HapR binding to its own promoter and autorepression (33).
Construction of RpoS-FLAG reporter strains.
To construct a strain expressing an RpoS-tagged protein from native transcription and translation initiation signals, we amplified the rpoS open reading frame (ORF) lacking its stop codon from strain C7258 genomic DNA using primers RpoS13 and RpoS994 and an Advantage 2 PCR kit (BD Biosciences Clontech). The resulting PCR fragment was confirmed by DNA sequencing and cloned as an EcoRI-BgLII fragment in pFLAG-CTC (Sigma-Aldrich) in frame with the FLAG epitope. The RpoS-FLAG fusion was then retrieved by PCR with primers RpoS3 and RpoS1020 and inserted into plasmid pTT3 (44) upstream of the rrnB T1T2 transcription terminator to yield pTT3RpoS-FLAG. Finally, the rpoS-FLAG-rrnB T1T2 cassette was transferred to the suicide vector pCVD442 to yield pCVDRpoS-FLAG. The resulting suicide vector was transferred from S17-1λpir (9) to V. cholerae strains C7258, AJB51 (ΔhapR), AJB501 (ΔluxO), HX03 (ΔhapRΔvpsT), and N16961 by conjugation, and exconjugants were selected on LB plates containing Amp and PolB. Integration by homologous recombination within the rpoS locus was confirmed by PCR using primer RpoSP189, which anneals to the rpoS promoter region (not present in pCVDRpoS-FLAG), and primer RpoS1020, which anneals to the FLAG epitope. The resulting PCR products were further confirmed by DNA sequencing, and the expression of RpoS-FLAG was established by Western blot analysis.
qRT-PCR.
Total RNA was isolated using an RNeasy kit (Qiagen Laboratories). The RNA samples were analyzed by quantitative real-time reverse-transcription-PCR (qRT-PCR) using an iScript two-step RT-PCR kit with SYBR green (Bio-Rad Laboratories) as described previously (30, 45). Relative expression values were calculated as 2−(CT target − CT reference), where CT is the fractional threshold cycle. The level of recA mRNA was used as a reference. Manipulation of the c-di-GMP pool did not affect recA mRNA levels. The following primer combinations were used: HapA1010 and HapA1551 for hapA mRNA, HapR589 and HapR1046 for hapR mRNA, RecA578 and RecA863 for recA mRNA, RpoS576 and RpoS1003 for rpoS mRNA, and VpsT56 and VpsT252 for vpsT mRNA. A control mixture lacking reverse transcriptase was run for each reaction to exclude chromosomal DNA contamination.
Manipulation of the c-di-GMP pool.
For manipulation of the c-di-GMP pool, the PDE VieA, the inactive PDE VieA(E170A), and the soluble DGC VCA0956 were overexpressed from the pBAD promoter of the araBAD (arabinose) operon. Plasmids encoding these enzymes (Table 1) were introduced into V. cholerae by electroporation as described in reference 36. For studying the effect of c-di-GMP on HA/protease production and its regulators, HapR and RpoS, V. cholerae transformants were grown in TSB to an optical density at 600 nm (OD600) of 2.0, the onset of HA/protease expression (2). At that time, cultures were divided in halves, and one half was induced with 0.2% l-arabinose and the second half was used as a control. Cultures were then incubated at 37°C with shaking for different times up to 16 h. Alternatively, plasmid pCMW75, encoding the V. harveyi DGC QrgB (54), was transformed into V. cholerae AJB503 and N16961-FLAG, and its expression was induced as described above but with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside).
Measurement of c-di-GMP content.
Manipulation of the c-di-GMP pool was confirmed by direct measurement of the c-di-GMP pool as described in reference 54. Briefly, pelleted biomass samples containing the same number of cells were washed with cold phosphate-buffered saline (PBS), pH 7.4, and extracted twice with 40% methanol–40% acetonitrile–0.1N formic acid. The supernatants from each extraction were combined and dried in an Eppendorf Vacufuge plus vacuum concentrator instrument. The extracted material was reconstituted in 30 μl of PBS, and c-di-GMP was measured by using liquid chromatography–tandem mass spectrometry (LC-MS-MS) with an Applied Biosystems API 3200 triple-quadrupole mass spectrometer instrument operating in positive turbo ion spray mode. The mass spectrometer and high-performance liquid chromatography (HPLC) were controlled with Analyst 1.4.2 software in multiple-reaction-monitoring mode. The mass/charge (m/z) transitions used for c-di-GMP were m/z 689.2 to 344, m/z 344 to 150, m/z 150 to 133, and m/z 133 to 79. The dwell time was 40 ms for all mass transitions, and the ion spray voltage was −4,500 V. For chromatography of c-di-GMP, gradients were generated with a Waters Acquity UPLC system consisting of a vacuum degasser, binary pump, and temperature-controlled autosampler. The injection volume was 10 μl, and the samples were maintained in the autosampler at 4°C. A 1.0- by 100-mm Waters Acquity UPLC BEH-C18 column was used. Mobile phases A and B were 10 mM tributylamine, pH 4.95, and methanol, respectively. The gradient was 0 to 1.0 min, 0% B; 1.0 to 2.5 min, 20% B; 2.5 to 4.0 min, 20% B; 4.0 to 7.0 min, 65% B; 7.0 to 7.5 min, 95% B; 7.5 to 9.0 min, 95% B; 9.0 to 9.1 min, 0% B; and 9.1 to 15.0 min, 0% B, and the flow rate used was 0.075 ml/min. The concentration of c-di-GMP was calculated using a standard curve for c-di-GMP (Biolog Life Science Institute, Bremen, Germany) and was standardized by biomass wet weight.
Motility measurement.
Manipulation of the c-di-GMP pool was also monitored by measuring flagellar motility. To this end, three independent cultures were stabbed into LB containing 0.3% agar (swarm agar) containing Cm with and without l-arabinose, and swarm diameters were measured after the plates were incubated at 30°C for 16 h.
