Abstract
Chemoreceptors such as Tsr, the serine receptor, function in trimer-of-dimer associations to mediate chemotactic behavior in Escherichia coli. The two subunits of each receptor homodimer occupy different positions in the trimer, one at its central axis and the other at the trimer periphery. Residue N381 of Tsr contributes to trimer stability through interactions with its counterparts in a central cavity surrounded by hydrophobic residues at the trimer axis. To assess the functional role of N381, we created and characterized a full set of amino acid replacements at this Tsr residue. We found that every amino acid replacement at N381 destroyed Tsr function, and all but one (N381G) of the mutant receptors also blocked signaling by Tar, the aspartate chemoreceptor. Tar jamming reflects the formation of signaling-defective mixed trimers of dimers, and in vivo assays with a trifunctional cross-linking reagent demonstrated trimer-based interactions between Tar and Tsr-N381 mutants. Mutant Tsr molecules with a charged amino acid or proline replacement exhibited the most severe trimer formation defects. These trimer-defective receptors, as well as most of the trimer-competent mutant receptors, were unable to form ternary signaling complexes with the CheA kinase and with CheW, which couples CheA to receptor control. Some of the trimer-competent mutant receptors, particularly those with a hydrophobic amino acid replacement, may not bind CheW/CheA because they form conformationally frozen or distorted trimers. These findings indicate that trimer dynamics probably are important for ternary complex assembly and that N381 may not be a direct binding determinant for CheW/CheA at the trimer periphery.
INTRODUCTION
Most chemotactic behaviors in Escherichia coli, as well as in other motile bacteria and archaea, depend on transmembrane chemoreceptors known as methyl-accepting chemotaxis proteins (MCPs) (16, 17). MCP molecules typically function as homodimers of ∼550-residue subunits. All MCPs share a defining, highly conserved cytoplasmic signaling domain, and most have an extracellular chemoeffector-sensing domain (Fig. 1A). E. coli has five MCP-family receptors (Aer, Tar, Tap, Trg, and Tsr), and they have been extensively characterized. Tsr detects serine, an attractant, through direct binding sites in the periplasmic domain (Fig. 1A). Tsr also mediates an attractant response to AI-2, a quorum signal common to many bacteria, most likely through the interaction of the periplasmic domain with LsrB, the periplasmic AI-2 binding protein (18).
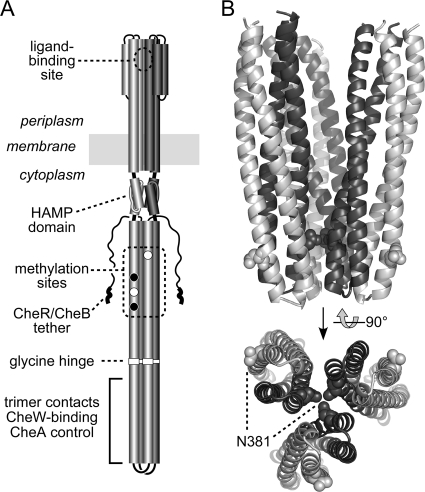
Fig. 1.
Structures of Tsr dimers and trimers of dimers. (A) Structure-function features of a Tsr homodimer. Cylindrical segments represent α-helices drawn approximately to scale. One subunit is shaded light gray; the other is dark gray. Methylation sites shown as white circles are translated as E residues; those shown as black circles are translated as Q residues and subsequently converted to E residues through CheB-mediated deamidation. A pentapeptide sequence (NWETF) at the C terminus of each subunit serves as a binding site for CheR and CheB, facilitating interaction with their substrate sites. Receptor dimers form trimers and ternary signaling complexes through interaction sites at the membrane-distal tip of the cytoplasmic four-helix bundle. (B) Trimer-of-dimer structure at the cytoplasmic tip. The tips of three Tsr dimers (residues 350 to 430) are shown in a trimer-of-dimer arrangement; atomic coordinates are from reference 22. Subunits at the trimer interface are dark gray, and subunits at the trimer periphery are light gray. Residue N381 (space-filled) resides in two different structural environments in the trimer, at the trimer axis (dark gray) and at the trimer periphery (light gray).
The membrane-distal tip of the MCP cytoplasmic domain contains binding determinants that promote higher-order interactions between MCP molecules, both of the same and different detection specificities, as well as interactions with the CheA and CheW proteins (3, 4, 22). Receptor-receptor interaction forms trimers of receptor dimers that in turn recruit CheA/CheW to assemble ternary complexes (25, 40). These receptor teams transmit control signals to the flagellar motors via the response regulator CheY (33, 44). CheA is a histidine autokinase (19), CheW couples CheA to chemoreceptor control (9, 14, 26), and CheY is an aspartate autokinase that obtains its phosphoryl groups from phospho-CheA (10, 32). Chemical stimuli, sensed as temporal changes in chemoeffector levels as the cell swims about, modulate CheA activity to influence the direction of flagellar rotation: counterclockwise (CCW) rotation, the default behavior, produces forward swimming, and clockwise (CW) rotation, elicited by the phosphorylated form of CheY, causes tumbling events that randomly reorient the cell.
The MCPs of E. coli operate in highly cooperative clusters at the cell poles (27, 37, 38). Trimer-of-dimer interactions between receptor molecules are believed to underlie array formation and to account for the large amplification factors of the receptor network. Experimental support for the trimer-of-dimer model comes from the crystal structure of the Tsr cytoplasmic domain (22), from in vivo receptor cross-linking studies (40, 42), from the signaling phenotypes of Tsr mutants with lesions at the trimer contact sites (4), from conformational suppression effects between receptor mutants with trimer contact alterations (2), and from studies of receptor molecules in nanodiscs (7, 24, 25).
The architectural details of ternary complexes and their mechanisms of CheA control remain obscure, in large part because the trimer-of-dimer organization confounds structure-function analyses. First, the trimer contact region in receptor molecules also contains CheA/CheW binding determinants needed for ternary complex assembly, so some tip residues probably have multiple functions. Second, the juxtaposition of receptor molecules in the trimer arrangement may create CheA/CheW docking surfaces that do not exist in receptor dimers. Third, the trimer geometry produces two very different structural environments for identical residues in each of the receptor dimers: one subunit resides at the trimer axis, the other at the trimer perimeter.
