Abstract
Clostridium difficile is an important human pathogen and one where the primary cause of disease is due to the transmission of spores. We have investigated the proteins found in the outer coat layers of C. difficile spores of pathogenic strain 630 (CD630). Five coat proteins, CotA, CotB, CotCB, CotD, and CotE, were shown to be expressed on the outer coat layers of the spore. We demonstrate that purified spores carry catalase, peroxiredoxin, and chitinase activity and that this activity correlates with the predicted functions of three spore coat proteins identified here, CotCB, CotD, and CotE. CotCB and CotD are putative manganese catalases, and CotE is a novel bifunctional protein with peroxiredoxin activity at its amino terminus and chitinase activity at its carboxy terminus. These enzymes could play an important role in coat assembly by polymerizing protein monomers in the coat. CotE, in addition to a role in macromolecular degradation, could play an important role in inflammation, and this may be of direct relevance to the development of the gastrointestinal symptoms that accompany C. difficile infection. Although specific enzyme activity has not yet been assigned to the proteins identified here, this work provides the first detailed study of the C. difficile spore coat.
INTRODUCTION
Clostridium difficile is the most common cause of nosocomial antibiotic-associated diarrhea in developed countries (32). Morbidity and mortality rates have been steadily increasing in recent years and probably result from the emergence of more virulent strains of C. difficile as well as the changing patterns of antibiotic usage. Recent estimates of the incidence of C. difficile-associated diarrhea (CDAD) in the United States suggest that there are as many as 500,000 cases per year, with up to 20,000 mortalities (29). C. difficile colonizes the intestinal tracts of infected patients, and antibiotic treatment can promote the overgrowth of this bacterium, which in turn leads to clinical symptoms of disease, from diarrhea to, in more severe cases, pseudomembranous colitis (32).
CDAD is caused by the secretion of two toxins, toxin A (TcdA) and toxin B (TcdB), both monoglucosyltransferases that are cytotoxic, enterotoxic, and proinflammatory (3). CDAD is particularly problematic to treat and to contain because of the ability of this bacterium to form robust endospores (spores) that can persist and be easily transferred from person to person in a hospital environment (9, 37). Currently, the only treatment for CDAD is the use of antibiotics such as vancomycin and metronidazole, and a relapse of CDAD (i.e., diarrhea recurring within 30 days after the first treatment) is a particular challenge in a hospital environment (9, 10). Moreover, evidence has now arisen showing that antibiotic treatment suppresses the diversity of resident intestinal microflora and promotes the growth and proliferation of highly infectious C. difficile spores (18). This “supershedder” state ends once antibiotic treatment is terminated, providing an important clue to both the transmission of C. difficile infection in humans in a hospital environment and the importance of the spore as the pathogenic agent.
With the advent of genomics and proteomics and by comparison with the extensive data available for unicellular differentiation in Bacillus subtilis, some invaluable information is now emerging on C. difficile sporulation. C. difficile strain QCD-32g58 has been found to contain 18 orthologues of B. subtilis spore coat proteins and 3 orthologues of proteins found in the exosporium of spores of Bacillus anthracis and Bacillus cereus (13). Bioinformatic analyses of the genome of C. difficile strain 630 (CD630) (30) coupled with recent studies of the spore proteome (19) have revealed only 18 orthologues of B. subtilis spore coat proteins (30). B. subtilis coats are comprised of about 70 different proteins, so it is probable that C. difficile and, indeed, other clostridial spore formers will have equivalent complexities, which in turn suggests that C. difficile spore coat proteins have diversified considerably.
In this work, we have made the first attempt at characterizing the spore coats of CD630. Using coat protein extractions, we have identified five coat (Cot) proteins, three of which, based on bioinformatics analysis, could have enzymatic activity (two catalases and one bifunctional peroxiredoxin-chitinase). We also show that these enzymes may be confined to the outermost coat layer, where they could play a key role in spore coat polymerization and maturation.
MATERIALS AND METHODS
General methods and strains.