Western blot analysis.
In all experiments, a volume containing 0.5 OD600 units of cells was centrifuged and the pellet was resuspended in 0.1 ml of Laemmli sample buffer (Bio-Rad Laboratories). The cell suspension was placed in a boiling-water bath for 10 min, and the cell debris was removed by centrifugation. Proteins were separated using Criterion precast 10% gels (Bio-Rad) and subsequently transferred to polyvinylidene difluoride (PVDF) membranes. Expression of RpoS-FLAG was determined with monoclonal anti-FLAG M2-peroxidase (Sigma-Aldrich) and a BM bioluminescence Western blotting kit (Roche Applied Science). In parallel, gels were directly stained with Coomassie blue as a loading control.
Enzyme assays.
Production of HA/protease was measured using an azocasein assay as described previously (2). One azocasein unit is the amount of enzyme that produces an increase of 0.01 optical density unit in the assay. β-Galactosidase activity was measured as described by Miller (39), using the substrate o-nitrophenyl-β-d-galactopyranoside (ONPG). Specific activities are given in Miller units {1,000[OD420/(t × v × OD600)]}, where t is reaction time and v is the volume of enzyme extract per reaction. For the determination of PDE activity, cells were disrupted by sonication in PBS containing 1 mM phenylmethylsulfonyl fluoride and the cell debris was removed by high-speed centrifugation at 4°C. Ten microliters of crude extract was added to 0.1 ml of reaction buffer consisting of 50 mM Tris (pH 8.0), 20 mM MgCl2, and 5 mM bis(p-nitrophenyl) phosphate. The enzymatic release of p-nitrophenol was followed by measurement of the OD410 every 5 min for 60 min in a Synergy microplate reader instrument at 37°C. One unit of activity was arbitrarily defined as the amount of enzyme causing an increase of 0.01 optical density unit and was standardized by protein concentration as determined with Bradford reagent (Bio-Rad Laboratories).
Protein purification and DNA binding assays.
HapR and VqmA proteins were purified using IMPACT and pMAL protein fusion and purification systems from New England BioLabs, respectively. Binding of the above proteins to the hapR promoters was analyzed using a DIG gel shift kit (Roche Applied Sciences). A more detailed description of this method is provided as supplemental material.
RESULTS
Cyclic di-GMP coordinately regulates the expression of HA/protease and motility.
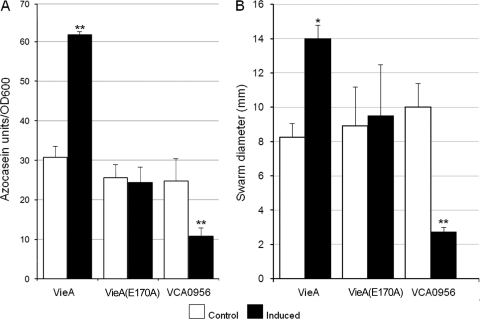
HA/protease and motility are both positively regulated by HapR and RpoS. The fact that c-di-GMP inhibits motility (5) prompted us to examine the effect of this second messenger on the expression of HA/protease. A major obstacle to this end is the redundancy of proteins involved in c-di-GMP biosynthesis and degradation. An approach pioneered by other investigators has been to examine the consequences of manipulating the intracellular c-di-GMP pool (5). In this study, we chose to overexpress the PDE VieA and DGC VCA0956 to diminish and increase the c-di-GMP pool, respectively. To confirm the effect of this manipulation. we measured c-di-GMP content by LC-MS-MS. The c-di-GMP content of C7258 grown to a high cell density was low, and as expected, the deletion of hapR had a positive effect (Table 3). Interestingly, the deletion of rpoS also positively enhanced the c-di-GMP pool, while inactivation of both rpoS and hapR had an additive effect (Table 3). Overexpression of VieA at high cell density further reduced c-di-GMP content 3.7-fold with respect to the uninduced control. This finding corresponded with an increase in PDE activity in crude extracts from 178 to 450 units/mg protein. Induction of the DGC VCA0956 resulted in a 60-fold increase in c-di-GMP content under the same conditions (Table 3). In Fig. 1A, we show that diminishing the c-di-GMP pool by overexpressing VieA enhanced HA/protease expression, while increasing c-di-GMP by overexpressing VCA0956 had the opposite effect. As expected, overexpression of the VieA(E170A) active-site mutant had no effect, indicating that the phenotypic consequence of overexpressing VieA in our El Tor biotype strain is attributable to its encoded PDE activity. Finally, cultures were stabbed into swarm agar with and without l-arabinose to confirm that changes in HA/protease production were accompanied by the expected effect on motility (Fig. 1B). These results show that both HA/protease and motility are negatively regulated by c-di-GMP.
Table 3.
Effect of PDE VieA and DGC VCA0956 induction on c-di-GMP content
| Strain | c-di-GMP content (pg/mg of wet wt)a |
|---|---|
| C7258 | 3.0 ± 0.6 |
| C7258 ΔhapR (AJB51) | 11.2 ± 2.3 |
| C7258 ΔrpoS (AJB50) | 10.8 ± 0.7 |
| C7258 ΔhapR ΔrpoS (AJB52) | 22.7 ± 1.9 |
| C7258 + VieA control | 2.6 ± 0.2 |
| C7258 + VieA induced | 0.7 ± 0.1 |
| C7258 + VCA0956 control | 2.9 ± 0.8 |
| C7258 + VCA0956 induced | 180.7 ± 9.7 |
Cultures were grown to an OD600 of 2.0 in TSB medium induced with l-arabinose as required; c-di-GMP content was measured after a 4-h incubation period.
Fig. 1.