Residue N381 of Tsr is a promising experimental subject with which to resolve some of these trimer-related complications (Fig. 1B). Of the 11 principal trimer contact residues (22), N381 is nearly invariant across the entire MCP superfamily (1). Unlike the other contact sites, which stabilize the trimer through hydrophobic or polar interactions with nonidentical partner residues, N381 forms a hydrogen-bonded network with its counterparts at the trimer central axis (Fig. 1B). Thus, one N381 residue in each dimer is buried at the trimer interface, and the other is solvent exposed on the outside of the trimer (Fig. 1B). Because multiple tip residues contribute to trimer stability (4, 22), amino acid replacements at N381 that introduce a smaller side chain might not disrupt trimer formation. In contrast, replacements that introduce charged or bulky side chains could destabilize the trimer enough to impair Tsr function. However, if N381 also plays an important role at the trimer periphery, for example as a major binding determinant for CheW or CheA, both sorts of structural lesions might impair Tsr function. Accordingly, we constructed and characterized a complete set of amino acid replacements at residue N381 of Tsr to determine (i) how critical this residue is to trimer formation, stability, and function; (ii) whether N381 plays additional roles in its location at the trimer periphery; and (iii) if trimer formation is, indeed, the key determinant for receptor clustering.
MATERIALS AND METHODS
Bacterial strains.
Strains were derivatives of E. coli K-12 strain RP437 (31). Their designations and relevant genotypes are the following: UU1250 [Δaer-1 ygjG::Gm Δtsr-7028 Δ(tar-tap)5201 Δtrg-100] (4), UU1535 [Δaer-1 ygjG::Gm Δtsr-7028 Δ(tar-cheB)2234 Δtrg-100] (6), UU1581 [Δ(flhD-flhA)4 Δtsr-7028 Δtrg-100] (40), UU1613 [tar(S364C) Δtsr-7028 Δtrg-100 Δ(tap-cheB)2234 Δ(cheA-cheW)2167] (42), UU1623 (Δtsr-7028 Δtap-3654 Δtrg-100) (5), UU2377 [tsr(R69E) Δaer-1 Δ(tar-tap)5201 Δtrg-4543 ΔrecA-SstII/EcoRI] (5), and UU2378 [tsr(T156K) Δaer-1 Δ(tar-tap)5201 Δtrg-4543 ΔrecA-SstII/EcoRI] (5).
Plasmids.
Plasmids were pRR48, a derivative of pBR322 (8) that confers ampicillin resistance and has an expression/cloning site with a tac promoter and an ideal (perfectly palindromic) lac operator under the control of a plasmid-encoded lacI repressor, which is inducible by isopropyl β-d-thiogalactopyranoside (IPTG) (42); and pRR53, a derivative of pRR48 that carries wild-type tsr under the control of IPTG (42).
The plasmids used in receptor-clustering assays were pVS49, a derivative of pACYC184 (13) that makes a functional fusion of CheZ to yellow fluorescent protein (YFP) under inducible arabinose control (39); pVS102, a relative of pVS49 that makes a functional YFP-CheR fusion protein under inducible arabinose control (21); and pPA801, a relative of pVS49 that makes a functional YFP-CheW fusion protein under inducible arabinose control (28).
Construction of Tsr mutants.
Mutations were generated in plasmid pRR53 by QuikChange PCR mutagenesis, using either degenerate codon primers (for N381 replacements) or site-specific primers (for S366C), as previously described (4, 5). QuikChange products were introduced into UU1250 by either electroporation or CaCl2 transformation and tested for the ability to support Tsr function on soft-agar plates (see below). Candidate plasmids were verified by sequencing the entire tsr coding region.
Chemotaxis assays.
Host strains carrying tsr plasmids were assessed for chemotactic ability on tryptone soft agar plates (29) containing ampicillin (50 μg/ml]) and 100 μM IPTG. Plates were incubated at 30°C for 7 to 10 h.
Expression levels of mutant Tsr proteins.
Tsr expression from pRR53 derivatives was analyzed in strain UU1535 to avoid multiple modification states. Cells were grown and lysates analyzed by SDS-PAGE as described previously (28).
Dominance, rescue, and jamming tests.
For dominance tests, mutant pRR53 derivatives were transferred into UU2377 and UU2378. For rescue and jamming tests, mutant plasmids were transferred into UU1623. Plasmid-containing cells were tested for Tsr and/or Tar function on tryptone soft-agar plates as described above.
Receptor clustering assays.
Mutant pRR53 derivatives were introduced into UU1250 or UU1581 cells harboring pVS49, pVS102, or pPA801. Cells containing each pair of compatible plasmids were grown at 30°C in tryptone broth containing 50 μg/ml ampicillin and 12.5 μg/ml chloramphenicol. Tsr expression from pRR53 derivatives was induced with 100 μM IPTG, YFP-CheZ (pVS49) was induced with 0.005% (wt/vol) l(+)-arabinose, YFP-CheR (pVS102) was induced with 0.01% (wt/vol) l(+)-arabinose, and YFP-CheW (pPA801) was induced with 0.004% (wt/vol) l(+)-arabinose. Cells were grown in tryptone broth at 30°C to mid-exponential phase and analyzed by fluorescence microscopy as previously described (4, 28).
Cross-linking assays for trimers of dimers.
All tests utilized the trifunctional thiol reagent tris-2-maleimidoethyl-amide (TMEA) (Pierce Chemical Co.). For direct cross-linking assays, mutant Tsr proteins carried an S366C reporter. Plasmid-containing UU1581 cells were grown to an optical density at 600 nm (OD600) of 0.7 at 30°C in tryptone broth containing 50 μg/ml ampicillin and 100 μM IPTG, treated with 50 μM TMEA for 30 s, and quenched with 10 mM N-ethyl maleimide (NEM), and cross-linking products were analyzed by SDS-PAGE and immunoblotting, as described previously (40, 41). For cross-linking competition assays, mutant plasmids encoding Tsr proteins without a cysteine reporter were transferred into strain UU1613. Cells were grown as described for direct cross-linking assays, except that IPTG inducer was added at 1 h to a final concentration of 200 μM. The remainder of the experimental protocol followed that for direct cross-linking. For mixed cross-linking tests, plasmids encoding Tsr proteins with the S366C reporter were expressed in strain UU1613. Cells were grown as in direct cross-linking tests, except with 40 μM IPTG and SDS-PAGE analysis on 11% low-bis gels (40, 42).
Flagellar rotation assays.
Flagellar rotation patterns of plasmid-containing UU1535 and UU1250 cells were studied by antibody tethering as described previously (26, 28, 35). We classified cells into five categories according to their pattern of flagellar rotation: exclusively CCW, CCW reversing, balanced CCW-CW, CW reversing, and exclusively CW.
Protein structural display.
Structure images were prepared with MacPyMOL software (http://www.pymol.org).
RESULTS
Construction and initial characterization of Tsr-N381 mutant receptors.
