Abstract
Chlorosomes are sac-like, light-harvesting organelles that characteristically contain very large numbers of bacteriochlorophyll (BChl) c, d, or e molecules. These antenna structures occur in chlorophototrophs belonging to some members of the Chlorobi and Chloroflexi phyla and are also found in a recently discovered member of the phylum Acidobacteria, “Candidatus Chloracidobacterium thermophilum.” “Ca. Chloracidobacterium thermophilum” is the first aerobic organism discovered to possess chlorosomes as light-harvesting antennae for phototrophic growth. Chlorosomes were isolated from “Ca. Chloracidobacterium thermophilum” and subjected to electron microscopic, spectroscopic, and biochemical analyses. The chlorosomes of “Ca. Chloracidobacterium thermophilum” had an average size of ∼100 by 30 nm. Cryo-electron microscopy showed that the BChl c molecules formed folded or twisted, sheet-like structures with a lamellar spacing of ∼2.3 nm. Unlike the BChls in the chlorosomes of the green sulfur bacterium Chlorobaculum tepidum, concentric cylindrical nanotubes were not observed. Chlorosomes of “Ca. Chloracidobacterium thermophilum” contained a homolog of CsmA, the BChl a-binding, baseplate protein; CsmV, a protein distantly related to CsmI, CsmJ, and CsmX of C. tepidum, which probably binds a single [2Fe-2S] cluster; and five unique polypeptides (CsmR, CsmS, CsmT, CsmU, and a type II NADH dehydrogenase homolog). Although “Ca. Chloracidobacterium thermophilum” is an aerobe, energy transfer among the BChls in these chlorosomes was very strongly quenched in the presence of oxygen (as measured by quenching of fluorescence emission). The combined analyses showed that the chlorosomes of “Ca. Chloracidobacterium thermophilum” possess a number of unique features but also share some properties with the chlorosomes found in anaerobic members of other phyla.
INTRODUCTION
Phototrophy is the process by which light energy is converted into chemical energy in the form of the proton motive force. Two mechanisms for this process are known. The first mechanism is retinal-dependent phototrophy, in which rhodopsin-like, transmembrane proteins that bind retinal act as light-activated proton gates (6, 32). The second mechanism, termed chlorophototrophy, employs chlorophyll (Chl)-protein complexes known as photochemical reaction centers (6, 26). All reaction centers contain a special pair of Chl or bacteriochlorophyll (BChl) molecules as well as bound cofactors, that include additional Chls, quinones and, in the case of type 1 reaction centers, iron-sulfur clusters (26). Photochemical reaction centers are usually surrounded by or connected to accessory light-harvesting complexes, which absorb and transfer excitation energy to the special pair of Chls of reaction centers. Nature exhibits great diversity in the structures of these antenna complexes, which include chlorosomes, phycobilisomes, and light-harvesting complexes I and II; each of these light-harvesting structures has distinctive protein and pigment compositions (2, 6).
Chlorosomes are sac-like, light-harvesting organelles that characteristically contain very large numbers of BChl c, d, or e molecules (>200,000 per chlorosome) within a protein-stabilized, lipid-containing envelope (7, 19, 41). These antenna structures have been found in members of three taxa that contain chlorophototrophs: the green sulfur bacteria (GSB; Chlorobiales); the filamentous anoxygenic phototrophs (FAPs) of the genera Chloroflexus, Chloronema, and Oscillochloris (green Chloroflexaceae); and one representative from the Acidobacteria, “Candidatus Chloracidobacterium thermophilum” (4–6, 19). Although most studies on the structure and biochemical properties of chlorosomes have been conducted with chlorosomes from the model organisms Chlorobaculum tepidum (GSB) and Chloroflexus aurantiacus (FAP) (7–11, 13–15, 17–20, 35, 36, 41, 42, 45–47, 52–54, 56–58), additional studies have been conducted on the chlorosomes of other green bacteria, including the FAPs “Candidatus Chlorothrix halophila” (64) and Chloronema giganteum (24) and other strains of GSB (39, 49, 51, 59, 65).
The BChls c, d, or e, which are actually chlorins and not bacteriochlorins, are unique among all (B)Chls, because they can self-aggregate and form supramolecular structures in chlorosomes without the aid of proteins (19, 27). This property is due to the absence of the 132-methylcarboxyl group and to the presence of the chiral 31-hydroxyl group that binds to the central magnesium atom of an adjacent BChl molecule (19, 20). Excitation energy absorbed by BChls moves quickly through these suprastructures due to strong excitonic coupling (48), and excitation energy is eventually transferred to BChl a molecules associated with the baseplate protein, CsmA (42, 46–49, 56). In the photosynthetic apparatus of GSB, an additional BChl a-binding protein, the Fenna-Matthews-Olson (FMO) protein, transfers excitation energy from CsmA to the core antenna molecules associated with the type 1 reaction centers found in the cytoplasmic membrane (4, 33, 44, 46, 62, 63, 69–71). High-resolution X-ray crystallographic studies of FMO have established that this protein binds eight BChl a molecules, rather than the seven that were previously suggested (33, 62, 70). Furthermore, mass spectrometry methods have been employed to map the orientation of FMO relative to the chlorosome baseplate (70, 71).
Chlorosomes are surrounded by a protein-stabilized, glycolipid-containing, monolayer envelope (19). In C. tepidum 10 Csm proteins (A, B, C, D, E, F, H, I, J, and X), which belong to four structure motif families, occur in the envelope (7–10, 18, 34–36, 66, 67). Although CsmA is essential for viability, mutants lacking one or up to four of the other proteins have been constructed, and the resulting chlorosomes have been characterized (18, 34, 36). Although the chlorosomes of C. aurantiacus have a protein composition distinct from that of C. tepidum chlorosomes, most of the proteins can be assigned to one of the four motif families found in C. tepidum (19, 67). C. aurantiacus chlorosomes contain a homolog of the BChl a-binding baseplate protein, CsmA, and they contain a [2Fe-2S] protein, CsmY, which is related to the CsmI/CsmJ/CsmX family. CsmM and CsmN are distantly related to CsmC and CsmD, and CsmO is a member of the CsmB/CsmF family. The only chlorosome protein in C. aurantiacus without a direct homolog in C. tepidum is CsmP, which seemingly is related to proteins found in Synechocystis sp. PCC 6803, Synechococcus sp. PCC 7002, and Halobacterium spp. (19).
Until recently, two models had been proposed to describe the structural organization of the BChls in chlorosomes: a rod/cylinder model (19, 45, 48, 54, 58, 59) and an “undulating lamellae” model (50). In the lamellar model described by Pšenčík and coworkers, the BChls were organized into multiple, parallel sheets separated by about 2 nm that were envisioned to run parallel to the long axis of the chlorosome (50). In the rod/cylinder model, BChls were organized into cylindrical structures with diameters of 5 to 10 nm, which also ran parallel to the long axis of the chlorosome (45, 58, 59). A recent study combined results from solid-state nuclear magnetic resonance, cryo-electron microscopy, and molecular modeling to solve the structure of the BChls in a bchQ bchR bchU mutant of C. tepidum as well as the corresponding wild type (20). These analyses showed that BChl d molecules in chlorosomes of the mutant and BChl c molecules in chlorosomes of the wild type were arranged in syn-anti stacks with an alternating syn-anti orientation of the esterifying tails with respect to the C-31 hydroxyl group. The sheet-like structures formed from these stacked BChls then form coaxial, cylindrical nanotubes. Interestingly, although the BChl c molecules in a bchQ bchR mutant formed parallel monomer stacks with an all-syn or an all-anti configuration, the BChl c molecules still formed coaxial, cylindrical nanotubes (S. Ganapathy, M. Reus, G. Oostergetel, P. K. Wawrzyniak, Y. Tsukatani, A. Gomez Maqueo Chew, F. Buda, D. A. Bryant, A. R. Holzwarth, and H. J. M. de Groot, unpublished data).
The acidobacterium “Candidatus Chloracidobacterium thermophilum” is the first known organism that synthesizes chlorosomes under oxic conditions (4, 5, 22). Moreover, it is also the first chlorosome-synthesizing organism that is not a GSB or a FAP. Because of these unique properties, chlorosomes from “Ca. Chloracidobacterium thermophilum” were isolated and analyzed with the goal of determining whether they might have unusual properties to enhance light harvesting under oxic conditions.
MATERIALS AND METHODS
Culture conditions.