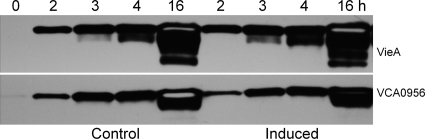
Regulation of HA/protease production by c-di-GMP. (A) Production of HA/protease. Strain C7258, containing plasmids pAT1568 (VieA, EAL domain), pAT1615 [VieA(E170A), inactive EAL domain], and pAT1662 (VCA0956, GGDEF domain), was grown in TSB medium, and PDE or DGC expression was induced as described in Materials and Methods. Cultures were incubated for 16 h, and the production of HA/protease was determined. Each value represents the mean for three independent cultures. Error bars indicate standard deviations. **, P < 0.01. (B) Motility assay. Overnight cultures of C7258 transformants were stabbed into swarm agar with or without (control) l-arabinose, and the plates were incubated at 30°C. Each of the resulting swarm diameters is the mean for three independent cultures. Error bars indicate standard deviations (*, P < 0.05; **, P < 0.01 for comparison of induced versus control cultures).
Cyclic di-GMP affects the expression of hapA, hapR, and rpoS mRNAs.
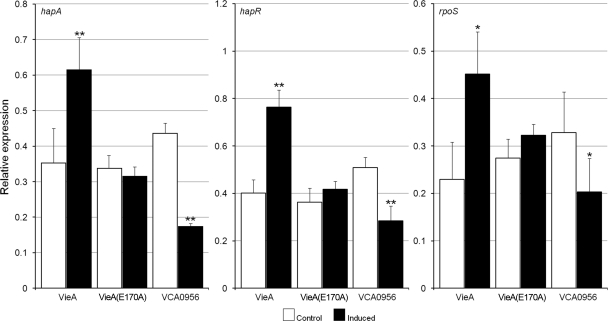
Since the azocasein assay measures the amount of HA/protease secreted to the culture medium, we used qRT-PCR to determine if c-di-GMP affects hapA transcription. As shown in Fig. 2, changes in the c-di-GMP pool exhibited the same effect on hapA mRNA and azocasein production, suggesting that it acts by modulating hapA transcription. Given that transcription of hapA requires HapR and RpoS, we also examined whether manipulation of c-di-GMP content affected the expression of these regulators. As shown in Fig. 2, expression of both hapR and rpoS mRNAs was affected by c-di-GMP, exhibiting the same pattern as that observed for hapA, though this effect was more modest for rpoS mRNA.
Fig. 2.
Effect of manipulating the c-di-GMP pool on hapA, hapR, and rpoS mRNA expression. Strain C7258, containing plasmids expressing VieA, VieA(E170A), and VCA0956, was grown in TSB medium, and PDE or DGC expression was induced as described in Materials and Methods. Total RNA was extracted, and hapA, hapR, and rpoS mRNAs were measured by qRT-PCR. Each value represents the mean for three independent cultures. Error bars indicate standard deviations (*, P < 0.05; **, P < 0.01 for comparison of induced versus control cultures).
Genetic analysis of HapR regulation by c-di-GMP.
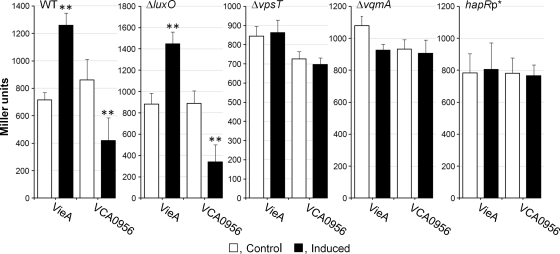
The results obtained in the qRT-PCR assay prompted us to determine if manipulation of c-di-GMP impacted HapR protein and activity. To this end, we introduced the VieA and VCA0956 expression vectors into strain SZS007, in which the chromosomal lacZ gene was replaced by a HapR-dependent luxC-lacZ transcriptional fusion (47). Manipulation of the c-di-GMP pool in this strain reproduced the same regulatory pattern as that observed in the azocasein and qRT-PCR assays (Fig. 3). To determine if the effect of c-di-GMP on HapR expression involved quorum sensing, we introduced the VieA and VCA0956 expression vectors into strain SZS012, which contains a deletion of luxO. As shown in Fig. 3, manipulation of the c-di-GMP pool continued to modulate HapR expression in this genetic background, indicating that this effect is independent of LuxO. The LuxR-type regulator VpsT binds c-di-GMP to regulate motility (27). Therefore, we introduced a vpsT deletion into strain SZS007 and examined the effect of c-di-GMP on HapR expression in the mutant. As shown in Fig. 3, c-di-GMP had no effect on HapR expression in a strain that does not make VpsT. Gene expression profiling has identified VpsT as a negative regulator of HapR, induced when c-di-GMP concentrations are artificially increased (5, 6). Thus, we used qRT-PCR to determine if induction of VCA0956 enhanced VpsT under our conditions. The relative expression of vpsT 4 h after the induction of VCA0956 was 2.10 ± 0.33, compared to 0.36 ± 0.09 for the control (n = 3). Two cases of LuxO-independent regulation of HapR expression have been reported. In the first case, the LuxR-type transcriptional regulator VqmA was shown to positively regulate the hapR promoter at a low cell density (34). In the second case, HapR was found to negatively autoregulate its expression by binding to its own promoter (33, 48). To determine if c-di-GMP affected the above regulatory mechanisms, we constructed the vqmA deletion mutant AJB505 in the genetic background of the luxC-lacZ reporter SZS007. As shown in Fig. 3, manipulation of the c-di-GMP pool had no effect on HapR expression in a strain that does not make VqmA. Furthermore, we constructed strain HX01, which harbors a A→G mutation in the hapR promoter (hapRp*) reported to abolish HapR binding and autorepression (33). Interestingly, a single-base change in the hapR promoter eliminated the effect of manipulating the c-di-GMP pool on HapR expression (Fig. 3). Since both VqmA and HapR have been shown to bind specific regions within the hapR promoter, we also examined the possibility that c-di-GMP affects the binding of these regulators. However, c-di-GMP had no effect on HapR binding to its own promoter (see Fig. S1 in the supplemental material). In addition, we could not demonstrate binding of VqmA to the hapR promoter in the absence or presence of c-di-GMP, suggesting that this regulator could require an unidentified ligand other than c-di-GMP for effective binding to the hapR promoter or could act indirectly (see Fig. S1 in the supplemental material).