Methods for the preparation of Bacillus spores were described previously (23). C. difficile 630 is a pathogenic strain that produces tcdA+ tcdB+ and was obtained from Neil Fairweather (Imperial College, United Kingdom). CD630 was routinely grown in vegetative culture by growth (10 ml) overnight at 37°C in TGY-vegetative medium (3% tryptic soy broth, 2% glucose, 1% yeast extract, 0.1% l-cysteine) (25). Streptococcus mutans GB1 was obtained from Phan Nghia (Hanoi University, Vietnam); B. subtilis strain PY79 is a prototrophic (Spo+) laboratory strain and a laboratory stock, as was Bacillus clausii O/C. Bacillus licheniformis strain HU14 was obtained from the Bacillus Genetic Stock Center (Columbus, OH).
Sporulation of C. difficile.
All manipulations were made in an anaerobic incubator (Don Whitley, United Kingdom). A single bacterial colony was grown on BHIS agar (brain heart infusion agar supplemented with 0.1% l-cysteine and 5 mg/ml yeast extract [31]) overnight at 37°C. One fresh single colony from the BHIS plate was inoculated into 10 ml of TGY medium (3% tryptic soy broth, 2% glucose, 1% yeast extract, 0.1% l-cysteine) (25) and incubated at 37°C overnight. One milliliter of TGY culture was then subcultured into SMC broth [90 g peptone, 5 g proteose peptone, 1 g (NH4)2SO4, 1.5 g Tris] containing 0.1% l-cysteine (modified from methods reported previously by Wilson et al. [38]), incubated until an optical density at 600 nm (OD600) of 0.4 to 0.7 was reached, and then plated onto SMC agar. After 7 days of incubation at 37°C, the sporulation efficiency was confirmed by phase-contrast microscopy and measurements of heat-resistant CFU, and spore crops were harvested immediately or after overnight incubation at 4°C.
Spore purification.
The methods used for spore purification were modified from those described previously by Lawley et al. (19). Spores were washed in water two times and then suspended in phosphate-buffered saline (PBS) containing 125 mM Tris, 200 mM EDTA, 0.3 mg/ml proteinase K (catalog number E00492; Fermentas), and 1% sarcosyl and incubated with gentle shaking at 37°C for 2 h. Spores were centrifuged (8,000 rpm for 10 min), and pellets were resuspended in water and washed a further 10 times. After the final suspension in water, spores were heat treated (60°C for 20 min) to kill any residual cells; aliquots were stored at 4°C until use. To calculate the spore CFU, aliquots were serially diluted in PBS and plated onto BHIS agar supplemented with 0.1% sodium taurocholate (Sigma, United Kingdom). Plates were incubated for 24 to 48 h before CFU were enumerated.
Spore coat extractions.
The spore coat extraction procedure was described previously (36), but in brief, 2 × 109 spores were suspended in 100 μl of freshly prepared sodium borate-sodium dodecyl sulfate (SDS)-dithiothreitol (DTT) buffer consisting of sodium borate (0.1 M; pH 10), 0.5% SDS, and 50 mM DTT, and then incubated at 68°C for 75 min with gentle agitation. After centrifugation (8,000 rpm for 15 min), the supernatant was removed, mixed with 4× SDS-PAGE loading buffer, and fractionated by SDS-PAGE. For B. subtilis and B. clausii spores, coat proteins were extracted by using an SDS-DTT buffer described previously (23).
Peptide fingerprinting.
Spore coat proteins were fractionated on 12.5% SDS-PAGE minigels, and bands were excised and digested with trypsin before analysis by matrix-assisted laser desorption ionization (MALDI) mass spectrometry. Digestions and analysis were conducted by the University of Cambridge Protein and Nucleic Acid Chemistry Facility (PNAC) (http://www.bioc.cam.ac.uk/pnac).
Antibody production.
pET28b expression vectors that express the complete cotA, cotB, cotCB, and cotD open reading frames (ORFs) were constructed by amplifying the respective DNAs by PCR from C. difficile 630 chromosomal DNA and ligating them into cleaved pET28b. For cotE, we were unable to clone the entire ORF, so a fragment encoding the N-terminal peroxiredoxin domain was cloned instead. Primers used for the construction of pET28b clones are shown in Table S1 in the supplemental material. High levels of expression were obtained upon isopropyl-β-d-thiogalactopyranoside (IPTG) induction and the purification of proteins by the passage of the cell lysate through a HiTrap chelating HP column on a Pharmacia Akta liquid chromatography system. Polyclonal antibodies were raised in mice immunized by the intraperitoneal route with 2 μg of purified recombinant proteins on days 1, 14, and 28. Antispore antibodies were made by treating spores with 2% formalin (2% [vol/vol] formaldehyde in PBS) overnight at 4°C. Spores were washed 5 times with PBS and were used to dose mice (2 × 108 spores/dose) on days 1 and 14.