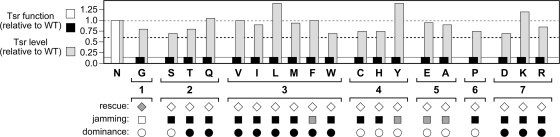
We used all-codon mutagenesis at the N381 codon of the tsr coding region in plasmid pRR53 to create a complete set of amino acid replacements at residue 381 of Tsr (5, 28, 48). Plasmid pRR53 produces very low Tsr expression levels without induction and fully complements UU1250, a strain lacking all MCP-family receptor genes (tsr, tar, tap, trg, and aer), for serine chemotaxis at 80 to 100 μM IPTG. These induction conditions were used for all physiological experiments in this study (42). At an optimal inducer concentration, all N381 mutant (designated N381*) receptors failed to promote chemotactic colony expansion in tryptone soft-agar plates. Colony sizes for cells expressing any of the mutant receptors were no larger than that of the tsr null control (vector plasmid pRR48) (Fig. 2). In a closely related receptorless host (UU1535), which also bears deletions in the genes for the MCP-modifying enzymes CheR and CheB, the mutant plasmids produced essentially normal levels of Tsr protein, ranging between 0.6- and 1.4-fold of the wild-type level (Fig. 2). Thus, all N381* receptors had nominally wild-type expression levels and stability but no discernible Tsr function in soft-agar assays.
Fig. 2.
Tsr function of N381 amino acid replacement mutants. Tsr-mediated chemotaxis was assessed by colony size and morphology in soft-agar assays (see Materials and Methods). Colony size is indicated relative to that of the wild type. The white bar indicates wild-type serine ring morphology (Tsr+), and black bars denote colonies with no serine ring (Tsr−). Gray bars indicate the relative expression levels of the mutant proteins. The dashed gray line (at 1.0) shows the wild-type value for comparison purposes. The dashed black line (at 0.6) indicates the cutoff criterion for essentially normal protein expression levels. The solid gray line (at 0.15) indicates the size of nonchemotactic colonies containing a vector control plasmid. The amino acid replacement in each mutant is indicated in single-letter code. Symbols at the bottom summarize mutant behavior in rescue (diamonds), jamming (squares), and dominance (circles) tests. Unfilled symbols indicate a lack of an effect; gray symbols indicate a partial effect; black symbols indicate a severe defect. On the basis of these and other function tests (see the text), the mutants were grouped into seven different phenotypic classes, as shown.
To determine whether any of the N381* receptors were capable of functional interaction with a heterologous chemoreceptor, we tested the mutant plasmids in strain UU1623, which expresses a wild-type aspartate receptor (Tar) but lacks other MCP genes (tsr, tap, and trg). One of the N381* proteins (N381G) regained the partial ability to promote serine chemotaxis in this host strain, an effect known as functional rescue (Fig. 2) (4). All other N381* proteins evinced no Tsr function in this test and, moreover, blocked Tar function, an effect known as functional jamming (Fig. 2) (5). Both rescue and jamming depend on the formation of mixed signaling complexes containing the mutant Tsr receptor and the heterologous Tar receptor, presumably through trimer-of-dimer associations (30). Rescuable receptors evidently regain proper conformation and dynamic behavior in association with normal signaling partners, whereas jamming receptors probably disrupt function by interfering with the proper assembly, geometry, or operation of mixed signaling teams.
To assess the structural severity of the lesions in N381* receptors, we tested their dominance properties in host strains expressing Tsr subunits that had a recessive lesion at the serine binding site (R69E or T156K). At equivalent expression levels of the N381* and binding-defective subunits, a majority of the Tsr molecules in the cells will be heterodimers that have one functional serine-binding site (46). R69E/N381* molecules will transmit serine-binding information across the membrane through the R69E subunit, whereas in T156K/N381* molecules, transmembrane signaling will occur through the N381* subunit (5, 48). Some N381 lesions proved to be recessive (A, C, E, G, H, P, S, and Y), while others were dominant (D, F, I, K, L, M, Q, R, T, V, and W) (Fig. 2). In all cases, each N381 defect behaved the same, either recessive or dominant, with both binding site lesions (data not shown).
On the basis of these initial function tests, the chemical properties of their amino acid replacements, and the additional tests described below, we assigned the N381* receptors to seven phenotypic classes (Fig. 2).
Trimer formation by N381* receptors.
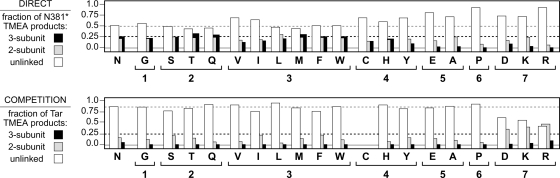
We expected that some, perhaps most, N381 lesions would disrupt trimer-of-dimer formation. However, except for N381G, all N381* receptors jammed wild-type Tar function, implying the formation of mixed trimer-based signaling teams. To better assess trimer formation by the N381* receptors, we examined their behaviors in three in vivo cross-linking tests: direct, competition, and mixed.
For the direct cross-linking test, an S366C reporter site was introduced into each of the N381* receptors. In trimers of dimers, residue 366 in the inner receptor subunits adopts a trigonal geometry at the trimer interface (22). Thus, S366C subunits can be efficiently cross-linked with the trifunctional thiol-reactive reagent TMEA, leading to roughly equivalent yields of two- and three-subunit cross-linking products (40). The outer receptor subunits in those trimers are not cross-linked by TMEA, yielding about 50% unlinked subunits (Fig. 3). For direct TMEA tests, mutant reporter plasmids were transferred to host strain UU1581, which does not express any chemotaxis-related gene products, to avoid CheB- and CheR-mediated modifications of receptor subunits and to preclude the assembly of CheA/CheW-containing ternary complexes. Under these conditions, wild-type receptor molecules reversibly associate as trimers of dimers (42). In the direct TMEA test, N381* receptors in groups 1 to 3 had essentially wild-type cross-linking patterns, whereas group 4 mutants had slightly reduced levels of both two- and three-subunit cross-linking products. Group 5 to 7 receptor mutants exhibited few (A, D, E, and K) or no (P and R) three-subunit cross-linking products; most (D, E, P, and R) also had reduced levels of two-subunit cross-linking products (Fig. 3). These results suggest that N381* receptors in group 4 (C, H, and Y) form unstable trimers, whereas receptors in groups 5 (E and A) and 6 (P) form very few complete trimers. Group 7 receptors (D, K, and R) also are defective in trimer formation, but to different extents. The least defective of this group is N381D, which presumably would be less prone to packing problems at the trimer interface than the N381K and N381R receptors, which have much larger mutant side chains.
Fig. 3.