“Ca. Chloracidobacterium thermophilum” was cultured in a HEPES-buffered, minimal salts medium supplemented with reduced organic carbon sources and ammonia as N source as previously described (5, 21, 22). Cultures were incubated at 53°C with moderate shaking and harvested 5 days after transfer to fresh medium, when BChl c absorption was maximal. C. tepidum and C. aurantiacus 396-1 were cultured under anoxic conditions as previously described (16, 38).
Chlorosome isolation.
Chlorosomes from C. tepidum were isolated as described previously (66, 67). For the isolation of chlorosomes from “Ca. Chloracidobacterium thermophilum” and C. aurantiacus Y-396, cells were harvested in chlorosome isolation buffer (CIB) containing 2.0 M NaSCN, 10 mM Tris-HCl (pH 7.5), 5 mM EDTA, 1.0 mM phenylmethylsulfonyl fluoride and 2.0 mM dithiothreitol, incubated with lysozyme (3 mg ml−1) for 20 min, and subjected to 3 passes through a chilled (∼4°C) French pressure cell at 138 MPa. Unbroken cells and debris were pelleted by low-speed centrifugation (2,000 × g), and the resulting supernatant was collected and brought to 30% (wt/vol) sucrose. A sucrose step gradient was generated with 45% (wt/vol) sucrose in CIB, followed by the low-speed supernatant fraction, 20% (wt/vol) sucrose (wt/vol) and 5% (wt/vol) sucrose, both in CIB. The gradients were centrifuged for 18 h at 250,000 × g. The chlorosome fraction appeared as a dark greenish-brown layer and at the top of the gradient. This band was collected and diluted 3-fold in chlorosome buffer (10 mM K-phosphate and 150 mM NaCl; pH 7.5); chlorosomes were then pelleted by high-speed ultracentrifugation at 220,000 × g. This last step was repeated once more, and the resulting chlorosome pellet was aliquoted and stored at 4°C for electron microscopy or stored at −80°C for other analyses.
Electron microscopy and cryo-electron microscopy.
Isolated chlorosomes were negatively stained with 1% (wt/vol) uranyl acetate and visualized in a JEOL 1200 EXII (Peabody, MA) or a Philips CM120 (Eindhoven, The Netherlands) transmission electron microscope. For electron microscopy of thin-sectioned cells, harvested “Ca. Chloracidobacterium thermophilum” cells were fixed with an osmium tetraoxide-potassium permanganate tandem fixation as described previously (29). Following fixation, cells were stained en bloc with 1% (wt/vol) uracil acetate and dehydrated with 25%, 50%, 70%, 80%, 90%, 95%, and 100% (vol/vol) ethanol and with acetonitrile. Dehydrated cells were embedded in Spurr resin according to standard protocols and allowed to polymerize in a 60°C oven overnight. Images were taken in a JEOL 1200 EXII transmission electron microscope (Peabody, MA). For cryo-electron microscopy, aliquots of purified chlorosomes were applied to holey carbon grids with a thin layer of carbon and were plunge-frozen in liquid ethane at 83 K with a Vitrobot vitrification system (FEI, Eindhoven, Netherlands). Electron microscopy was performed with a Tecnai G2 Polara electron microscope (FEI, Eindhoven, The Netherlands), which was equipped with a Gatan energy filter at 115,000× magnification and a specimen temperature of 80 K. Images were recorded in the zero-loss imaging mode, using a slit width of 20 eV, with a slow-scan charge-coupled device camera at 1-μm underfocus, to have optimal phase contrast transfer at 300 kV for details with a periodicity of about 2 nm. For tomography, dual-axis tilt series of a negatively stained chlorosome specimen were recorded in a Tecnai G2 Polara electron microscope at a magnification of 63,250×, with tilt angles from −66° to +58°, using a 2° increment. Tomograms were calculated using IMOD software (31). To produce the final tomograms, the volumes derived from the two single-axis tilts were aligned to each other and were combined and denoised with 20 iterations by nonlinear anisotropic diffusion (12).
Polyacrylamide gel electrophoresis.
Chlorosome proteins were analyzed by polyacrylamide gel electrophoresis (PAGE) in the presence of sodium dodecyl sulfate (SDS) using a Tris-Tricine buffer system (55). The stacking gel was 3% monomer and 3.3% cross-linker, and the resolving gel was 15% monomer and 3.3% cross-linker. Briefly, samples containing ∼20 μg of BChl c were incubated at 98°C in 1× loading buffer (0.1 M Tris-Cl buffer [pH 6.8], 8% [vol/vol] glycerol, 2% [wt/vol] SDS, 5% [vol/vol] mercaptoethanol, and trace amounts of bromophenol blue) for about 2 min and electrophoresed for 16 h at constant voltage. Proteins were visualized through silver staining (3) or Coomassie blue staining. Formaldehyde in the initial fixation step in the silver staining protocol was sometimes omitted (see Results and Discussion).
Amino-terminal sequencing and mass spectrometric analyses of chlorosome proteins.
Chlorosome proteins separated by SDS-PAGE were transferred to a PVDF membrane in a Bio-Rad semidry electroblotting apparatus using a 25 mM Tris, 192 mM glycine, 20% (vol/vol) methanol, and 0.1% (wt/vol) SDS transfer buffer according to the instructions of the manufacturer. Following electrophoretic transfer, blots were rinsed with distilled water, and transferred proteins were visualized by staining with Ponceau S (0.1% [wt/vol] Ponceau S in 5% [vol/vol] acetic acid). Bands were excised and sequenced at the Proteomics and Mass Spectrometry Core Facility, Huck Institutes of the Life Sciences, The Pennsylvania State University, Hershey, PA. For tryptic peptide mass fingerprinting analyses, SDS-PAGE gels were stained with silver, and visualized bands were excised and analyzed at the Proteomics and Mass Spectrometry Core Facility, Huck Institutes of the Life Sciences, The Pennsylvania State University, University Park, PA. Peptides produced by tryptic digestion were identified using the search engine MASCOT (Matrix Science).
Absorbance and fluorescence emission spectroscopy.
Room temperature absorbance spectra were measured with a GENESYS 10 spectrophotometer (Thermo Electron Corp., Rochester, NY). Fluorescence emission spectra were measured with an SLM-Aminco model 8000C spectrofluorometer, modified by OLIS, Inc., for computerized data acquisition and equipped with a liquid nitrogen Dewar when required for acquisition of spectra at 77 K. Data were analyzed using Igor-Pro (Wavemetrics, Inc., Lake Oswego, OR). For fluorescence emission measurements, cell and chlorosome fractions were thawed and diluted with 10 mM Tris-HCl buffer, pH 7.2, to an absorbance of 0.1 at the Qy maximum for BChl c. Except for measurements recorded under oxic conditions, sodium dithionite (final concentration, 25 mM) was added, and the samples were incubated in the dark for ∼2 h prior to measurements.
X-band EPR spectroscopy.
Low-temperature electron paramagnetic resonance (EPR) spectroscopy was performed using a Bruker ECS-106 X-band spectrometer equipped with an Oxford liquid helium cryostat and temperature controller. Spectra are the averages of 8 scans recorded under the following conditions: frequency, 9.48 GHz; gain, 20,000; modulation amplitude, 10 G at 100 kHz; power and temperature, as indicated in the figure legend. Samples contained 9.2 mg BChl c ml−1 and were reduced with 50 mM dithionite in 100 mM glycine, pH 10.
RESULTS
Electron microscopy of “Ca. Chloracidobacterium thermophilum” cells and isolated chlorosomes.
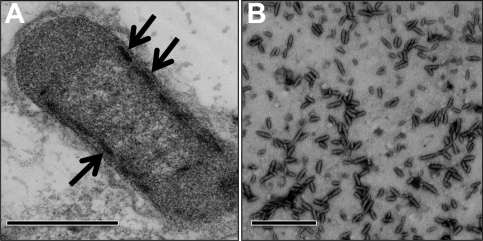
Figure 1A shows a thin-section electron micrograph of a “Ca. Chloracidobacterium thermophilum” cell. Electron-opaque, chlorosome-like structures were observed, which were tightly appressed to the inner surface of the cytoplasmic membrane. On average, the chlorosomes in cells were ∼100 nm in length and 25 to 30 nm in width. Interestingly, chlorosomes were only observed on the cytoplasmic membrane surfaces along the lateral walls of the cells, and chlorosomes were never observed near the poles of cells. A capsule or glycocalyx-like layer surrounded the cells.
Fig. 1.
Electron micrographs of a “Ca. Chloracidobacterium thermophilum” cell and a field of negatively stained chlorosomes. (A) Chlorosomes (arrows) are visible as electron-opaque rod-like objects appressed to the lateral inner leaflet of the cell membrane. Thin sections were stained with 2% (wt/vol) uranyl acetate prior to visualization. (B) Chlorosomes were stained with 1% (wt/vol) uranyl acetate. Bars, 500 nm.