Confocal microscopy.
Spores were labeled with mouse anti-Cot serum (1:1,000 dilution), followed by an anti-mouse IgG-tetramethyl rhodamine isocyanate (TRITC) conjugate. Images were taken by using a Nikon Eclipse fluorescence microscope equipped with a Bio-Rad Radiance 2100 laser scanning system.
TEM.
A transmission electron microscopy (TEM) methodology using suspensions of purified CD630 spores (7 days old) was described previously for B. subtilis spores (14).
Spore germination.
Spores (1 × 108) were suspended in 100 μl of 0%, 3%, and 5% sodium taurocholate in PBS (pH 7.4). Spore suspensions were routinely agitated, and absorbance (A580) readings were determined until no further change in absorbance [OD580 (tn) − OD580 (t0)] could be detected.
Catalase assay.
A catalase assay was performed as described previously (1). Spores or vegetative cells (1 × 107 CFU) were pelleted and resuspended in 60 μl of 50 mM potassium phosphate buffer (pH 7.0). H2O2 (1.94 ml) was added to the mixture to start the reaction at room temperature (RT). Samples were centrifuged, and the OD240 of supernatants was measured immediately.
Peroxiredoxin assay.
The peroxiredoxin assay was described previously (11, 20). Spores or vegetative cells (1 × 108) were pelleted by centrifugation and suspended in a reaction buffer that included H2O2. Reactions were made at 37°C, and after 15 min, cells were pelleted, and the OD340 of supernatants was measured.
Chitinase assay.
The chitinase activity was determined by using a presupplied kit (catalog number CS0980; Sigma) using spore or cell suspensions (1 × 108 CFU/assay) in water. 4-Nitrophenyl-N-acetyl-β-d-glucosaminide (1 mg/ml) was used as a substrate, and after the reaction was stopped (200 μl of 0.04 g/ml sodium carbonate), the suspension was centrifuged, and the OD405 of supernatants was measured. The reaction time was 3 h at 37°C. The assay was also performed on spores that had been pregerminated or following sonication. For germination, 1 × 108 spores were suspended in 100 μl of 0%, 3%, and 5% sodium taurocholate in PBS (pH 7.4) for 30 min at 37°C, after which the chitinase activity was determined. For sonication, 1 × 108 spores were suspended in 100 μl PBS (pH 7.4). The solution was sonicated for 2 or 7 times (3-mm microtip probe and 10% amplitude for 30 s) (S-450D sonicator; Branson), after which the assay was performed.
RESULTS
C. difficile spore formation.
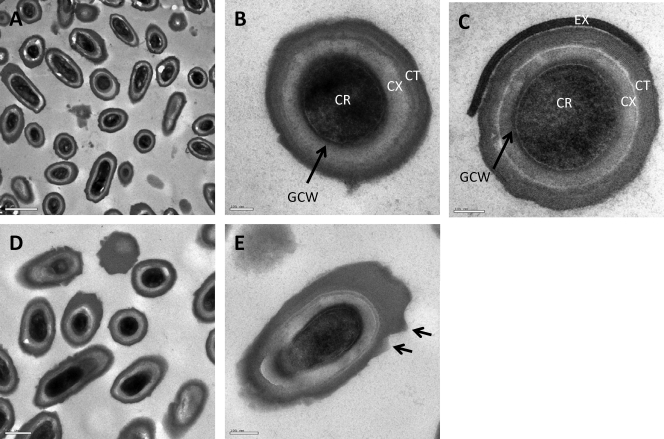
Using an empirical approach, we adapted existing methods (38) to generate high levels of spore formation on a solid medium using C. difficile strain 630 (tcdA+ tcdB+), which we refer to here as CD630. After 7 days of growth on agar, we routinely obtained >75% sporulation (see Fig. S1 in the supplemental material), with crops consisting of mature, released spores, which were then purified, further providing suspensions devoid of viable vegetative cells. TEM analysis of spores (Fig. 1A and B) revealed a structure common to those produced by the majority of Gram-positive spore formers (13), namely, an inner core surrounded by a primordial germ cell wall (peptidoglycan derived from vegetative cell walls) and a thick cortical layer (loosely cross-linked peptidoglycan specific to the spore) (Fig. 1B). Finally, above the cortex, a thick, more electron-dense layer was present on all spores, this being the spore coat. A closer examination revealed further definition to this layer, including laminations resembling the striated outer coats of Bacillus spores (13). In other work, spores of C. difficile were reported to carry an exosporium (19, 24), a loose-fitting saclike structure enveloping the mature spore (13). In our studies, we have observed an exosporial layer resembling that described by previous studies, and an example is shown in Fig. 1C. However, we found observations of this structure to be inconsistent. The exosporium was apparent only in samples harvested and processed immediately (Fig. 1C), and for those spores where it was detected, the layer was only partially attached. In contrast, the images shown in Fig. 1A and B were from CD630 spore preps that had been left overnight at 4°C before spore purification.