Trimer-forming abilities of N381 mutant receptors. The upper panel shows the behavior of N381 mutant receptors in direct TMEA cross-linking tests. Each Tsr subunit bears the S366C TMEA cross-linking reporter. The histogram shows the relative levels of unlinked, two-subunit, and three-subunit TMEA products for each N381 mutant. For comparison purposes, the dashed gray line shows the proportion of unlinked subunits (white bars) for wild-type Tsr (0.54 ± 0.11; means and standard deviations from eight independent experiments), and the dashed black line (at 0.25) shows the approximate wild-type proportions of the two-subunit (gray bars) and three-subunit (black bars) TMEA products. Note that the scale begins below 0.0 (gray line) to emphasize mutants with near-zero values. The lower panel shows the behavior of N381 mutant receptors in TMEA competition assays. In these tests, the host cells express Tar-S364C, whereas the coexpressed Tsr molecules do not bear a TMEA reporter. An excess of trimer-competent Tsr molecules dilutes pure Tar trimers, reducing the TMEA cross-linking signals. For comparison purposes, the dashed gray line shows the fraction of unlinked Tar reporter subunits (white bars) in an excess of wild-type Tsr molecules (0.85 ± 0.08; means and standard deviations from two experiments), and the dashed black line (at 0.25) indicates the expected level of two-subunit (gray bars) and three-subunit (black bars) products in the absence of competition effects. Note that the scale begins below 0.0 (gray line) to emphasize mutants with near-zero values.
Trimer competition experiments assess the ability of a mutant Tsr receptor to form mixed trimers with Tar molecules that carry a TMEA reporter (S364C). In this test, the N381* receptors did not carry a reporter site, so TMEA could only cross-link the Tar-S364C subunits. Competition tests were performed by transferring N381* plasmids to strain UU1613, which expresses Tar-S364C from the chromosome but lacks other receptor genes as well as the cheA, cheW, cheR, and cheB genes. At stoichiometric excess, wild-type Tsr molecules essentially abolish three-subunit Tar cross-linking products and drastically reduce two-subunit products by eliminating Tar-only trimers (42). In these tests, N381* receptors in groups 1 to 3 competed as effectively as wild-type Tsr (Fig. 3). Mutant receptors in groups 4, 5, and 6 also competed effectively (Fig. 3). (Note that N381C was not tested, because its cysteine residue confounded the TMEA cross-linking patterns.) However, group 7 receptors (D, K, and R) were only partially effective as competitors. Those mutant molecules eliminated three-subunit Tar cross-linking products but caused elevated levels of two-subunit Tar products (Fig. 3). This competition behavior suggests that only one group 7 mutant molecule can be accommodated in a mixed trimer.
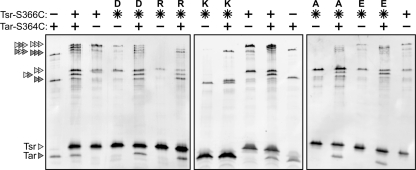
The direct TMEA cross-linking patterns indicate that N381* receptors in groups 5 to 7 have substantial defects in trimer formation. However, all of these mutant receptors exhibited partial or full competition ability, suggesting that the mutant molecules readily join or disrupt Tar trimers. We explored this predicted behavior with mixed cross-linking tests, in which both Tsr-N381* and wild-type Tar carried TMEA reporter sites (Fig. 4). At about 5-fold stoichiometric excess, N381* receptors in groups 5 to 7 could be trapped in mixed cross-linking products with Tar molecules. The examples in Fig. 4 show that N381A (group 5) and N381D (group 7) formed both two- and three-subunit mixed products with Tar-S364C. N381P molecules (group 6) exhibited a similar cross-linking profile (data not shown). N381E (group 5) and N381R receptors (group 7) yielded low levels of mixed three-subunit products and higher levels of mixed two-subunit products. The N381K receptor (group 7) appeared to form neither three- nor two-subunit products with the Tar reporter, but in this gel system Tsr-N381K subunits migrated anomalously faster than wild-type Tsr subunits, so we cannot exclude the possibility that mixed two-subunit N381K products comigrated with Tar-only bands (Fig. 4).
Fig. 4.
Examples of TMEA cross-linking results with N381 mutant receptors. SDS-PAGE (11% acrylamide, 7.5% bis-acrylamide) and Western analysis of TMEA cross-linking products (see Materials and Methods). Tsr-N381 mutant receptors bearing the S366C reporter were expressed either alone or in cells expressing Tar-S364C. Triangles at the left indicate the position and subunit composition of cross-linking products (Tsr subunits, light gray; Tar subunits, dark gray). Note that Tsr-N381K subunits migrate considerably faster than wild-type Tsr subunits and slightly faster than Tar subunits. During the preparation of cell extracts, some Tsr molecules are cleaved near the C terminus by a periplasmic protease (P. Ames, unpublished results). Those cleavage products give rise to background bands near the two- and three-subunit cross-linking products, e.g., the Tsr wild type in lane 3.
Ternary complex and cluster formation by N381 mutant receptors.
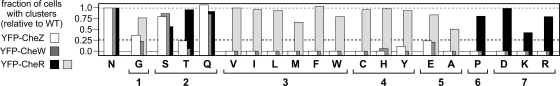
Trimer-of-dimer formation by chemoreceptors appears to underlie their ability to assemble ternary signaling complexes and clusters (4, 40). The trimer defects in group 5 to 7 N381* receptors, evident in cross-linking tests, predict that these mutant receptors should be defective in ternary complex and cluster formation. We tested these properties by fluorescence microscopy in strain UU1250, which contains CheA and CheW, using three different clustering reporters: YFP-CheW binds directly to the trimer contact region of receptor molecules (12), YFP-CheR binds to the NWETF pentapeptide sequence at the C terminus of each receptor subunit (34, 39, 45), and YFP-CheZ binds to CheAS, which is an alternate cheA translation product and a component of ternary signaling complexes (11, 36, 39). The results of these clustering assays are summarized in Fig. 5.
Fig. 5.
Cluster formation by N381 mutant receptors. Clustering assays were performed in UU1250 cells containing a Tsr-N381 mutant plasmid and a compatible plasmid expressing a YFP-tagged clustering reporter (CheZ, CheW, and CheR; see Materials and Methods). Under these conditions, wild-type Tsr forms polar spots with all three reporters (white, dark gray, and black bars, respectively). Most Tsr-N381 mutant receptors defective in ternary complex assembly and CheW binding (e.g., classes 3, 4, and 5) formed polar caps with the YFP-CheR reporter (light gray bars), but several (classes 6 and 7) formed tight polar spots with the YFP-CheR reporter (black bars). For comparison purposes, the dashed gray line (at 1.0) shows the normalized wild-type fraction of cells with one or more clusters; the dashed black line (at 0.25) indicates the cutoff criterion for defective clustering ability. Note that the scale begins below 0.0 (gray line) to emphasize mutants with near-zero values.