Fig. 3.
Effect of c-di-GMP on HapR expression in different genetic backgrounds. Strain SZS007, containing a HapR-dependent luxC-lacZ fusion (WT), and the isogenic SZS012 (ΔluxO), HX02 (ΔvpsT), AJB505 (ΔvqmA), and HX01 (hapRp*) mutants were transformed with plasmids expressing VieA and VCA0956. Transformants were grown in TSB medium, PDE or DGC expression was induced as described in Materials and Methods, and β-galactosidase activity was determined. Each value represents the mean for three independent cultures. Error bars indicate standard deviations (**, P < 0.01 for comparison of induced versus control cultures).
Since expression of hapA requires both HapR and RpoS (42), we measured the production of HA/protease in the ΔluxO, ΔvpsT, ΔvqmA, and hapRp* genetic backgrounds. Regulation of HA/protease by c-di-GMP was also found to be independent of LuxO (data not shown). Interestingly, mutations that abolished c-di-GMP regulation of HapR (Fig. 3) also eliminated its effect on HA/protease production (data not shown), even though induction of VCA0956 was found to diminish rpoS mRNA in the wild-type background (Fig. 2). These results suggest that the observed effect of c-di-GMP on the expression of RpoS could be an indirect consequence of c-di-GMP modulation of HapR. Thus, we further examined the role of HapR and c-di-GMP in the expression of RpoS.
Effect of HapR and c-di-GMP on the expression of RpoS protein.
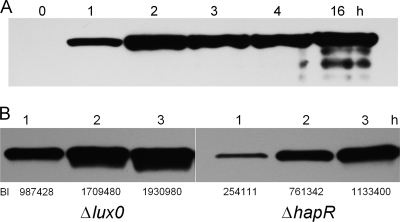
Since in Escherichia coli the expression of RpoS is subject to complex regulation at the level of transcription, translation, and protein stability (20), we devised an RpoS reporter that would allow us to monitor the expression of RpoS in V. cholerae at the protein level. To this end, we constructed strain AJB502, which expresses a C-terminal FLAG-tagged RpoS protein. The tagged rpoS allele was integrated by homologous recombination into the rpoS locus so that the expression of the FLAG epitope would reflect the production of RpoS from native transcription and translation initiation signals. The reporter system allowed us to determine the kinetics of RpoS expression. As shown in Fig, 4A, RpoS could be detected 1 h after the culture reached an OD600 of 2.0 in TSB medium. To determine if the expression of HapR could influence RpoS biosynthesis, we compared the expression of RpoS-FLAG in the isogenic ΔluxO (HapR constitutive) and ΔhapR strains. As shown in Fig. 4B, approximately 4 times less RpoS was detected in the ΔhapR mutant than in the ΔluxO strain at the 1 h time point. Expression of RpoS was approximately 50% reduced in the ΔhapR mutant at the later time points, suggesting that in the absence of HapR, induction of RpoS is delayed. A parallel Coomassie-stained gel was run to confirm that each well received a similar protein load (see Fig. S2 in the supplemental material).
Fig. 4.
Induction of RpoS in quorum-sensing mutants. (A) Strain AJB502, containing a chromosomally integrated rpoS-FLAG reporter, was grown to an OD600 of 2.0 (time zero), and samples were taken at different time points for Western blot analysis of RpoS-FLAG expression. (B) Strains AJB504 (ΔluxO) and AJB503 (ΔhapR), containing a chromosomally integrated rpoS-FLAG reporter, were grown to an OD600 of 2.0, and cells were collected at different time points for Western blot analysis of RpoS-FLAG expression. Band intensities (BI) were determined using Kodak 1D image analysis software.
Next, we examined the effect of directly manipulating the c-di-GMP pool on the expression of RpoS-FLAG. To this end, we grew strain AJB502, which contains the VieA and VCA0956 expression vectors, to an OD600 of 2.0 and added l-arabinose to diminish or increase the c-di-GMP pool, respectively. As shown in Fig. 5, although manipulation of the c-di-GMP pool affected rpoS expression in the qRT-PCR assay, it had a minor impact at the protein level measured using the RpoS-FLAG reporter. This result suggests that the positive effect of HapR on RpoS expression observed at high cell density (Fig. 4B) is not due to changes in c-di-GMP content.
Fig. 5.
Effect of c-di-GMP on RpoS expression at a high cell density. Strain AJB502, which produces rpoS-FLAG, was transformed with plasmids expressing VieA and VCA0956. Transformants were grown in TSB medium to an OD600 of 2.0 (time zero), and PDE or DGC expression was induced as described in Materials and Methods. Induction of RpoS-FLAG was determined by Western blotting.