Fig. 1.
Ultrastructure of C. difficile 630 spores. (A to C) Representative images of CD630 spores after 7 days of incubation on solid medium. Panel B shows the basic structural features found in a mature endospore. CR, core; GCW, germ cell wall; CX, cortex; CT, coats. Panel C shows a spore containing a partially attached exosporium (EX). (D and E) Seven-day-old spores subjected to 10 cycles of sonication. Panel E shows angular projections found to be more abundant in sonicated samples.
Identification of C. difficile spore coat proteins.
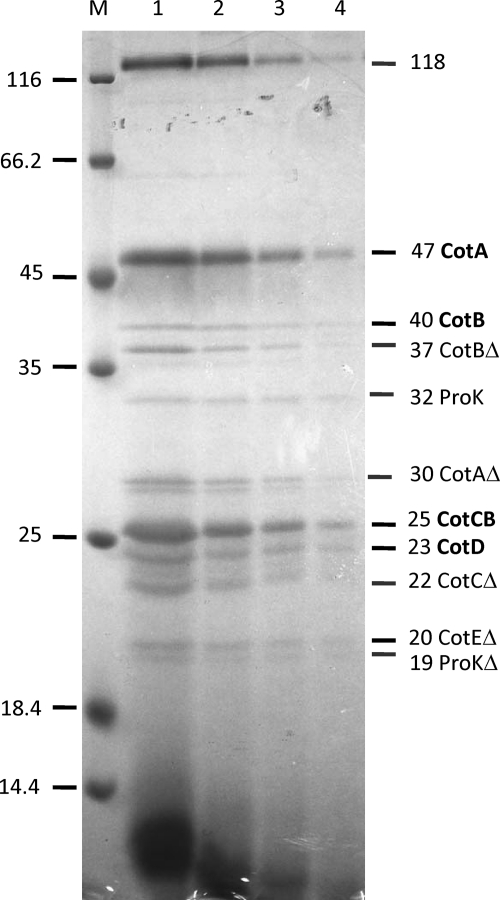
Coat proteins were extracted from freshly prepared spores of CD630 grown on solid medium using a sodium borate-SDS-DTT buffer and fractionated by SDS-PAGE (Fig. 2). Since spores were processed immediately, we reasoned that they may carry some residual exosporial material, as shown in Fig. 1C. Eleven protein bands were excised from Coomassie-stained gels and subjected to peptide mass fingerprinting using trypsin digestion and MALDI mass spectrometry. This analysis revealed that a number of protein bands corresponded to truncated breakdown products (Table 1). One high-molecular-mass species of 118 kDa could not be identified and may be an aggregate. Two further bands were chain E of proteinase K, which was a contaminant derived from the spore purification process. The remaining eight protein species corresponded to five different proteins, which we refer to as CotA, CotB, CotCB, CotD, and CotE, and we refer to their genes as cotA to cotE (Fig. 3), based on the nomenclature used for B. subtilis (13) (orthologues are shown in Fig. S2 in the supplemental material). CotA shared no homology with other proteins in existing databases, but CotB had orthologues in a number of bacilli and clostridia. CotCB and CotD were homologous with both each other (70% conserved residues) and manganese catalases, including the CotJC inner spore coat protein (and putative catalase) found in B. subtilis (see Fig. S3 in the supplemental material). As will be discussed below, the 25-kDa protein is most probably encoded by the second cistron of an operon, so we refer to the gene and protein as cotCB and CotCB, respectively. CotE, based on its amino acid sequence, corresponded to a novel bifunctional protein with amino-terminal peroxiredoxin (1-Cys peroxiredoxin) and carboxy-terminal manganese chitinase activities (Fig. S4). The predicted molecular mass of this protein was 81 kDa, although the full-length protein was not clearly discernible in our SDS-PAGE fractionations; rather, a 20-kDa truncated species was found. CotE had orthologues in a number of spore formers (Fig. S2). As a single bifunctional protein, no orthologues were found for other bacilli or clostridia, but matches were found with either the peroxiredoxin or chitinase domain carried in CotE. These included a putative peroxiredoxin, YkuU, in B. subtilis (BS938810) and a number of putative chitinases from exosporium-containing species, including Bacillus anthracis, B. cereus, Bacillus thuringiensis, Bacillus clausii, and Bacillus halodurans (Fig. S2).