UU1250 cells containing wild-type Tsr molecules formed polar clusters detectable with each of the three fluorescently tagged reporter proteins. In contrast, most of the N381* receptors exhibited aberrant clustering behavior with one or more of the reporters (Fig. 5). The group 2 mutants (S, T, and Q) showed the most normal clustering patterns, although N381T clusters were less evident with the CheW and CheZ reporters, indicative of compromised ternary complex formation (Fig. 5). N381G also formed ternary complexes with low efficiency but formed diffuse clusters detectable with the CheR reporter at high efficiency, which is consistent with this mutant receptor's ability to form trimers of dimers (Fig. 3 and 5). Similarly, mutant receptors in groups 3, 4, and 5 formed diffuse clusters detectable with the CheR reporter (Fig. 5, light gray bars). These clusters (with the possible exception of N381E) were not detectable with the CheW and CheZ reporters, indicating that these mutant receptors have severe defects in CheW binding and ternary complex formation (Fig. 4). The group 6 (P) and group 7 (D, K, and R) receptors evinced no clusters with the CheW and CheZ reporters, but unlike mutant receptors in groups 3 to 5, they formed tight clusters that were detectable with the CheR reporter (Fig. 4, black bars). Given the significant trimer formation defects of group 6 and 7 receptors, these clusters most likely form by a trimer-independent mechanism.
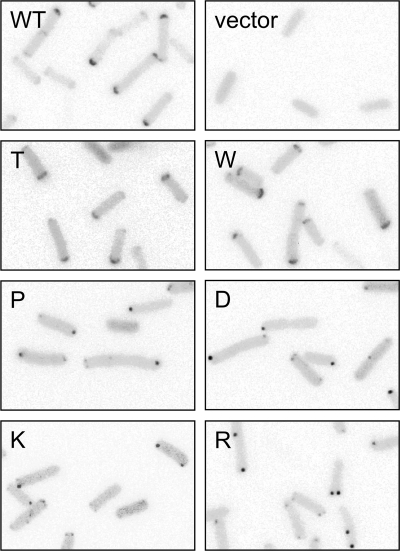
Anomalous clustering of some N381 mutant receptors.
In cells lacking CheA and CheW and thus precluding ternary complex formation, trimer-competent receptor molecules cluster as diffuse polar caps (20, 21, 28) (Fig. 6). In the presence of CheA and CheW, receptors that are able to form ternary signaling complexes cluster as compact polar spots, presumably owing to CheA/CheW-promoted binding interactions between receptor trimers (4). The tight clusters formed by group 6 and 7 N381* receptors were seen initially in cells containing CheA and CheW, which might have been responsible for their compact appearance. To test this possibility, we examined clustering by group 6 and 7 mutant receptors in UU1581 cells, which lack CheA and CheW. In this host, all four mutant receptors (N381P, N381D, N381K, and N381R) still formed compact polar spots detectable with a YFP-CheR reporter (Fig. 6). In contrast, a wild-type receptor formed more diffuse polar caps, as did the N381W receptor, which is defective in ternary complex formation (Fig. 6). Thus, the tight clusters formed by group 6 and 7 mutant receptors do not require CheA/CheW binding interactions or trimer-of-dimer formation.
Fig. 6.
Examples of anomalous clustering by N381 mutant receptors. Clustering assays were performed with the YFP-CheR reporter as in Fig. 5 except that UU1581 cells were used, which contain no CheA, CheW, or any other chemotaxis proteins. Under these conditions, wild-type Tsr molecules cannot assemble ternary signaling complexes but still associate as trimers of dimers, leading to polar caps. Each panel shows an inverted, gray-scale portion of the original fluorescence image.
The N381T receptor, which appeared to be somewhat defective in ternary complex assembly in clustering tests with the CheW and CheZ reporters, nevertheless efficiently formed compact polar spots detectable with the CheR clustering reporter in cells containing CheA and CheW (Fig. 5). In cells lacking CheA and CheW, the N381T receptor formed polar caps, demonstrating that its ability to form compact clusters must depend on binding interactions with CheA/CheW. Unlike the group 6 and 7 mutant receptors, which formed tight clusters in the absence of CheA and CheW, the N381T receptor exhibited a normal trimer-forming ability in the direct and competition cross-linking tests (Fig. 3).
Defective ternary complex formation by some N381 mutant receptors.
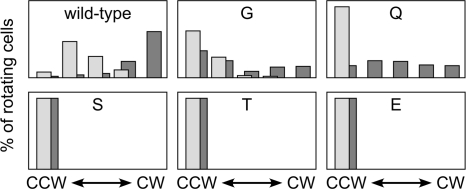
Most N381* receptors appeared to be defective in ternary complex assembly. They formed receptor clusters that were detectable with the CheR reporter but not with the CheW or CheZ reporter. In contrast, the N381E receptor (group 5) and the mutant receptors of group 1 (G) and group 2 (S, T, and Q) bound the CheW and CheZ clustering reporters to various extents, implying some assembly of ternary signaling complexes (Fig. 5). Because the latter mutant receptors cannot mediate chemotaxis to serine (Fig. 2), they most likely form defective signaling complexes. To test this proposition, we examined their ability to activate the CheA kinase in flagellar rotation assays. N381* plasmids were transferred to two receptorless host strains, one lacking the CheR and CheB enzymes (UU1535) and the other competent for sensory adaptation (UU1250). In CheR− CheB− cells, wild-type Tsr receptors assemble ternary complexes with high CheA activity, resulting in nearly incessant clockwise (CW) rotation of their flagellar motors (Fig. 7). In CheR+ CheB+ cells, the signaling complexes of wild-type Tsr receptors produce less CW output, reflecting the low-CheA-activity set point of the sensory adaptation system. The N381S, N381T, and N381E receptors produced no CW output in either host; the N381G and N381Q receptors produced low levels of CW output in the adaptation-deficient host that were further attenuated in the adaptation-proficient host (Fig. 7). Thus, all five N381* receptors that assembled ternary complexes exhibited substantially impaired CW signal output, which is indicative of defects in CheA activation.
Fig. 7.
Flagellar rotation patterns elicited by N381 mutant receptors. Each panel shows the rotation profile of UU1250 cells (CheR+ CheB+; light gray bars) and UU1535 cells (CheR− CheB−; dark gray bars) expressing the indicated Tsr N381 amino acid replacement (G, Q, S, T, or E). At least 100 antibody-tethered cells were observed for 15 s each and assigned to one of five rotation categories, from exclusively CCW (left extreme) to exclusively CW (right extreme).
DISCUSSION
Receptor dynamics and signaling.