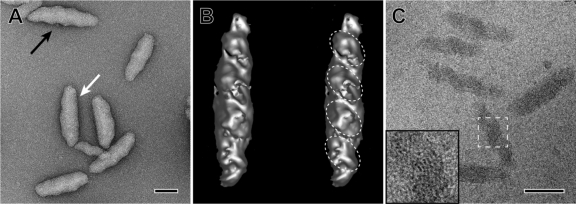
Highly purified chlorosome preparations could be obtained by only slight modification of the method used to purify chlorosomes from C. tepidum (66, 67) (see Materials and Methods). A chaotrophic agent, 2.0 M NaSCN, efficiently released the chlorosomes from the membranes. Electron micrographs of isolated, negatively stained chlorosomes revealed that they were rod-shaped, cylindrical objects. In good agreement with their apparent size in thin sections of cells, their average length was 99.6 ± 19 nm (mean ± standard deviation; range, 65 to 129 nm), and their average width was 30.8 ± 2.0 nm (range, 24 to 32 nm) (Fig. 1B). Irregularly spaced ridges and grooves were observed on the surfaces of a large proportion of the isolated chlorosomes (Fig. 2A). Additionally, a flattened surface could be observed in many chlorosomes (Fig. 2A). This surface probably includes the baseplate, which attaches the chlorosomes to FMO on the cytoplasmic membrane.
Fig. 2.
High-resolution electron micrograph, tomographic reconstruction of a negatively stained chlorosome, and cryo-electron microscopic images of chlorosomes of “Ca. Chloracidobacterium thermophilum.” (A) Negatively stained chlorosomes. The black arrow points to the ridges and grooves visible on one of the surfaces of the chlorosomes. The white arrow points to the flattened surface area that presumably includes the CsmA-containing baseplate structure. (B) Stereo image of a negatively stained chlorosome as an iso-surface representation from a tomographic reconstruction. The internal domains described in Results are indicated by the dashed ellipses. (C) Cryo-electron microscopic image of isolated chlorosomes embedded in vitreous ice. Note the twisted or folded lamellae. (Insert) Enlarged view of the boxed area, showing the curved lamellae with 2.3-nm spacing. The lamellar spacing was determined from a Fourier transformation of the data (see references 20 and 45 for further details). Bars, 50 nm.
Figure 2B shows a stereo image of a 3-dimensional reconstruction of a single negatively stained chlorosome from a tilt series of micrographs. This reconstruction also showed that the chlorosome surface was highly irregular, with both ridges and grooves being apparent. This image, as well as images obtained with unstained cryopreserved specimens (Fig. 2C), suggested that the “Ca. Chloracidobacterium thermophilum” chlorosomes are constructed from roughly ellipsoidal domains, which varied in size from about 15 to 20 nm by 25 to 30 nm, with the long axis of these ellipsoidal domains occurring at an angle of approximately 45° relative to the long axis of the chlorosome (Fig. 2B). In cryo-electron microscopy images, surfaces with a lamellar spacing of ∼2.1 to 2.3 nm (determined by Fourier transformation; data not shown, but see references 20 and 45) were observed (Fig. 2C, inset). These spacings were similar to the spacings of the cylindrical and arched lamellae in the chlorosomes of C. tepidum (20, 45, 50; Ganapathy et al., unpublished). The lamellae were arranged in irregular arcs or possibly folded or twisted sheets (Fig. 2C). The lamellar sheets did not form concentric nanocylinders, as previously described for C. tepidum (20), but the precise organization of the supramolecular structure formed by the BChls in these chlorosomes is currently unknown and will require further study.
Spectroscopic properties of chlorosomes from “Ca. Chloracidobacterium thermophilum.”
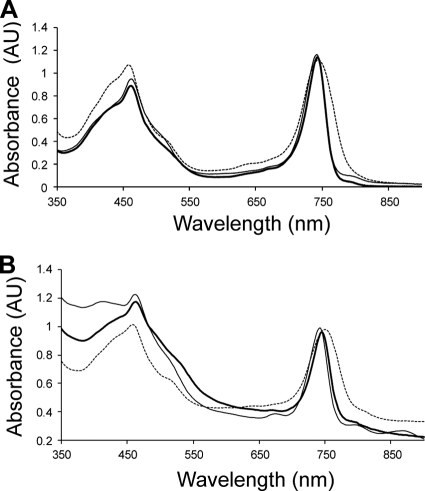
Figure 3A shows the absorption spectrum of a typical chlorosome preparation from “Ca. Chloracidobacterium thermophilum,” which had maxima due to BChl c at 461 nm (Soret) and 743 nm (Qy) and a small absorption maximum due to BChl a from the baseplate at ∼790 nm. The Qy absorption band was much narrower than that of chlorosomes from C. tepidum and was also blue-shifted by about 10 nm relative to that of C. tepidum. However, the absorption spectrum was very similar to that of chlorosomes of C. aurantiacus (Fig. 3A). The absorbance spectrum of aggregated BChl c molecules in intact cells was very similar to that in isolated chlorosomes and had maxima at 462 nm and 744 nm (Fig. 3B). The exact values for these absorption maxima varied by 1 to 2 nm and seemed to depend on the physiological state of the cells.
Fig. 3.
Absorption spectra of chlorosomes and cells of chlorosome-containing organisms. (A) Absorption spectra of chlorosomes of “Ca. Chloracidobacterium thermophilum” (thick solid line), C. tepidum (dashed line), and C. aurantiacus (thin solid line). (B) Absorption spectra of whole cells of “Ca. Chloracidobacterium thermophilum” (thick solid line), C. tepidum (dashed line), and C. aurantiacus (thin solid line).
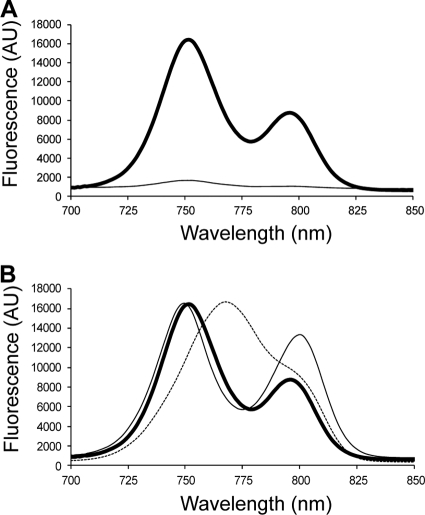
The fluorescence emission spectrum of isolated, dithionite-reduced chlorosomes from “Ca. Chloracidobacterium thermophilum” exhibited two well-separated emission bands, with maxima at 752 nm and 796 nm (Fig. 4A). These emission bands corresponded to those for the BChl c aggregates and BChl a associated with CsmA in the baseplate, respectively (1, 7, 17, 41, 42, 46–49, 53, 56, 65). In agreement with the absorption spectra (Fig. 3), the fluorescence emission spectrum of “Ca. Chloracidobacterium thermophilum” chlorosomes most closely resembled that of C. aurantiacus and differed significantly from that of C. tepidum (Fig. 4B). The fluorescence emission amplitude from “Ca. Chloracidobacterium thermophilum” chlorosomes decreased by a factor of 17 when the spectra were recorded in the presence of oxygen and without the addition of dithionite (Fig. 4A). This behavior suggested that, like the chlorosomes of C. tepidum, the chlorosomes of “Ca. Chloracidobacterium thermophilum” have a redox-dependent quenching mechanism that blocks energy transfer when chlorosomes (or cells) are exposed to oxic conditions (1, 13–15, 68, 72).
Fig. 4.
Fluorescence spectra for chlorosomes of “Ca. Chloracidobacterium thermophilum” under oxidizing and reducing conditions. (A) Fluorescence emission spectra of isolated chlorosomes from “Ca. Chloracidobacterium thermophilum” under reducing (thick solid line) and oxidizing (thin solid line) conditions. (B) Fluorescence emission spectra of isolated chlorosomes from “Ca. Chloracidobacterium thermophilum” (thick solid line), C. tepidum (dashed line), and C. aurantiacus (thin solid line) under reducing conditions. The excitation wavelength for these measurements was 440 nm. To produce oxic conditions, chlorosomes were diluted into air-saturated buffer; to produce reducing conditions, 25 mM sodium dithionite was added to the chlorosomes, which were incubated in the dark for ∼2 h prior to recording the emission spectra.