Fig. 2.
Proteins extracted from CD630 spores. Proteins were extracted from CD630 spores (7-day-old cultures grown on solid medium) by using a sodium borate-SDS-DTT extraction buffer. Proteins were fractionated by SDS-PAGE (12.5% gel), and samples were loaded as dilutions. Lane 1, no dilution; lane 2, 1/2 dilution; lane 3, 1/4 dilution; lane 4, 1/8 dilution. M, markers. Alongside the gel, the identities of the bands excised and analyzed by mass spectrometry are shown. Partially truncated proteins (Δ) are indicated.
Table 1.
SDS-PAGE and MALDI peptide fingerprint analysis of C. difficile 630 spore coat proteins
| Fragment molecular mass (kDa) | Protein descriptiona | Coding sequenceb | Predicted molecular mass (kDa) | Assigned gene |
|---|---|---|---|---|
| 118 | ND | |||
| 47 | Hypothetical protein | CD1613 | 34 | cotA |
| 40 | Hypothetical protein | CD1511 | 35 | cotB |
| 37 | Hypothetical protein | CD1511 | 35 | cotB |
| 32 | Proteinase K (contaminant from purification steps) | |||
| 30 | Hypothetical protein | CD1613 | 34 | cotA |
| 25 | Putative spore coat protein; manganese catalase; similar to CotJC of B. subtilis | CD0598 | 21 | cotCB |
| 23 | Putative spore coat protein; manganese catalase; similar to CotJC of B. subtilis | CD2401 | 21 | cotD |
| 22 | Putative spore coat protein; manganese catalase; similar to CotJC of B. subtilis | CD0598 | 21 | cotCB |
| 20 | Putative bifunctional protein, peroxiredoxin/chitinase | CD1433 | 81 | cotE |
| 19 | Proteinase K (contaminant from purification steps) |
Based on peptide mass fingerprinting of tryptic digestions. ND, no determination.
Coding sequences are described in reference 30.
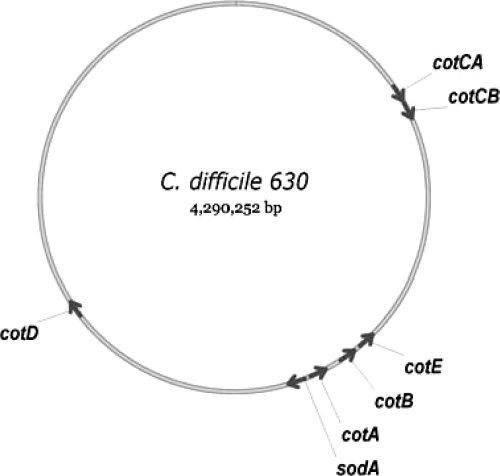
Fig. 3.
Spore coat genes. The chromosomal positions of genes referred to in this work are shown.
Immunoanalysis of spore coat proteins.