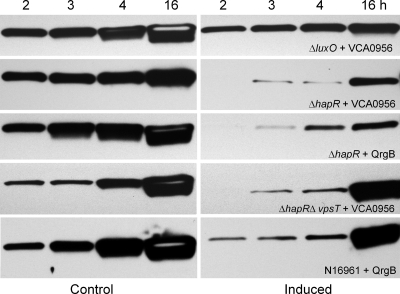
The above result prompted us to consider if c-di-GMP could affect RpoS biosynthesis in the absence of quorum sensing. Thus, we compared the effect of increasing c-di-GMP on RpoS-FLAG expression in strains AJB504 (ΔluxO) and AJB503 (ΔhapR) locked in regulatory stages mimicking high and low cell densities, respectively. As shown in Fig. 6, increasing c-di-GMP in the ΔluxO mutant had a minor negative effect on RpoS expression. In contrast, increasing c-di-GMP in the ΔhapR mutant strongly inhibited RpoS expression. To exclude the possibility of VCA0956 inhibiting RpoS expression in this genetic background by a mechanism unrelated to its DGC activity, we introduced plasmid pCMW75, which encodes DGC QrgB, into strain AJB503 (ΔhapR). The V. cholerae genome does not encode a homolog of this DGC. Again, induction of QrgB by the addition of IPTG strongly inhibited expression of RpoS-FLAG (Fig. 6). We also examined the effect of c-di-GMP on RpoS expression in a ΔhapR ΔvpsT double mutant. In contrast to c-di-GMP regulation of HapR, significant c-di-GMP-mediated repression of RpoS in the strain lacking hapR and vpsT was observed (Fig. 6). Finally, we modified strain N16961, which contains a natural frameshift mutation in hapR, to express the RpoS-FLAG reporter. As shown in Fig. 6, overexpression of QrgB in strain N16961-FLAG resulted in a substantial delay in full RpoS induction. Taken together, our results suggest that under conditions in which HapR is not expressed, c-di-GMP negatively regulates the expression of RpoS.
Fig. 6.
Regulation of RpoS expression by c-di-GMP in the absence of HapR. Strains AJB504 (ΔluxO), AJB503 (ΔhapR), and HX03 (ΔhapR ΔvpsT), containing the rpoS-FLAG reporter and the VCA0956 expression vector, as well as strains AJB503 and N16961-FLAG, containing the QrgB expression vector, were grown to an OD600 of 2.0 (time zero). At this stage, cultures were divided in halves; one half was induced by adding l-arabinose for VCA0956 or IPTG for QrgB, and the other half was used as a control. Samples were taken at different time points for Western blot analysis of RpoS-FLAG expression.
DISCUSSION
In this study, we examined the role of c-di-GMP in the regulation of HA/protease, known to affect cholera pathogenesis in animal models and to contribute to live-vaccine reactogenicity (3, 43). As is the case with motility, expression of HA/protease increased upon lowering of the c-di-GMP pool and decreased upon overexpression of a DGC. This result suggests that HA/protease and motility are coordinately regulated by c-di-GMP. HA/protease and motility have been proposed to facilitate V. cholerae detachment from the intestinal mucosa (4, 14, 40, 43, 44). The coordinate regulation of these phenotypes by CRP, HapR, RpoS, and c-di-GMP suggests that they could be simultaneously expressed in the host to modulate pathogenesis. V. cholerae possesses a single polar flagellum and is unlikely to swim in a highly viscous medium (1). It has been reported that the cholera bacterium loses its flagellum in a viscous mucin-containing medium (35). We have proposed that production of extracellular proteases could facilitate preservation of the flagellum of V. cholerae during detachment by decreasing the viscosity of the medium (44). This could result from HA/protease degradation of preexisting mucin (13) and cleavage of the GbpA adhesin, which has also been reported to enhance the production of intestinal mucins (7, 25). Since it is well established that motility and low c-di-GMP content promote intestinal colonization (28, 50), the coordinate expression of HA/protease and motility could facilitate the detachment of motile-stage V. cholerae cells, which could readily swim toward and colonize additional sites along the gastrointestinal tract.
The expression of HapR is fine-tuned through the action of numerous regulators and regulatory loops that can function under different physiological conditions. For instance, in addition to regulation by small RNAs (sRNAs) (29, 48), HapR levels can be modulated by autorepression, VqmA, VpsT, and VpsR (6, 33, 34, 54, 58). In this study, we report that expression of HapR measured by either qRT-PCR or by using an HapR-dependent luxC-lacZ fusion is negatively modulated by c-di-GMP content. Our data are consistent with the finding that a deletion mutant lacking the PDE CdgC (expected to exhibit elevated c-di-GMP content) expressed a lower level of HapR (32). We found that c-di-GMP acts downstream of LuxO to negatively affect HapR expression. Deletion of vqmA or a single-base change in the hapR promoter that abolished autorepression (33) eliminated the negative effect of c-di-GMP on HapR expression. Importantly, we found that c-di-GMP regulation of HapR was eliminated by the deletion of VpsT, known to directly sense c-di-GMP (27). Thus, our data suggest that c-di-GMP may repress HapR expression indirectly by binding to VpsT and activating it (5, 27). VpsT could then act through VqmA or other regulators in a LuxO-independent manner to diminish HapR expression. We also considered the possibility that c-di-GMP directly interacts with HapR, known to contain a binding pocket for an unknown ligand (10), and/or with VqmA to regulate hapR expression. However, we did not find c-di-GMP to influence binding of HapR to its own promoter, nor could we demonstrate VqmA binding to this promoter in the absence or presence of c-di-GMP (see the supplemental material). The relationship between HapR autorepression and HapR regulation by c-di-GMP is intriguing. Conceivably, binding of HapR to its own promoter might be part of a more complex regulation involving additional factors modulated by c-di-GMP content.
Deletion of vqmA did not significantly impact the expression of HapR in uninduced cultures. This result was expected, given that our studies were conducted at a high cell density, when VqmA has no effect on hapR transcription (34). In contrast to previous studies (33, 48), elimination of HapR autorepression in strain C7258 did not result in elevated HapR levels in the stationary phase. However, we did confirm that HapR purified from C7258 binds to the wild-type promoter but not to the mutant promoter resistant to autorepression (see the supplemental material). A possible explanation for this difference is that the luxC-lacZ fusion used in our study reflects the accumulation of active HapR protein, while previous studies used hapR-lacZ and hapR-GFP promoter and translational fusions (33, 48). Recently, it was shown that HapR is additionally regulated at the level of activity (53).