As described below, one of the proteins (CsmV) of the chlorosome envelope of “Ca. Chloracidobacterium thermophilum” contains an adrenodoxin domain and is distantly related to the CsmI, CsmJ, and CsmX motif family of C. tepidum and CsmY of C. aurantiacus, all of which are known or suspected to bind [2Fe-2S] clusters (19, 34, 66). The EPR spectrum recorded at 40 K of chlorosomes that had been chemically reduced with dithionite displayed a prominent axial signal with g values of 2.017 and 1.936 (Fig. 5). A weaker isotropic signal with a g value of 2.002 was also observed. Nearly identical spectra were obtained regardless of the identity of the chaotrope (sodium iodide or sodium thiocyanate) used during the isolation procedure (data not shown). A very similar set of resonances were previously reported for chemically reduced chlorosomes isolated from C. tepidum; in those studies, the axial signal was attributed to a [2Fe-2S] cluster and the isotropic signal to an organic radical, which could possibly be derived from a semiquinone species (66).
Fig. 5.
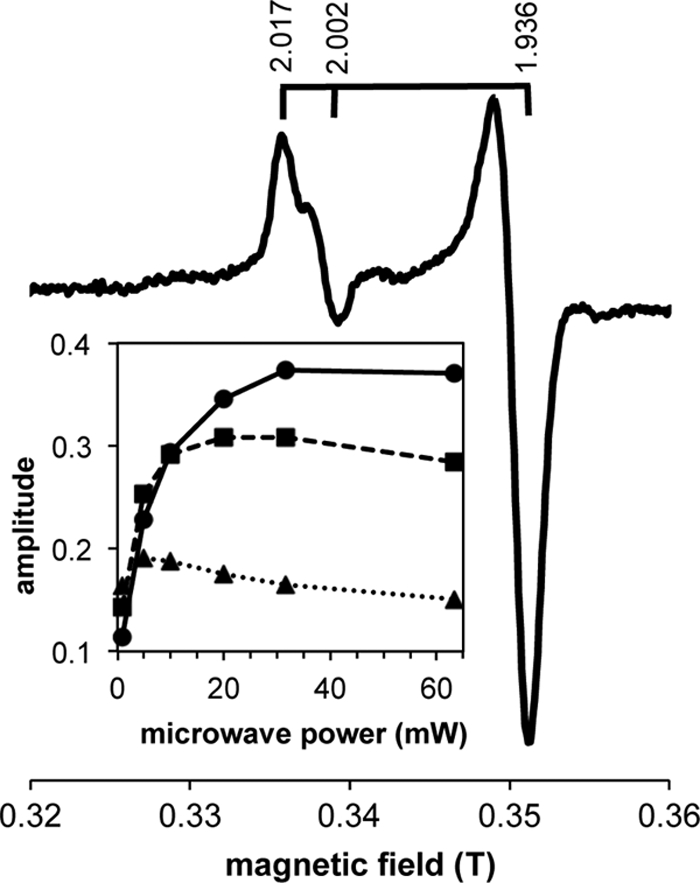
EPR spectrum of chemically reduced “Ca. Chloracidobacterium thermophilum” chlorosomes recorded at 20, 30, and 40 K and 10 mW microwave power. Other conditions were as indicated in Materials and Methods. Brackets above the spectrum indicate g values. (Inset) Power saturation curve for the g = 1.936 component of the axial signal measured at 40 K (circles and solid line), 30 K (squares and dashed line), and 20 K (triangles and dotted line).
In order to resolve the origins of the two signals in the chlorosomes of “Ca. Chloracidobacterium thermophilum” better, the EPR properties of these signals were studied as a function of microwave power and temperature. Under the conditions examined, the amplitude of the axial signal was maximal at 40 K, whereas the amplitude of the isotropic signal continued to increase with increasing temperature (data not shown). The power saturation behaviors for the g = 1.936 feature of the axial signal at 20 K, 30 K, and 40 K are shown in the inset in Fig. 5. These power saturation and temperature dependencies are consistent with the assignment of the axial signal (g = 2.017 and 1.936) to a reduced Fe-S cluster and the isotropic signal (g = 2.002) to an organic radical. Because [4Fe-4S] clusters typically saturate at power values higher than observed here and are rarely detected above 40 K (43), the axial signal is probably derived from a [2Fe-2S] cluster.
Analysis of the proteins of the chlorosome envelope.
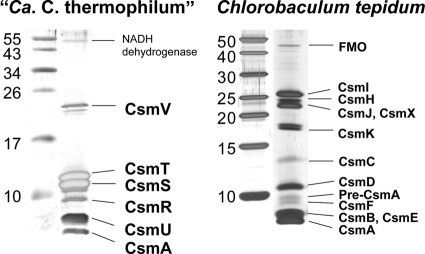
SDS-PAGE analyses of the chlorosomes of “Ca. Chloracidobacterium thermophilum,” in combination with N-terminal amino acid sequencing and tryptic peptide mass fingerprinting, led to the identification of seven polypeptides in the chlorosome envelopes of “Ca. Chloracidobacterium thermophilum” (Fig. 6). These proteins, the gene loci that encode them, and some properties of the proteins (predicted mass, pI, and predicted number of transmembrane α-helices) are summarized in Table 1. Only two of the proteins, CsmA and CsmV, have orthologs in the envelopes of chlorosomes from C. tepidum and C. aurantiacus. CsmA (the product of Cabther_A1765) from “Ca. Chloracidobacterium thermophilum” is highly divergent in sequence from other CsmA proteins (Fig. 7), but the predicted protein is similar in length (75 amino acids) and includes the conserved histidine residue that is thought to provide the ligand for binding BChl a (46, 47). Previous studies have shown that CsmA is proteolytically processed at its C terminus during its maturation and assembly into the chlorosome baseplate (8, 35, 46, 52, 61). All amino acids from 1 to 56 (including the N-terminal methionine residue) were detected by peptide mass fingerprinting of tryptic peptides derived from “Ca. Chloracidobacterium thermophilum” CsmA in chlorosomes. However, no peptides were detected for the 19 amino acids at the C terminus (data not shown, but see Fig. 7). Together with electrophoretic mobility data, which indicated “Ca. Chloracidobacterium thermophilum” CsmA was similar in molecular weight to mature CsmA of C. tepidum (Fig. 6), these data provide very strong evidence that CsmA is synthesized as a precursor protein and proteolytically processed during chlorosome biogenesis. CsmV, which is the product of open reading frame (ORF) Cabther_A2114, apparently migrated as three species with masses of 22 to 24 kDa on SDS-PAGE. Two of these three polypeptides had peptide fingerprints for CsmV, and no other protein was identified from this region of the gel. This electrophoretic behavior may be due to the formation of disulfide bonds between Cys residues, which normally ligate the Fe-S cluster in this protein, in the oxidizing environment of the polyacrylamide gels employed for electrophoresis.
Fig. 6.
SDS-PAGE analyses showing the protein composition of isolated chlorosomes from “Ca. Chloracidobacterium thermophilum” and C. tepidum. Left lanes, molecular mass standards (sizes are in kDa). Right lanes, chlorosomes from the indicated organism. Proteins corresponding to 20 μg of BChl c were loaded on the gels and were visualized by silver staining. Each protein is identified on the right.
Table 1.
Properties of chlorosome proteins from “Ca. Chloracidobacterium thermophilum”
| Protein | Locus taga | Predicted mass (kDa) | Predicted no. of transmembrane domainsb | Predicted pIb | Comment |
|---|---|---|---|---|---|
| CsmA | A1765 | 8.15 | None | 6.55 | BChl a-binding baseplate protein |
| CsmU | B0831 | 9.82 | None | 6.72 | Function unknown |
| CsmR | B0542 | 10.27 | 1 | 7.99 | Function unknown |
| CsmS | A1839 | 13.07 | 2 | 8.89 | Function unknown |
| CsmT | A0992 | 13.48 | 2 | 4.72 | Function unknown |
| CsmV | A2114 | 29.34 | 1 | 7.67 | Contains [2Fe-2S]-binding motif |
| NADH dehydrogenase | B0793 | 48.53 | 4 | 8.56 | Reduction of quencher? |
Locus tags beginning with A indicate genes encoded on chromosome 1; locus tags beginning with B indicate genes encoded on chromosome 2.
Predicted by using the ExPASY Proteomics server (23).
Fig. 7.