Conformational changes at the cytoplasmic hairpin tip regulate the signaling activity of MCP molecules (Fig. 1). The nature of those signaling changes is not altogether clear, but recent studies suggest that the dynamic behavior of the tip controls receptor signal output. Stimulus inputs modulate the HAMP domain, whose packing stability may be oppositionally coupled to that of the four-helix methylation bundle (48). The methylation helix bundle, in turn, appears to have an opposed stability relationship with the signaling tip (43). Accordingly, attractant stimuli may inhibit kinase activity by stabilizing the receptor tip, whereas repellents may elevate kinase activity by destabilizing the tip.
Receptor dynamic behavior also influences the assembly of ternary signaling complexes (23, 47). Although the receptor dimer is the unit of stimulus detection and transmembrane signaling, MCP molecules must form trimers of dimers to generate and regulate output signals (24, 25). Receptor molecules at either extreme of the dynamic range appear to be poor trimer formers (23, 47). Once formed, receptor trimers bind the CheW and CheA proteins to assemble ternary signaling complexes, which in turn organize into large hexagonal arrays at the cell poles. It appears that two receptor trimers and two CheW molecules are necessary and sufficient to control one CheA dimer (24). Presumably, each trimer in the minimal signaling unit binds one CheW molecule. Binding could occur at surface determinants that exist in each dimer or at sites that are unique to the trimer, for example at the cleft between adjacent dimers (Fig. 1B). In either scenario, the dynamic behavior of the trimer should influence CheW binding. Both very stable and very unstable trimers could spend little time in the proper high-affinity binding conformation, thereby retarding ternary complex assembly and promoting nonproductive binding interactions between, for example, CheW and receptor dimers (12).
Most of the N381 mutant receptors described in this report were defective in binding CheW and in forming ternary signaling complexes. However, the evidence and reasoning discussed below suggest that N381 may not be an important direct binding determinant for CheW, rather N381 lesions affect trimer stability and geometry in ways that impair ternary complex assembly.
Trimer-deficient N381 mutants.
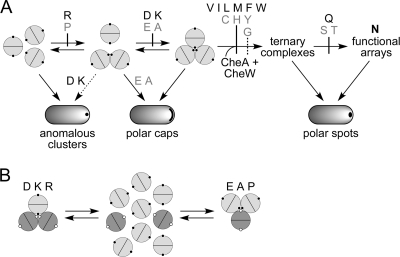
Some amino acid replacements at Tsr residue N381 interfered with trimer formation. As mutant homodimers, the N381P and N381R receptors could not initiate trimer formation, and N381A/D/E/K receptors formed partial trimer assemblies (dimers of dimers) (Fig. 8A). All of these mutant receptors were able to form mixed trimers with wild-type Tar molecules, suggesting that interaction with a normal partner could ameliorate, to some extent, their trimer defect. Mixed trimers accommodated up to two A/E/P dimers but no more than one D/K/R dimer. We suggest that the D, K, and R lesions impede trimer-forming interactions through charge repulsion effects and side chain steric clashes (K and R) at the trimer axis. The structural perturbations of the A, E, and P lesions, whose defects were recessive in mutant/wild-type heterodimers, evidently are less drastic. N381P most likely destabilizes the main trimer contact helix. In heterodimers, intersubunit helix packing interactions could alleviate that structural instability; in mixed trimers, interdimer packing interactions could enhance N381P stability in an analogous fashion (Fig. 8B).
Fig. 8.
Trimer formation defects of N381 mutant receptors. (A) Steps in trimer assembly, ternary complex, and cluster formation affected by different N381 lesions. Tsr dimers are represented as conjoined semicircles; small black circles indicate the position of residue 381 in each subunit. Early steps in trimer assembly are reversible but become irreversible upon binding CheW/CheA to form ternary signaling complexes. The N381G lesion has a less severe effect on complex assembly than other N381 receptors blocked at the same step. Although the D, E, A, and K replacements seem to affect the same step (completion of trimer assembly), the D and K lesions lead to anomalous clustering, whereas the E and A lesions do not. Amino acid replacements that cause recessive defects are listed in gray, and dominant lesions are in black. (B) Behavior of N381 mutant receptors in trimer competition assays. Tsr dimers are shown as light gray circles with small black circles representing residue 381 in each subunit; Tar dimers are shown as dark gray circles with small white circles representing the 381-equivalent positions. In cells expressing both Tar and Tsr, the D, K, and R mutant receptors can drive Tar molecules only to dimers of dimers. In contrast, the E, A, and P mutant receptors fully dissipate Tar trimers of dimers.
The severe trimer formation defects of the N381D/K/P/R receptors revealed a pathway for anomalous cluster formation that probably accounts for some of their functional defects (Fig. 8A). Similar behavior was previously reported for another Tsr trimer contact lesion, R388P (28). Like normal receptor clusters, anomalous clusters formed mainly at the cell poles. They probably are not inclusion bodies, because (i) the mutant receptor molecules were expressed at physiological levels, (ii) their C-terminal binding site for the YFP-CheR clustering reporter was accessible, and (iii) the mutant receptors both jam Tar signaling and form mixed cross-linking products with Tar molecules, demonstrating that they are not sequestered from normal receptor molecules in the same cell. Perhaps anomalous clusters form through nonspecific association of the hydrophobic tips of receptor molecules (Fig. 9). Such binding interactions could lead to polar clusters by the same stochastic diffusion-interaction mechanism that forms trimer-based polar clusters (15). In trimer-proficient receptors, trimer interactions probably block or outcompete anomalous clustering interactions; such aberrant interactions would be unopposed in profoundly trimer-deficient receptors. Anomalous clustering conceivably could account for the dominance of the N381D/K/R defects.
Fig. 9.
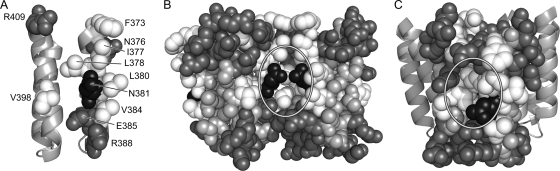
Two structural environments of N381 in receptor trimers of dimers. (A) Face-on view of trimer contact residues in the Tsr tip. Most of the trimer-stabilizing contacts (F373-R388) are located in the N-terminal helix of each subunit; only one set of contact residues in the dimer is shown: hydrophobic residues (white), polar residues (dark gray), and N381 (black). (B) Packing of trimer contact residues at the trimer interface. Two dimers of the trimer are shown; the third, which would be in front of these two, has been removed to show the cavity surrounding N381 at the trimer axis. All tip residues are space filled: hydrophobic (V, I, L, M, F, Y, and W, white), polar (D, E, N, Q, H, K, and R, dark gray, N381 (black), and small and/or nonpolar (G, A, S, and T, light gray). There are no C or P residues in the Tsr tip. (C) Trimer contact residues at the trimer periphery. An outside view of the trimer of dimers is shown. The dimer in front is space filled, using the same shading conventions as those for panel B. The two dimers behind are shown as helical backbones. The region surrounding N381 (circled) may not be a critical docking site for ternary complex assembly (see the text).