Both lowering and increasing the c-di-GMP pool affected rpoS expression in the qRT-PCR assay. To monitor RpoS protein expression, we constructed a reporter strain expressing RpoS-FLAG. The kinetics of RpoS-FLAG expression closely paralleled the time course of HA/protease expression in the same medium (2). Furthermore, induction of RpoS-FLAG biosynthesis closely mimicked the induction of native RpoS in genetically unmodified C7258 when a polyclonal RpoS antiserum was used (data not shown). Differences in rpoS mRNA levels induced by c-di-GMP did not result in different RpoS protein levels in the Western blot analysis. The lack of correlation between mRNA and protein levels is not completely unexpected in this case, due to the complex regulation of RpoS at the levels of transcription, translation, and protein stability (20). Consistent with c-di-GMP having no effect on RpoS levels in the wild-type strain at a high cell density, elimination of c-di-GMP modulation of HapR was sufficient to abolish its regulation of RpoS-dependent HA/protease production. Nevertheless, we did show that expression of HapR had a positive effect on RpoS biosynthesis, though this regulation is not likely to involve c-di-GMP signaling. This finding is in agreement with a previous study showing that an rpoS-lacZ fusion exhibited reduced expression in a hapR mutant (23). In the absence of quorum sensing, increasing the c-di-GMP content severely inhibited RpoS biosynthesis. Interestingly, deletion of vpsT in the hapR generic background did not abolish c-di-GMP repression of RpoS, suggesting that c-di-GMP modulation of HapR and RpoS involves distinct mechanisms. We suggest that expression of HapR in the wild-type genetic background could oppose the negative effect of high c-di-GMP content on RpoS expression in several ways. For instance, as shown above, HapR has a positive effect on the expression of RpoS protein at a high cell density that is independent of c-di-GMP. In addition, HapR could repress a component of the c-di-GMP signaling pathway that affects RpoS expression or indirectly prevent binding of an unknown regulator activated by c-di-GMP to the rpoS promoter. As expected, increasing c-di-GMP inhibited RpoS expression in strain N16961, which contains a natural frameshift mutation in hapR. Taken together, our results suggest that c-di-GMP functions as a negative regulator of RpoS expression when HapR is not expressed or in V. cholerae strains exhibiting dysfunctional quorum-sensing systems. V. cholerae strains with dysfunctional quorum-sensing systems are very common among environmental and clinical isolates and have been shown to arise with high frequency under laboratory conditions (16, 24).
It has been reported that expression of HapR diminishes the c-di-GMP pool (17, 54). In this study, we showed that expression of RpoS also contributes to lowering of the intracellular c-di-GMP content. In Escherichia coli, many GGDEF and EAL domain proteins are regulated by RpoS (19). In V. cholerae, VCA0080, VC1986 (GGDEF domain family), VC1087, and VC2340 (HD-GYP domain family) were found to be differentially expressed in an rpoS mutant (40). The elevated c-di-GMP content observed in our ΔrpoS mutant is consistent with the fact that rpoS mutants are less motile than the wild type and express lower levels of numerous genes involved in flagellar motility and chemotaxis (40, 45).
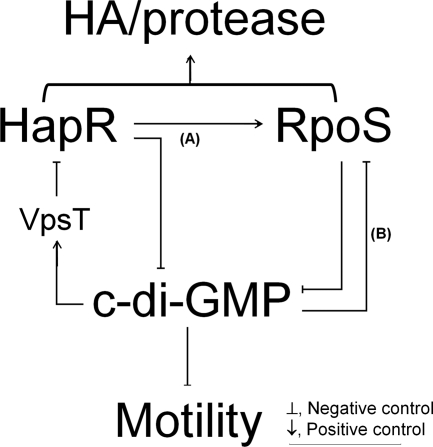
Since HapR acts to diminish the intracellular c-di-GMP pool (17, 54), our findings suggest the occurrence of a double-negative regulatory loop mediated by VpsT, which enhances the expression of HA/protease at a high cell density (Fig. 7). Briefly, (i) expression of HapR at a high cell density results in lower c-di-GMP content (17, 54); (ii) the lowering of c-di-GMP content further enhances HapR, generating a double-negative regulatory loop; (iii) enhanced expression of HapR positively impacts the biosynthesis of RpoS (pathway A); (iv) expression of RpoS feeds into the regulatory loop by diminishing c-di-GMP content; and (v) concurrent activation of HapR and RpoS results in elevated expression of HA/protease and motility. Cyclic di-GMP also inhibits RpoS expression in the absence of HapR (pathway B). Pathway B could be significant under conditions in which V. cholerae cells could enter the stationary phase prior to activation of quorum sensing (i.e., nutrient limitation) or in strains with dysfunctional quorum-sensing systems. We propose that the above model for the coordinate expression of HA/protease and motility by HapR, RpoS, and c-di-GMP could contribute to the dissemination of colonizing V. cholerae cells along the gastrointestinal tract in terms of multiple cycles of detachment and recolonization.
Fig. 7.
Model for the interplay between c-di-GMP, HapR, and RpoS in the regulation of HA/protease expression. The second messenger c-di-GMP negatively affects the expression of HapR. This regulation is independent of LuxO but requires the c-di-GMP binding protein VpsT and VqmA. At a high cell density, HapR enhances RpoS (pathway A) and both regulators diminish the c-di-GMP pool, generating a double-negative regulatory loop that further enhances the expression of HapR, HA/protease production, and motility (see the text for details). We propose that in the absence of quorum sensing, c-di-GMP also inhibits the expression of RpoS (pathway B).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grants R21AI081039 and GM008248 to A.J.S.