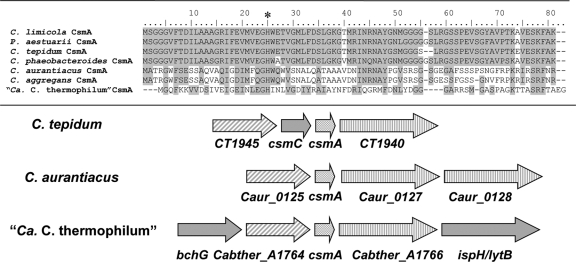
Alignment of CsmA sequences from various green bacteria. CsmA sequences from “Ca. Chloracidobacterium thermophilum,” C. aurantiacus, Chloroflexus aggregans, C. tepidum, Chlorobium limicola, Chlorobium phaeobacteroides, and Prosthecochloris aestuarii were aligned. The star indicates the conserved histidine thought to ligate BChl a. The arrows below the sequence alignment show the arrangement of genes in the vicinity of csmA in green bacteria belonging to three phyla. Open reading frames CT1945, Caur_0125, and Cabther_A1764 are similar in sequence and are probably orthologous. Ct1940, Caur_0127, and Cabther_A1766 are also similar in sequence and are probably orthologous. Caur_0127 and Caur_0128 are paralogous and appear to have arisen by gene duplication.
The other five proteins identified had not previously been detected in chlorosomes (Table 1), and only one of them was similar to other proteins in GenBank. The largest protein (∼55 kDa) was identified as a homolog of type II NADH dehydrogenase (product of Cabther_B0793), which interestingly was most similar to some type II NADH dehydrogenases in cyanobacteria. The other four proteins, denoted CsmR (Cabther_B0542), CsmS (Cabther_A1839), CsmT (Cabther_A0992), and CsmU (Cabther_B0831), were not similar to other proteins in chlorosomes or to any proteins in GenBank. CsmS and CsmT are 43% identical in sequence to one another (Fig. 8) and clearly arose through a gene duplication event, but these two proteins have pI values that differ by four pH units (Table 1). Surprisingly, each of these proteins is predicted to contain two transmembrane α-helices. CsmR and especially CsmU were difficult to detect, because these proteins diffused out of the polyacrylamide gel during staining unless the proteins had previously been fixed with formaldehyde (data not shown). This behavior is reminiscent of that of CsmB from C. tepidum (9) and possibly indicates that CsmR and CsmU might have properties and functions similar to CsmB, although these proteins do not share any sequence homology.
Fig. 8.
Sequence comparison of CsmS and CsmT. Identical amino acids and conservative replacements are shaded in gray. The bars below the sequence indicate the predicted transmembrane α-helices.
DISCUSSION
The data presented in this study describe the isolation, morphology, spectroscopic properties, and protein composition of the chlorosomes of “Ca. Chloracidobacterium thermophilum,” currently the only chlorophototrophic member of the phylum Acidobacterium. An isolation procedure similar to those employed for members of the Chlorobiales and Chloroflexaceae was used to isolate chlorosomes from “Ca. Chloracidobacterium thermophilum,” and although some specific differences were found, in general these chlorosomes resemble those found in other green bacteria (4, 5, 19).
Chlorosomes from “Ca. Chloracidobacterium thermophilum” have a striking morphology that suggests that the BChl c molecules form a suprastructure comprising internal ellipsoidal domains made up of folded or twisted sheets with a lamellar spacing of ∼2.3 nm. The structure formed differs from that in chlorosomes of C. tepidum, which are mostly arranged in concentric nanotubes formed from stacks of syn-anti BChl dimers (20). The suprastructure also appears to be different from the concentric nanotubes formed in a bchQ bchR mutant of C. tepidum, in which the BChl c molecules are arranged in all-syn or all-anti parallel monomer stacks (Ganapathy et al., unpublished). However, in “Ca. Chloracidobacterium thermophilum,” the BChl c molecules seem to form a suprastructure that resembles folded or twisted lamellar sheets that are clearly unlike structures observed in other chlorosomes studied to date (19, 20, 45, 50–52; Ganapathy et al., unpublished). Thus, our data, and those of others, suggest that the BChl molecules of chlorosomes can form distinctly different suprastructures in different organisms. The significance and functional differences for these different molecular arrangements of the BChls are currently unknown, but these structural differences are probably related to differences in the absorption and fluorescence properties observed for chlorosomes from various organisms (Fig. 3 and 4).
Although this is the first report describing this type of substructural domains in chlorosomes, “Ca. Chloracidobacterium thermophilum” may not be the only chlorosome-containing chlorophototroph with this arrangement of BChls. A previous study on the surface topography of chlorosomes isolated from representative green bacteria from both the Chlorobiales and the Chloroflexi revealed two morphologies, described as “rough” and “smooth,” of chlorosome surfaces (39). Although those authors attributed these differences to the lipid and protein composition of the chlorosome envelope, our findings lead us to propose that the “rough chlorosomes” described in that report have BChl molecules arranged in ellipsoidal domains similar to the ones observed in the chlorosomes of “Ca. Chloracidobacterium thermophilum” and that these domains are responsible for the “rough” surface topology. Similarly, Pšenčík et al. (51) proposed that the “rough” surface topology in these organisms is probably due to small domains of parallel lamellae positioned at random angles from the long axis of the chlorosome (51). Interestingly, whether an organism synthesizes chlorosomes that are “rough” or “smooth” appears to be a property associated with differences in BChl suprastructures inside chlorosomes (nanotubes or ellipsoidal domains) (36, 39). These structural differencs are independent of phylogenetic affiliation, because members of the same phylum (e.g., C. tepidum and Chlorobium phaeobacterioides) or even mutants of one organism (e.g., C. tepidum) can have different chlorosomes shapes, surfaces, and suprastructures (36, 39).
The chlorosomes of both “Ca. Chloracidobacterium thermophilum” and green FAPs have similar, narrow Qy absorption peaks. This observation suggests that the BChl c molecules in these chlorosomes may form similar suprastructures (see discussion above and references 52 and 58). These absorption properties differ from those of the BChl c aggregates in chlorosomes from C. tepidum, which have a broad Qy absorbance (Fig. 3). C. tepidum mutants lacking BchQ and BchR lack the ability to methylate the C-8 and C-12 positions of BChl c, respectively, and these mutants (especially the latter) produce chlorosomes with narrow Qy absorption bands (28), which resemble those of C. aurantiacus and “Ca. Chloracidobacterium thermophilum” chlorosomes. C. aurantiacus naturally lacks the BchQ and BchR methyltransferases (60) and thus can only synthesize BChl c with an ethyl side chain at C-8 and a methyl group at C-12 (25, 27). In contrast, “Ca. Chloracidobacterium thermophilum” has both BchQ and BchR (22), and like C. tepidum and other GSB, produces a heterogeneous mixture of BChl c molecules as a result (40), but the resulting BChl suprastructure has absorption properties more similar to those of chlorosomes of C. aurantiacus. Interestingly, despite the differences in their BChl c methylation patterns at the C-82 and C-121 positions, both C. aurantiacus and “Ca. Chloracidobacterium thermophilum” synthesize BChl c homologs which predominantly have saturated, straight-chain alcohols, principally octadecanol and hexadecanol, esterifed to the C-172 propionate group (4, 5, 21, 25). These combined observations suggest that the chemical nature of the esterifying alcohol might be a critical factor in establishing the spectroscopic properties of BChl aggregates in some chlorosomes.
In agreement with the absorbance results, the fluorescence emission spectrum of “Ca. Chloracidobacterium thermophilum” chlorosomes was also more similar to that of C. aurantiacus chlorosomes than to that of C. tepidum chlorosomes. Unlike C. aurantiacus, however, both in whole cells and in isolated chlorosomes of “Ca. Chloracidobacterium thermophilum,” the fluorescence emission amplitude was enhanced by a factor of ca. 17-fold by the addition of the reducing agent sodium dithionite, which showed that a redox-dependent quenching mechanism for energy transfer occurs in these chlorosomes (1, 13–15, 68, 72). “Ca. Chloracidobacterium thermophilum” is an aerobic microorganism that produces chlorosomes in the presence of oxygen (5). However, these fluorescence emission results imply that “Ca. Chloracidobacterium thermophilum” chlorosomes will not transfer light energy efficiently to reaction centers under oxidizing conditions. These data clearly show that, in this respect, the chlorosomes of “Ca. Chloracidobacterium thermophilum” resemble those of the strict anaerobe C. tepidum, which also exhibits enhanced fluorescence emission under reducing conditions after the addition of sodium dithionite (1). The redox dependence of the fluorescence emission amplitude suggested that a redox-sensitive quencher must be present in the chlorosomes, and the quencher has tentatively been identified as menaquinone 8 (21; A. M. Garcia Costas et al., unpublished data).