Trimer-proficient N381 mutants.
Many N381 mutants made trimers and mixed trimers with ostensibly wild-type proficiency (Fig. 8A). However, only a few of them (N381Q/S/T) efficiently assembled ternary signaling complexes; most of the mutant trimers were defective in recruiting CheW (and CheA). On one hand, the binding-defective N381 lesions (V/I/L/M/F/W, C/H/Y, and G) might directly alter a critical CheW binding determinant at the trimer periphery. On the other hand, they might affect trimer stability or geometry and thereby influence CheW binding. We favor the latter explanation, mainly because the binding-defective receptors seem to fall into two very different trimer stability classes. The N381G and the N381C/H/Y receptors may make trimers that are less stable than wild-type ones. Their functional defects are recessive in heterodimers, implying that the wild-type subunits can promote stable trimer formation by preferentially orienting at the trimer axis. In contrast, N381* receptors with a hydrophobic replacement (V/I/L/M/F/W) may make trimers that are more stable than wild-type ones. Their functional defects are dominant in heterodimers, implying that wild-type subunits cannot exclude mutant ones from the trimer axis.
Structural features of the trimer interface support these disparate stability assessments. The N381 residues at the trimer axis reside in a water-filled cavity, about 200 Å3 in volume, that is lined with hydrophobic trimer contacts (Fig. 9B). The N381 side chains form a hydrogen-bonded network at the center of the cavity that contributes to trimer stability. Hydrophobic amino acids at the N381 position likely would contribute to trimer packing forces as well, whereas glycine would not. The N381 cavity has ample space to accommodate a variety of side chain volumes, but bulky residues with polar character (H and Y) could distort the packing geometry without contributing significantly to packing stability. In contrast, large aromatics (F and W) might still contribute hydrophobic interactions that stabilize the trimer interface despite altering its packing geometry.
The CheW binding defects of mutant receptors with presumptive trimer stability changes are less easily explained by the N381 structural environment around the outside of the trimer. At the trimer periphery, N381 lies in a shallow depression on the face of each dimer, surrounded by mainly hydrophobic residues (Fig. 9C). If the hydrophobic hollow adjacent to N381 were part of a CheW docking site, larger side chain replacements at N381 might be expected to occlude the site and interfere with CheW binding. In contrast, small side chain replacements might have little or no effect on CheW binding. This appears not to be the case, because some small replacements (e.g., G, A, and V) impaired CheW binding, whereas a larger side chain replacement (Q) did not. If, instead, N381 itself were a critical binding determinant, then most amino acid replacements might be expected to impair binding, either through steric hindrance or through the elimination of side chain binding contacts. The N381* behaviors are more consistent with this scenario, although side chain character appears to be more critical than size. Thus, some mutant receptors with polar amino acid replacements (S, T, and Q) retained CheW binding, whereas nonpolar (G, A, and V) or charged (D and E) replacements of about the same size prevented CheW binding.
In conclusion, residue N381 of Tsr is critical for signaling; no other amino acid at this position can support function. All N381 mutant receptors except N381G disrupted the function of a heterologous receptor, Tar. There may be two different jamming mechanisms: some N381* receptors probably destabilize mixed trimers, while others probably overstabilize or distort the trimer interfaces, locking in a nonproductive conformation. Although most N381 lesions abrogated ternary complex formation, N381 may not be a direct binding determinant for CheW. Rather, the CheW binding defects of N381* receptors could stem from trimer assemblies that have incorrect geometry or dynamic behavior. The CheW docking surface on receptor molecules remains to be identified.
ACKNOWLEDGMENTS
We thank Martin Horvath (University of Utah) for calculating the volume of the N381 cavity in the Tsr trimer and Kika Kitanovic (University of Utah) for comments on the manuscript. This work was supported by research grant GM19559 from the National Institute of General Medical Sciences. Y.Z. received support from the Bioscience Undergraduate Research Program, Department of Biology, University of Utah.
DNA sequencing and primer synthesis were carried out by the Protein-DNA Core Facility at the University of Utah, which receives support from National Cancer Institute grant CA42014 to the Huntsman Cancer Institute.
Footnotes
Published ahead of print on 30 September 2011.
REFERENCES
- 1. Alexander R. P., Zhulin I. B. 2007. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc. Natl. Acad. Sci. U. S. A. 104:2885–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ames P., Parkinson J. S. 2006. Conformational suppression of inter-receptor signaling defects. Proc. Natl. Acad. Sci. U. S. A. 103:9292–9297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ames P., Parkinson J. S. 1994. Constitutively signaling fragments of Tsr, the Escherichia coli serine chemoreceptor. J. Bacteriol. 176:6340–6348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ames P., Studdert C. A., Reiser R. H., Parkinson J. S. 2002. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:7060–7065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ames P., Zhou Q., Parkinson J. S. 2008. Mutational analysis of the connector segment in the HAMP domain of Tsr, the Escherichia coli serine chemoreceptor. J. Bacteriol. 190:6676–6685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bibikov S. I., Miller A. C., Gosink K. K., Parkinson J. S. 2004. Methylation-independent aerotaxis mediated by the Escherichia coli Aer protein. J. Bacteriol. 186:3730–3737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boldog T., Grimme S., Li M., Sligar S. G., Hazelbauer G. L. 2006. Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties. Proc. Natl. Acad. Sci. U. S. A. 103:11509–11514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bolivar F., et al. 1977. Construction and characterization of new cloning vehicles. Gene 2:95–113 [PubMed] [Google Scholar]
- 9. Borkovich K. A., Kaplan N., Hess J. F., Simon M. I. 1989. Transmembrane signal transduction in bacterial chemotaxis involves ligand-dependent activation of phosphate group transfer. Proc. Natl. Acad. Sci. U. S. A. 86:1208–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bourret R. B., Hess J. F., Simon M. I. 1990. Conserved aspartate residues and phosphorylation in signal transduction by the chemotaxis protein CheY. Proc. Natl. Acad. Sci. U. S. A. 87:41–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cantwell B. J., et al. 2003. CheZ phosphatase localizes to chemoreceptor patches via CheA-short. J. Bacteriol. 185:2354–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cardozo M. J., Massazza D. A., Parkinson J. S., Studdert C. A. 2010. Disruption of chemoreceptor signalling arrays by high levels of CheW, the receptor-kinase coupling protein. Mol. Microbiol. 75:1171–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chang A. C. Y., Cohen S. N. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the p15A cryptic miniplasmid. J. Bacteriol. 134:1141–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gegner J. A., Graham D. R., Roth A. F., Dahlquist F. W. 1992. Assembly of an MCP receptor, CheW, and kinase CheA complex in the bacterial chemotaxis signal transduction pathway. Cell 70:975–982 [DOI] [PubMed] [Google Scholar]