We are grateful to Andrew Camilli (Tufts University School of Medicine) for providing expression vectors encoding the PDE VieA and the DGC VCA0956 and to Christopher Waters (Michigan State University) for providing the DGC QrgB expression vector. We are grateful to Doyle Ray Moore II from the University of Alabama at Birmingham Targeted Metabolomics and Proteomics Laboratory for his assistance in c-di-GMP quantitation by LC/MS/MS.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 30 September 2011.
REFERENCES
- 1. Atsumi T., et al. 1996. Effect of viscosity on swimming by the lateral and polar flagella of Vibrio alginolyticus. J. Bacteriol. 178:5024–5026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Benitez J. A., Silva A. J., Finkelstein R. A. 2001. Environmental signals controlling production of hemagglutinin/protease in Vibrio cholerae. Infect. Immun. 69:6549–6553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Benítez J. A., et al. 1999. Preliminary assessment of the safety and immunogenicity of a new CTXφ-negative hemagglutinin/protease-defective El Tor strain as a cholera vaccine candidate. Infect. Immun. 67:539–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Benítez J. A., et al. 1997. Adherence of Vibrio cholerae to cultured differentiated human intestinal cells: an in vitro colonization model. Infect. Immun. 65:3474–3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beyhan S., Tischler A. D., Camilli A., Yildiz F. H. 2006. Transcriptome and phenotypic responses of Vibrio cholerae to increased cyclic di-GMP level. J. Bacteriol. 188:3600–3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beyhan S., Bilecen K., Salama S. R., Casper-Lindley C., Yildiz F. H. 2007. Regulation of rugosity and biofilm formation in Vibrio cholerae: comparison of VpsT and VpsR regulons and epistasis analysis of vpsT, vpsR, and hapR. J. Bacteriol. 189:388–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bhowmick R., et al. 2008. Intestinal adherence of Vibrio cholerae involves a coordinated interaction between colonization factor GbpA and mucin. Infect. Immun. 76:4968–4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. D'Argenio D. A., Miller S. I. 2004. Cyclic di-GMP as a bacterial second messenger. Microbiology 150:2497–2502 [DOI] [PubMed] [Google Scholar]
- 9. de Lorenzo V., Eltis L., Kessler B., Timmis K. N. 1993. Analysis of the Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene 123:17–24 [DOI] [PubMed] [Google Scholar]
- 10. De Silva R. S., et al. 2007. Crystal structure of the Vibrio cholerae quorum-sensing regulatory protein HapR. J. Bacteriol. 189:5683–5691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Donnenberg M. S., Kaper J. B. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Finkelstein R. A. 1992. Cholera enterotoxin (choleragen): a historical perspective, p. 155–187 In Barua D., Greenough W. B. (ed.), Cholera. Plenum Medical Book Company, New York, NY [Google Scholar]
- 13. Finkelstein R. A., Boesman-Finkelstein M., Holt P. 1983. Vibrio cholerae hemagglutinin/lectin/protease hydrolyzes fibronectin and ovomucin: F.M. Burnet revisited. Proc. Natl. Acad. Sci. U. S. A. 80:1092–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Finkelstein R. A., Boesman-Finkelstein M., Chang Y., Häse C. C. 1992. Vibrio cholerae hemagglutinin/protease, colonial variation, virulence, and detachment. Infect. Immun. 60:472–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Galperin M. Y. 2004. Bacterial signal transduction network in a genomic perspective. Environ. Microbiol. 6:552–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hammer B. K., Bassler B. L. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50:101–114 [DOI] [PubMed] [Google Scholar]
- 17. Hammer B. K., Bassler B. L. 2009. Distinct sensory pathways in Vibrio cholerae El Tor and classical biotypes modulate cyclic dimeric GMP levels to control biofilm formation. J. Bacteriol. 191:169–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Häse C. C., Finkelstein R. A. 1991. Cloning and nucleotide sequence of the Vibrio cholerae hemagglutinin/protease (HA/protease) gene and construction of an HA/protease-negative strain. J. Bacteriol. 173:3311–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7:263–273 [DOI] [PubMed] [Google Scholar]
- 20. Hengge-Aronis R. 2002. Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Herrington D. A., et al. 1988. Toxin, toxin co-regulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J. Exp. Med. 168:1487–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jobling M. G., Holmes R. K. 1997. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol. Microbiol. 26:1023–1034 [DOI] [PubMed] [Google Scholar]
- 23. Joelsson A., Kan B., Zhu J. 2007. Quorum sensing enhances the stress response in Vibrio cholerae. Appl. Environ. Microbiol. 73:3742–3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Joelsson A., Liu Z., Zhu J. 2006. Genetic and phenotypic diversity of quorum-sensing systems in clinical and environmental isolates of Vibrio cholerae. Infect. Immun. 74:1141–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jude B. A., Martinez R. M., Skorupski K., Taylor R. K. 2009. Levels of the secreted Vibrio cholerae attachment factor GbpA are modulated by quorum-sensing-induced proteolysis. J. Bacteriol. 191:6911–6917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kaper J. B., Morris G., Jr., Levine M. M. 1995. Cholera. Clin. Microbiol. Rev. 8:48–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krasteva P. V., et al. 2010. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science 327:866–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee S. H., Butler S. M., Camilli A. 2001. Selection for in vivo regulators of bacterial virulence. Proc. Natl. Acad. Sci. U. S. A. 98:6889–6894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lenz D. H., et al. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118:69–82 [DOI] [PubMed] [Google Scholar]
- 30. Liang W., Pascual-Montano A., Silva A. J., Benitez J. A. 2007. The cyclic AMP receptor protein modulates quorum sensing, motility and multiple genes that affect intestinal colonization in Vibrio cholerae. Microbiology 153:2964–2975 [DOI] [PubMed] [Google Scholar]