“Ca. Chloracidobacterium thermophilum” naturally occurs in microbial mats of thermal features in Yellowstone National Park, including Mushroom and Octopus Springs (30). These mats are subject to very high light intensities and become supersaturated with oxygen during the day. However, recent metatranscriptomic studies of samples collected from the mats of Mushroom Spring at different times in the diel cycle revealed that “Ca. Chloracidobacterium themophilum” exhibits high transcriptional activity at dusk and dawn, when light intensity and oxygen concentrations are low (37). These observations strongly suggest that “Ca. Chloracidobacterium thermophilum” may not use chlorosomes during the mid-day hours but may principally use chlorosomes to provide additional light energy for cyclic photophosphorylation when light conditions are not optimal in the mats. During periods of high light intensity, the BChl a-containing FMO and core antenna of the reaction centers may provide sufficient light-harvesting capacity to drive the photochemical reactions.
The lipid envelope of the chlorosomes of “Ca. Chloracidobacterium thermophilum” has a unique protein composition that includes homologs of CsmA and CsmI/CsmJ/CsmX of C. tepidum in addition to several novel proteins. CsmA, the BChl a-binding, chlorosome baseplate protein, has been found in all chlorosomes characterized to date. CsmA is usually the most abundant of the chlorosome proteins (7, 19), and it is the only chlorosome protein that is essential for viability in C. tepidum (8, 10, 18). Previous studies have shown that CsmA is proteolytically processed at its C terminus during maturation and assembly, and cross-linking studies and electron microscopy have shown that CsmA forms a paracrystalline array in the chlorosome baseplate (7, 8, 35, 47, 52, 57–59, 61). Our data show that CsmA in the chlorosomes of “Ca. Chloracidobacterium thermophilum” has a similar molecular mass and probably plays a similar role as in other chlorosome-containing microorganisms. It contains the critical histidine residue for binding BChl a and, similar to other chlorosomes, it is one of the most abundant proteins in the chlorosomes (Fig. 6). “Ca. Chloracidobacterium thermophilum” CsmA is apparently produced as a precursor protein of 75 amino acids and then is proteolytically processed by removal of 19 amino acids at the C terminus of the protein. It is interesting that this processing mechanism is conserved in all chlorosome-containing organisms, including members of the Chlorobiales, Chloroflexaceae, and now Acidobacteria (8, 18, 19, 34, 35, 42, 61). CsmA from “Ca. Chloracidobacterium thermophilum” is more closely related in sequence to CsmA from C. aurantiacus and C. aggregans (Fig. 7). ORFs adjacent to the csmA gene in “Ca. Chloracidobacterium thermophilum” are also most similar to ORFs adjacent to csmA of both GSB and FAPs (Fig. 7). These ORFs of unknown function flank the csmA genes in other chlorosome-containing microorganisms, and in C. tepidum (and presumably other green bacteria) they seem to be essential for viability (19; Y. Tsukatani and D. A. Bryant, unpublished results).
The chlorosomes of “Ca. Chloracidobacterium thermophilum” also contain an Fe-S protein, denoted CsmV, that is distantly related to CsmJ of the GSB Chloroherpeton thalassium. This protein contains an adrenodoxin-like [2Fe-2S] domain, and it is similar in molecular weight to other [2Fe-2S] proteins found in various chlorosomes. It seems reasonable to hypothesize that this protein plays a role in reduction of the oxidized quencher, as is the case for CsmI and CsmJ in C. tepidum (34). EPR analyses of isolated chlorosomes resulted in data consistent with the presence of [2Fe-2S] clusters. The EPR properties of these clusters in “Ca. Chloracidobacterium thermophilum” were similar to those in the chlorosomes of C. tepidum (66). The results also suggested that the Fe-S clusters of CsmV are stable under oxic conditions, like the [2Fe-2S] clusters of CsmI and CsmJ in the chlorosomes of C. tepidum (66).
In addition to CsmA and CsmV, the chlorosomes of “Ca. Chloracidobacterium thermophilum” contained two additional abundant proteins, CsmS and CsmT, which exhibited no sequence similarity to other known chlorosome proteins, nor did they exhibit any particular sequence motifs that might provide clues about their function. CsmS and CsmT are highly similar to one another in size and sequence (Fig. 8), and these proteins undoubtedly arose by gene duplication. Surprisingly, these two polypeptides are predicted to have two transmembrane α-helical domains and are the first chlorosome proteins to have this property. Although an algorithm designed for lipid bilayer membranes rather than a monolayer chlorosome membrane was used to predict these α-helices (23), no chlorosome protein from any other green bacterium has been predicted to contain such α-helical domains. It should also be noted that the predicted helices include two conserved proline residues, which could prevent helix formation. The difference in isoelectric points for these two peptides might be indicative of different functions, or it could indicate that they interact to form a heterodimeric complex stabilized by complementary electrostatic interactions. An interesting possibility is that the unusual morphology of the chlorosomes of “Ca. Chloracidobacterium thermophilum” (discussed above) could partly derive from these novel components of the envelope.
A homolog of type II NADH dehydrogenase was associated with the chlorosome envelope of “Ca. Chloracidobacterium thermophilum.” Type II NADH dehydrogenases are flavoproteins that transfer electrons from NAD(P)H to quinones, and thus this enzyme, as discussed above for CsmV, is probably involved in the reduction and possibly the oxidation of the menaquinones of the chlorosome, which are postulated to be the redox-sensitive quenching agents in these chlorosomes (21; Garcia Costas et al., unpublished data). In GSB such as C. tepidum, which contains high concentrations of low-potential ferredoxins (4), these redox reactions are mostly catalyzed by Fe-S proteins CsmI, CsmJ and, to a smaller extent, CsmX (34). However, high concentrations of ferredoxins are unlikely to occur in cells of the aerobe “Ca. Chloracidobacterium thermophilum,” suggesting that another enzyme, such as type II NADH dehydrogenase, could perform this function. Interestingly, a gene encoding a putative type II NADH dehydrogenase also occurs in an operon containing several chlorosome-related genes, immediately downstream from the gene for a major chlorosome envelope protein, CsmN, in the genome of C. aurantiacus J-10-fl (60). We hypothesize that this type II NADH dehydrogenase homolog may also occur in the chlorosome envelopes of C. aurantiacus and that it may have a similar role in redox regulation of energy transfer.
Chlorosomes of “Ca. Chloracidobacterium thermophilum” additionally contained two low-molecular-mass, low-abundance proteins, which we have designated CsmR and CsmU. Interestingly, these proteins were usually not observed, or were only very faintly visible, after SDS-PAGE when the gels were stained without formaldehyde fixation of the proteins. This phenomenon was previously observed with CsmB of C. tepidum and suggested a possible functional similarity of these proteins (9). Single, double, and triple mutants in C. tepidum which lacked CsmB and distantly related proteins with similar structure motifs, CsmF and CsmH, produced chlorosomes that differed in total pigment content and shape (36). Based on these studies, these proteins were postulated to influence the overall shape of chlorosomes. They may also influence carotenoid distribution/import into chlorosomes and may determine the orientation of the BChl c monomer stacks relative to the long axis of the chlorosomes (36). Despite the lack of apparent sequence homology, CsmR and CsmU in “Ca. Chloracidobacterium thermophilum” might play a similar role(s), because their properties are similar to those of CsmB. Further studies on CsmR, CsmU, and the other chlorosome proteins of “Ca. Chloracidobacterium thermophilum” will help elucidate their topological arrangements as well as their roles in chlorosome structure and function.
The combined spectroscopic and biochemical analyses of isolated chlorosomes from “Ca. Chloracidobacterium thermophilum” presented here show that, although this is an aerobic microorganism, its chlorosomes are remarkably similar in terms of BChl c arrangement, absorption properties, quenching behavior, and basic protein composition to those of other green bacteria, all of which produce and utilize these organelles under anoxic conditions (4, 19). Interestingly, the chlorosomes of “Ca. Chloracidobacterium thermophilum” share some spectroscopic properties with those of members of both the GSB and the FAPs. Significantly, “Ca. Chloracidobacterium thermophilum” synthesizes several unique chlorosome proteins, and further studies on those, e.g., the type II NADH dehydrogenase homolog, might provide additional clues to how this organism and others regulate energy transfer and protect this large organelle from oxidative damage.
ACKNOWLEDGMENTS
This research was supported by grant DE-FG02-94ER20137 from the U.S. Department of Energy to D.A.B. and National Science Foundation grants MCB-0519743 and MCB-1021725 to J.H.G. and D.A.B.
Footnotes
Published ahead of print on 30 September 2011.