- 15. Greenfield D., et al. 2009. Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy. PLoS Biol. 7:e1000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hazelbauer G. L., Falke J. J., Parkinson J. S. 2008. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem. Sci. 33:9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hazelbauer G. L., Lai W. C. 2010. Bacterial chemoreceptors: providing enhanced features to two-component signaling. Curr. Opin. Microbiol. 13:124–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hegde M., et al. 2011. Chemotaxis to the quorum-sensing signal AI-2 requires the Tsr chemoreceptor and the periplasmic LsrB AI-2-binding protein. J. Bacteriol. 193:768–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hess J. F., Bourret R. B., Simon M. I. 1988. Histidine phosphorylation and phosphoryl group transfer in bacterial chemotaxis. Nature 336:139–143 [DOI] [PubMed] [Google Scholar]
- 20. Kentner D., Sourjik V. 2006. Spatial organization of the bacterial chemotaxis system. Curr. Opin. Microbiol. 9:619–624 [DOI] [PubMed] [Google Scholar]
- 21. Kentner D., Thiem S., Hildenbeutel M., Sourjik V. 2006. Determinants of chemoreceptor cluster formation in Escherichia coli. Mol. Microbiol. 61:407–417 [DOI] [PubMed] [Google Scholar]
- 22. Kim K. K., Yokota H., Kim S. H. 1999. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature 400:787–792 [DOI] [PubMed] [Google Scholar]
- 23. Kitanovic S., Ames P., Parkinson J. S. 2011. Mutational analysis of the control cable that mediates transmembrane signaling in the E. coli serine chemoreceptor. J. Bacteriol. 193:5062–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li M., Hazelbauer G. L. 2011. Core unit of chemotaxis signaling complexes. Proc. Natl. Acad. Sci. U. S. A. 108:9390–9395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li M., Khursigara C. M., Subramaniam S., Hazelbauer G. L. 2011. Chemotaxis kinase CheA is activated by three neighbouring chemoreceptor dimers as effectively as by receptor clusters. Mol. Microbiol. 79:677–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu J. D., Parkinson J. S. 1989. Role of CheW protein in coupling membrane receptors to the intracellular signaling system of bacterial chemotaxis. Proc. Natl. Acad. Sci. U. S. A. 86:8703–8707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maddock J. R., Shapiro L. 1993. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science 259:1717–1723 [DOI] [PubMed] [Google Scholar]
- 28. Mowery P., Ostler J. B., Parkinson J. S. 2008. Different signaling roles of two conserved residues in the cytoplasmic hairpin tip of Tsr, the Escherichia coli serine chemoreceptor. J. Bacteriol. 190:8065–8074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Parkinson J. S. 1976. cheA, cheB, and cheC genes of Escherichia coli and their role in chemotaxis. J. Bacteriol. 126:758–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Parkinson J. S., Ames P., Studdert C. A. 2005. Collaborative signaling by bacterial chemoreceptors. Curr. Opin. Microbiol. 8:116–121 [DOI] [PubMed] [Google Scholar]
- 31. Parkinson J. S., Houts S. E. 1982. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 151:106–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sanders D. A., Gillece-Castro B. L., Stock A. M., Burlingame A. L., Koshland D. E., Jr 1989. Identification of the site of phosphorylation of the chemotaxis response regulator protein, CheY. J. Biol. Chem. 264:21770–21778 [PubMed] [Google Scholar]
- 33. Segall J. E., Ishihara A., Berg H. C. 1985. Chemotactic signaling in filamentous cells of Escherichia coli. J. Bacteriol. 161:51–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shiomi D., Zhulin I. B., Homma M., Kawagishi I. 2002. Dual recognition of the bacterial chemoreceptor by chemotaxis-specific domains of the CheR methyltransferase. J. Biol. Chem. 277:42325–42333 [DOI] [PubMed] [Google Scholar]
- 35. Slocum M. K., Parkinson J. S. 1985. Genetics of methyl-accepting chemotaxis proteins in Escherichia coli: null phenotypes of the tar and tap genes. J. Bacteriol. 163:586–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Smith R. A., Parkinson J. S. 1980. Overlapping genes at the cheA locus of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 77:5370–5374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sourjik V. 2004. Receptor clustering and signal processing in E. coli chemotaxis. Trends Microbiol. 12:569–576 [DOI] [PubMed] [Google Scholar]
- 38. Sourjik V., Berg H. C. 2004. Functional interactions between receptors in bacterial chemotaxis. Nature 428:437–441 [DOI] [PubMed] [Google Scholar]
- 39. Sourjik V., Berg H. C. 2000. Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol. Microbiol. 37:740–751 [DOI] [PubMed] [Google Scholar]
- 40. Studdert C. A., Parkinson J. S. 2004. Crosslinking snapshots of bacterial chemoreceptor squads. Proc. Natl. Acad. Sci. U. S. A. 101:2117–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Studdert C. A., Parkinson J. S. 2007. In vivo crosslinking methods for analyzing the assembly and architecture of chemoreceptor arrays. Methods Enzymol. 423:414–431 [DOI] [PubMed] [Google Scholar]
- 42. Studdert C. A., Parkinson J. S. 2005. Insights into the organization and dynamics of bacterial chemoreceptor clusters through in vivo crosslinking studies. Proc. Natl. Acad. Sci. U. S. A. 102:15623–15628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Swain K. E., Gonzalez M. A., Falke J. J. 2009. Engineered socket study of signaling through a four-helix bundle: evidence for a yin-yang mechanism in the kinase control module of the aspartate receptor. Biochemistry 48:9266–9277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wolfe A. J., Conley M. P., Kramer T. J., Berg H. C. 1987. Reconstitution of signaling in bacterial chemotaxis. J. Bacteriol. 169:1878–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wu J., Li J., Li G., Long D. G., Weis R. M. 1996. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation. Biochemistry 35:4984–4993 [DOI] [PubMed] [Google Scholar]
- 46. Yang Y., Park H., Inouye M. 1993. Ligand binding induces an asymmetrical transmembrane signal through a receptor dimer. J. Mol. Biol. 232:493–498 [DOI] [PubMed] [Google Scholar]
- 47. Zhou Q., Ames P., Parkinson J. S. 2011. Biphasic control logic of HAMP domain signalling in the Escherichia coli serine chemoreceptor. Mol. Microbiol. 80:596–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhou Q., Ames P., Parkinson J. S. 2009. Mutational analyses of HAMP helices suggest a dynamic bundle model of input-output signalling in chemoreceptors. Mol. Microbiol. 73:801–814 [DOI] [PMC free article] [PubMed] [Google Scholar]