- 31. Liang W., Sultan S. Z., Silva A. J., Benitez J. A. 2008. Cyclic AMP post-transcriptionally regulates the biosynthesis of a major bacterial autoinducer to modulate the cell density required to activate quorum sensing. FEBS Lett. 582:3744–3750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lim B., Beyhan S., Yildiz F. H. 2007. Regulation of Vibrio polysaccharide synthesis and virulence factor production by CdgC, a GGDEF-EAL domain protein, in Vibrio cholerae. J. Bacteriol. 189:717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lin W., Kovacikova G., Skorupski K. 2005. Requirements for Vibrio cholerae HapR binding and transcriptional repression at the hapR promoter are distinct from those at the aphA promoter. J. Bacteriol. 187:3013–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu Z., Hsiao A., Joelsson A., Zhu J. 2006. The transcriptional regulator VqmA increases expression of the quorum-sensing activator HapR in Vibrio cholerae. J. Bacteriol. 188:2446–2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu Z., et al. 2008. Mucosal penetration primes Vibrio cholerae for host colonization by repressing quorum sensing. Proc. Natl. Acad. Sci. U. S. A. 105:9769–9774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marcus H., Ketley J. M., Kaper J. B., Holmes R. K. 1990. Effects of DNase production, plasmid size, and restriction barriers on transformation of Vibrio cholerae by electroporation and osmotic shock. FEMS Microbiol. Lett. 56:149–154 [DOI] [PubMed] [Google Scholar]
- 37. Mel S. F., Fullner K. J., Wimer-Mackin S., Lencer W. I., Mekalanos J. J. 2000. Association of protease activity in Vibrio cholerae vaccine strains with decreases in transcellular epithelial resistance of polarized T84 intestinal cells. Infect. Immun. 68:6487–6492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Merrell D. S., et al. 2002. Host-induced epidemic spread of the cholera bacterium. Nature 417:642–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miller J. H. 1971. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 40. Nielsen A. T., et al. 2006. RpoS controls the Vibrio cholerae mucosal escape response. PLoS Pathog. 2:e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Robert A., et al. 1996. Tagging a Vibrio cholerae El Tor candidate vaccine strain by disruption of its hemagglutinin/protease gene using a novel reporter enzyme: Clostridium thermocellum endoglucanase A. Vaccine 14:1517–1522 [DOI] [PubMed] [Google Scholar]
- 42. Silva A. J., Benitez J. A. 2004. Transcriptional regulation of Vibrio cholerae hemagglutinin/protease by the cyclic AMP receptor protein and RpoS. J. Bacteriol. 186:6374–6382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Silva A. J., Leitch G. J., Camilli A., Benitez J. A. 2006. Contribution of hemagglutinin/protease and motility to the pathogenesis of El Tor biotype cholera. Infect. Immun. 74:2072–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Silva A. J., Pham K., Benitez J. A. 2003. Haemagglutinin/protease expression and mucin gel penetration in El Tor biotype Vibrio cholerae. Microbiology 149:1883–1891 [DOI] [PubMed] [Google Scholar]
- 45. Silva A. J., Sultan S. Z., Liang W., Benitez J. A. 2008. Role of the histone-like nucleoid structuring protein in the regulation of rpoS and RpoS-dependent genes in Vibrio cholerae. J. Bacteriol. 190:7335–7345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Simm R., Morr M., Kader A., Nimtz M., Römling U. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53:1123–1134 [DOI] [PubMed] [Google Scholar]
- 47. Sultan S. Z., Silva A. J., Benitez J. A. 2010. The PhoB regulatory system modulates biofilm formation and stress response in El Tor biotype Vibrio cholerae. FEMS Microbiol. Lett. 302:22–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Svenningsen S. L., Waters C. M., Bassler B. L. 2008. A negative feedback loop involving small RNAs accelerates Vibrio cholerae's transition out of quorum-sensing mode. Genes Dev. 22:226–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tamayo R., Pratt J. T., Camilli A. 2007. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu. Rev. Microbiol. 61:131–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tamayo R., Schild S., Pratt J. S., Camilli A. 2008. Role of cyclic di-GMP during El Tor biotype Vibrio cholerae infection: characterization of the in vivo-induced cyclic di-GMP phosphodiesterase CdpA. Infect. Immun. 76:1617–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tischler A. T., Camilli A. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53:857–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tsou A. M., Cai T., Liu Z., Zhu J., Kulkarni R. V. 2009. Regulatory targets of quorum sensing in Vibrio cholerae: evidence for two distinct HapR-binding motifs. Nucleic Acids Res. 37:2747–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tsou A. M., Liu Z., Cai T., Zhu J.10 March 2011. The VarS/VarA two-component system modulates the activity of the Vibrio cholerae quorum sensing transcriptional regulator HapR. Microbiology [Epub ahead of print.] doi:10.1099/mic.0.046235-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Waters C. M., Lu W., Rabinowitz J. D., Bassler B. L. 2008. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J. Bacteriol. 190:2527–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wu Z., Milton D., Nybom P., Sjö A., Magnusson K. E. 1996. Vibrio cholerae hemagglutinin/protease (HA/protease) causes morphological changes in cultured epithelial cells and perturbs their paracellular barrier function. Microb. Pathog. 21:111–123 [DOI] [PubMed] [Google Scholar]
- 56. Wu Z., Nybom P., Magnusson K. E. 2000. Distinct effects of Vibrio cholerae haemagglutinin/protease on the structure and localization of the tight junction-associated proteins occludin and ZO-1. Cell. Microbiol. 2:11–17 [DOI] [PubMed] [Google Scholar]
- 57. Yildiz F. H., Schoolnik G. K. 1998. Role of rpoS in stress survival and virulence of Vibrio cholerae. J. Bacteriol. 180:773–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yildiz F. H., Liu X. S., Heydorn A., Schoolnik G. K. 2004. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol. Microbiol. 53:497–515 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.