REFERENCES
- 1. Blankenship R. E., et al. 1993. Redox regulation of energy transfer efficiency in antennas of green photosynthetic bacteria. Photochem. Photobiol. 57:103–107 [DOI] [PubMed] [Google Scholar]
- 2. Blankenship R. E. 2002. Molecular mechanisms of photosynthesis. Blackwell Science Ltd., Oxford, England [Google Scholar]
- 3. Blum H., Beier H., Gross H. 1987. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8:93–99 [Google Scholar]
- 4. Bryant D. A., et al. 2011. Comparative and functional genomics of anoxygenic green bacteria from the taxa Chlorobi, Chloroflexi, and Acidobacteria, p. 47–102 In Burnap R. L., Vermaas W. (ed.), Advances in photosynthesis and respiration, vol. 33 Functional genomics and evolution of photosynthetic systems. Springer, Dordrecht, The Netherlands [Google Scholar]
- 5. Bryant D. A., et al. 2007. Candidatus Chloracidobacterium thermophilum: an aerobic phototrophic acidobacterium. Science 317:523–526 [DOI] [PubMed] [Google Scholar]
- 6. Bryant D. A., Frigaard N.-U. 2006. Prokaryotic photosynthesis and phototrophy illuminated. Trends Microbiol. 14:488–496 [DOI] [PubMed] [Google Scholar]
- 7. Bryant D. A., Vassilieva E. V., Frigaard N.-U., Li H. 2002. Selective protein extraction from Chlorobium tepidum chlorosomes using detergents. Evidence that CsmA forms multimers and binds bacteriochlorophyll a. Biochemistry 41:14403–14411 [DOI] [PubMed] [Google Scholar]
- 8. Chung S., Frank G., Zuber H., Bryant D. A. 1994. Genes encoding two chlorosome components from the green sulfur bacteria Chlorobium vibrioforme strain 8327D and Chlorobium tepidum. Photosynth. Res. 41:261–275 [DOI] [PubMed] [Google Scholar]
- 9. Chung S., Bryant D. A. 1996. Characterization of csmB genes, encoding a 7.5-kDa protein of the chlorosome envelope, from the green sulfur bacteria Chlorobium vibrioforme 8327D and Chlorobium tepidum. Arch. Microbiol. 166:234–244 [DOI] [PubMed] [Google Scholar]
- 10. Chung S., Shen G., Ormerod J., Bryant D. A. 1998. Insertional inactivation studies of the csmA and csmC genes of the green sulfur bacterium Chlorobium vibrioforme 8327: the chlorosome protein CsmA is required for viability by CsmC is dispensible. FEMS Microbiol. Lett. 164:353–361 [DOI] [PubMed] [Google Scholar]
- 11. Feick R. G., Fuller R. C. 1984. Topography of the photosynthetic apparatus of Chloroflexus aurantiacus. Biochemistry 23:3693–3700 [Google Scholar]
- 12. Frangakis A. S., Hegerl R. 2001. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J. Struct. Biol. 135:239–250 [DOI] [PubMed] [Google Scholar]
- 13. Frigaard N.-U., Takaichi S., Hirota M., Shimada K., Matsuura K. 1997. Quinones in chlorosomes of green sulfur bacteria and their role in the redox-dependent fluorescence studied in chlorosome-like bacteriochlorophyll c aggregates. Arch. Microbiol. 167:343–349 [Google Scholar]
- 14. Frigaard N.-U., Matsuura K., Hirota M., Miller M., Cox R. P. 1998. Studies of the location and function of isoprenoid quinones in chlorosomes from green sulfur bacteria. Photosynth. Res. 58:181–190 [Google Scholar]
- 15. Frigaard N.-U., Tokita S., Matsuura K. 1999. Exogenous quinones inhibit photosynthetic electron transfer in Chloroflexus aurantiacus by specific quenching of the excited bacteriochlorophyll c antenna. Biochim. Biophys. Acta 1413:108–116 [DOI] [PubMed] [Google Scholar]
- 16. Frigaard N.-U., Bryant D. A. 2001. Chromosomal gene inactivation in the green sulfur bacterium Chlorobium tepidum by natural transformation. Appl. Environ. Microbiol. 67:2538–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Frigaard N.-U., Voigt G. D., Bryant D. A. 2002. Chlorobium tepidum mutant lacking bacteriochlorophyll c made by inactivation of the bchK gene, encoding bacteriochlorophyll c synthase. J. Bacteriol. 184:3368–3376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Frigaard N. U., Li H., Milks K. J., Bryant D. A. 2004. Nine mutants of Chlorobium tepidum each unable to synthesize a different chlorosome protein still assemble functional chlorosomes. J. Bacteriol. 186:646–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Frigaard N.-U., Bryant D. A. 2006. Chlorosomes: antenna organelles in photosynthetic green bacteria, p. 79–114 In Shively J. M. (ed.), Complex structures in prokaryotes, vol. 2 Springer, Berlin, Germany [Google Scholar]
- 20. Ganapathy S., et al. 2009. Alternating syn-anti bacteriochlorophylls form concentric helical nanotubes in chlorosomes. Proc. Natl. Acad. Sci. U. S. A. 106:8525–8530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Garcia Costas A. M. 2010. Isolation and characterization of Candidatus Chloracidobacterium thermophilum. Ph.D. thesisThe Pennsylvania State University, University Park, PA [Google Scholar]
- 22. Garcia Costas A. M., et al. Complete genome of Candidatus Chloracidobacterium thermophilum, a chlorophyll-based photoheterotroph belonging to the phylum Acidobacteria. Environ. Microbiol., in press [DOI] [PubMed] [Google Scholar]
- 23. Gasteiger E., et al. 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31:3784–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gich F., et al. 2003. Characterization of the chlorosome antenna of the filamentous anoxygenic phototrophic bacterium Chloronema sp. strain UdG9001. Arch. Microbiol. 180:417–426 [DOI] [PubMed] [Google Scholar]
- 25. Gloe A., Risch N. 1978. Bacteriochlorophyll cs, a new bacteriochlorophyll from Chloroflexus aurantiacus. Arch. Microbiol. 118:153–156 [DOI] [PubMed] [Google Scholar]
- 26. Golbeck J. H. 1993. Shared thematic elements in photochemical reaction centers. Proc. Natl. Acad. Sci. U. S. A. 90:1642–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gomez Maqueo Chew A., Bryant D. A. 2007. Chlorophyll biosynthesis in bacteria: the origins of structural and functional diversity. Annu. Rev. Microbiol. 53:89–104 [DOI] [PubMed] [Google Scholar]
- 28. Gomez Maqueo Chew A., Frigaard N.-U., Bryant D. A. 2007. Bacteriochlorophyllide c C-8(2) and C-12(1) methyltransferases are essential for adaptation to low light in Chlorobaculum tepidum. J. Bacteriol. 189:6176–6184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hohmann-Marriott M. F., Blankenship R. E., Roberson R. W. 2005. The ultrastructure of Chlorobium tepidum chlorosomes revealed by electron microscopy. Photosynth. Res. 86:145–154 [DOI] [PubMed] [Google Scholar]
- 30. Klatt C. G., et al. 2011. Metagenomic analyses of phototrophic hot spring microbial mat communities. ISME J. 5:1262–127821697961 [Google Scholar]
- 31. Kremer J. R., Mastronarde D. N., McIntosh J. R. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76 [DOI] [PubMed] [Google Scholar]
- 32. Lanyi J. K. 2006. Proton transfers in the bacteriorhodopsin photocycle. Biochim. Biophys. Acta 1757:1012–1018 [DOI] [PubMed] [Google Scholar]
- 33. Larson C. R., et al. 2011. The three-dimensional structure of the FMO protein from Pelodictyon phaeum and the implications for energy transfer. Photosynth. Res. 107:139–150 [DOI] [PubMed] [Google Scholar]
- 34. Li H. 2006. Organization and function of chlorosome proteins in the green sulfur bacterium Chlorobium tepidum. Ph.D. thesis The Pennsylvania State University, University Park, PA [Google Scholar]
- 35. Li H., Frigaard N.-U., Bryant D. A. 2006. Molecular contacts for chlorosome envelope proteins revealed by cross-linking studies with chlorosomes from Chlorobium tepidum. Biochemistry 45:9095–9103 [DOI] [PubMed] [Google Scholar]
- 36. Li H., Bryant D. A. 2009. Envelope proteins of the CsmB/CsmF and CsmC/CsmD motif families influence the size, shape, and composition of chlorosomes in Chlorobaculum tepidum. J. Bacteriol. 191:7109–7120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu Z., et al. 2011. Metatranscriptomic analyses of chlorophototrophs of a hot-spring microbial mat. ISME J. 5:1279–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Madigan M. T., Petersen S. R., Brock T. D. 1974. Nutritional studies on Chloroflexus, a filamentous photosynthetic, gliding bacterium. Arch. Microbiol. 100:97–103 [Google Scholar]
- 39. Martinez-Planells A., et al. 2002. Determination of the topography and biometry of chlorosomes by atomic force microscopy. Photosynth. Res. 71:83–90 [DOI] [PubMed] [Google Scholar]
- 40. Masse A. M., Airs R. L., Keely B. J., de Wit R. 2004. The impact of different intensities of green light on the bacteriochlorophyll homologue composition of the Chlorobiaceae: Prosthecochloris aestuarii and Chlorobium phaeobacteroides. Microbiology 150:2555–2564 [DOI] [PubMed] [Google Scholar]
- 41. Montaño G. A., et al. 2003. Characterization of Chlorobium tepidum chlorosomes: a calculation of bacteriochlorophyll c per chlorosome and oligomer modeling. Biophys. J. 85:2560–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Montaño G. A., Wu H. M., Lin S., Brune D. C., Blankenship R. E. 2003. Isolation and characterization of the B798 light-harvesting baseplate from the chlorosomes of Chloroflexus aurantiacus. Biochemistry 42:10246–10451 [DOI] [PubMed] [Google Scholar]
- 43. Ohnishi T. 1998. Iron-sulfur clusters/semiquinones in complex 1. Biochim. Biophys. Acta 1364:186–206 [DOI] [PubMed] [Google Scholar]
- 44. Olson J. M. 2004. The FMO protein. Photosynth. Res. 80:181–187 [DOI] [PubMed] [Google Scholar]
- 45. Oostergetel G. T., et al. 2007. Long-range organization of bacteriochlorophyll in chlorosomes of Chlorobium tepidum investigated by cryo-electron microscopy. FEBS Lett. 581:5435–5439 [DOI] [PubMed] [Google Scholar]
- 46. Pedersen M. Ø., Borch J., Højrup P., Cox R. P., Miller M. 2006. The light-harvesting antenna of Chlorobium tepidum: interactions between the FMO protein and the major chlorosome protein CsmA studied by surface plasmon resonance. Photosynth. Res. 89:63–69 [DOI] [PubMed] [Google Scholar]
- 47. Pedersen M. Ø., Linnanto J., Frigaard N.-U., Nielsen N. C., Miller M. 2010. A model of the protein-pigment baseplate complex in chlorosomes of photosynthetic green bacteria. Photosynth. Res. 104:233–243 [DOI] [PubMed] [Google Scholar]
- 48. Prokhorenko V. I., Steensgaard D. B., Holzwarth A. R. 2003. Exciton theory for supramolecular chlorosomal aggregates. 1. Aggregate size dependence of the linear spectra. Biophys. J. 85:3173–3186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pšenčík J., Ma Y. Z., Arellano J. B., Hála J., Gillbro T. 2003. Excitation energy transfer dynamics and excited-state structure in chlorosomes of Chlorobium phaeobacteroides. Biophys. J. 84:1161–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pšenčík J., et al. 2004. Lamellar organization of pigments in chlorosomes, the light harvesting complexes of green photosynthetic bacteria. Biophys. J. 87:1165–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pšenčík J., et al. 2006. Internal structure of chlorosomes from brown-colored Chlorobium species and the role of carotenoids in their assembly. Biophys. J. 91:1433–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pšenčík J., et al. 2009. Structure of chlorosomes from the green filamentous bacterium Chloroflexus aurantiacus. J. Bacteriol. 191:6701–6708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Saga Y., et al. 2002. Spectral heterogeneity in single light-harvesting chlorosomes from green sulfur photosynthetic bacterium Chlorobium tepidum. Photochem. Photobiol. 75:433–436 [DOI] [PubMed] [Google Scholar]
- 54. Saga Y., Tamiaki H. 2006. Transmission electron microscopic study on supramolecular nanostructures of bacteriochlorophyll self-aggregates in chlorosomes of green photosynthetic bacteria. J. Biosci. Bioeng. 102:118–123 [DOI] [PubMed] [Google Scholar]
- 55. Schagger H., Jagow G. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel eleoctrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368–379 [DOI] [PubMed] [Google Scholar]
- 56. Shibata Y., Saga Y., Tamiaki H., Itoh S. 2006. Low-temperature fluorescence from single chlorosomes, photosynthetic antenna complexes of green filamentous and sulfur bacteria. Biophys. J. 91:3787–3796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sprague S. G., Staehelin L. A., DiBartolomeis M. J., Fuller R. C. 1981. Isolation and development of chlorosomes in the green bacterium Chloroflexus aurantiacus. J. Bacteriol. 147:1021–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Staehelin L. A., Golecki J. R., Fuller R. C., Drews G. 1978. Visualization of the supramolecular architecture of chlorosomes (Chlorobium type vesicles) in freeze-fractured cells of Chloroflexus aurantiacus. Arch. Microbiol. 119:269–277 [Google Scholar]
- 59. Staehelin L. A., Golecki J. R., Drews G. 1980. Supramolecular organization of chlorosomes (Chlorobium vesicles) and of their membrane attachment sites in Chlorobium limicola. Biochim. Biophys. Acta 589:30–45 [DOI] [PubMed] [Google Scholar]
- 60. Tang K.-H., et al. 2011. Complete genome sequence of the filamentous anoxygenic phototrophic bacterium Chloroflexus aurantiacus. BMC Genomics 12:334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Theroux S. J., Redlinger T. E., Fuller R. C., Robinson S. J. 1990. Gene encoding the 5.7-kilodalton chlorosome protein of Chloroflexus aurantiacus: regulated message levels and a predicted carboxy-terminal protein extension. J. Bacteriol. 172:4497–4504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tronrud D. E., Wen J., Gay L., Blankenship R. E. 2009. The structural basis for the difference in absorbance spectra for the FMO antenna protein from various green sulfur bacteria. Photosynth. Res. 100:79–87 [DOI] [PubMed] [Google Scholar]
- 63. Tsukatani Y., Wen J., Blankenship R. E., Bryant D. A. 2010. Characterization of the bacteriochlorophyll a-binding, Fenna-Matthews-Olson protein from Candidatus Chloracidobacterium thermophilum. Photosynth. Res. 104:201–209 [DOI] [PubMed] [Google Scholar]
- 64. van de Meene A. M., Le Olson T., Collins A. M., Blankenship R. E. 2007. Initial characterization of the photosynthetic apparatus of “Candidatus Chlorothrix halophila,” a filamentous, anoxygenic photoautotroph. J. Bacteriol. 189:4196–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. van Dorssen R. J., Gerola P. D., Olson J. M., Amesz J. 1986. Optical and structural properties of chlorosomes of the photosynthetic green sulfur bacterium Chlorobium limicola. Biochim. Biophys. Acta 848:77–82 [Google Scholar]
- 66. Vassilieva E. V., et al. 2001. Electron transfer may occur in the chlorosome envelope: the CsmI and CsmJ proteins of chlorosomes are 2Fe-2S ferredoxins. Biochemistry 40:464–473 [DOI] [PubMed] [Google Scholar]
- 67. Vassilieva E. V., et al. 2002. Subcellular localization of chlorosome proteins in Chlorobium tepidum and characterization of three new chlorosome proteins: CsmF, CsmH and CsmX. Biochemistry 41:4358–4370 [DOI] [PubMed] [Google Scholar]
- 68. Wang J., Brune D. C., Blankenship R. E. 1990. Effects of oxidants and reductants on the efficiency of excitation transfer in green photosynthetic bacteria. Biochim. Biophys. Acta 1015:457–463 [DOI] [PubMed] [Google Scholar]
- 69. Wen J., et al. 2011. Structural and spectroscopic insights of the FMO antenna protein of the aerobic chlorophototroph Candidatus Chloracidobacterium thermophilum. Biochim. Biophys. Acta 1807:157–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wen J., Zhang H., Gross M. L., Blankenship R. E. 2009. Membrane orientation of the FMO antenna protein from Chlorobaculum tepidum as determined by mass spectrometry-based footprinting. Proc. Natl. Acad. Sci. U. S. A. 106:6134–6139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wen J., Zhang H., Gross M. L., Blankenship R. E. 2011. Native electrospray mass spectrometry reveals the nature and stoichiometry of pigments in the FMO photosynthetic antenna protein. Biochemistry 50:3502–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhou W., LoBrutto R., Lin S., Blankenship R. E. 1994. Redox effects on the bacteriochlorophyll a-containing Fenna-Matthews-Olson protein from Chlorobium tepidum. Photosynth. Res. 41:89–96 [DOI] [PubMed] [Google Scholar]