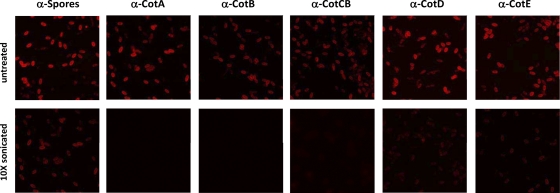
Polyclonal antibodies to recombinant Cot proteins were raised in mice. In the case of CotE, we used the amino-terminal peroxiredoxin domain of CotE to generate antibodies. Using confocal imaging of antibody-labeled C. difficile spores (purified after overnight incubation at 4°C), we observed uniform surface decoration using all antisera (Fig. 4), while naive serum gave no labeling (not shown). Since, as mentioned above, these spores lacked an exosporium, our data suggest that each of the five coat proteins must be exposed on the outermost layers of the spore and must be components of the spore coat rather than the exosporium.
Fig. 4.
Surface display of CotA, CotB, CotC, CotD, and CotE using confocal imaging of suspensions of CD630 spores (7-day-old cultures grown on solid medium) labeled with mouse serum (1:1,000 dilution) raised against each of the five Cot proteins. CD630 spores labeled with preimmune serum served as a control and showed no labeling (not shown). Spores labeled with antispore serum are also shown (“Spores”). An anti-mouse IgG-TRITC conjugate was used for secondary labeling. Images were taken by using a Nikon Eclipse fluorescence microscope equipped with a Bio-Rad Radiance 2100 laser scanning system (image size, 37 by 37 μm). The top row shows the labeling of untreated spores, and the bottom row shows the labeling of spores that had been sonicated 10 times.
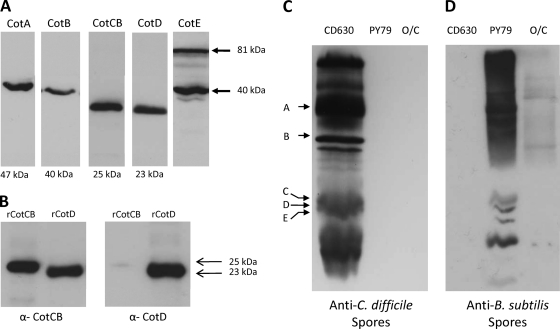
These antibodies were used in Western blot analyses to probe spore coat protein extractions (Fig. 5A). CotA, CotB, and CotD were present as single bands of 47, 40, and 23 kDa, respectively, corresponding to the predicted molecular masses of each of these proteins. CotE antisera identified two strongly reacting bands of 81 and 40 kDa, but the 20-kDa species (identified as a peptide fragment in Fig. 2) was not observed. The most likely explanation is that this 20-kDa species is a C-terminal fragment that is not recognized by the polyclonal CotE antibodies that were raised against the N terminus of CotE. For CotCB, when probed with anti-CotCB antibody, we could sometimes discern two bands of 25 and 23 kDa, although this is not apparent in Fig. 5A. Since CotCB and CotD were homologous, we wondered whether these proteins shared related epitopes. Using recombinant proteins (recombinant CotCB [rCotCB] and rCotD), we probed each one with anti-CotCB and anti-CotD sera. As shown in Fig. 5B, CotD was recognized by both anti-CotCB and anti-CotD sera. On the other hand, CotCB was detectable by using anti-CotCB antibodies but only very weakly by using anti-CotD antibody.
Fig. 5.
Immunoanalysis of spore coats. (A) Spore coats of CD630 were extracted, and separate lanes were probed with polyclonal (mouse) antibodies to CotA to CotE. Molecular masses of the relevant bands are shown. For CotE, two principal bands of 81 and 40 kDa were found. Serum from unimmunized mice did not react with C. difficile spore coat proteins. (B) Purified recombinant CotC and CotD proteins (2 μg) were fractionated on SDS-PAGE gels and probed with either CotC or CotD antibodies at 1/1,500 and 1/3,000 dilutions, respectively. Positions of the CotC (25 kDa) and CotD (23 kDa) bands are indicated. (C) Coat proteins extracted from spores of CD630, B. subtilis PY79, and B. clausii O/C were fractionated and probed with antiserum to formalin-inactivated CD630 spores. Positions of CotA to CotE are shown. (D) Same as panel C except that proteins were probed with antiserum to formalin-inactivated B. subtilis PY79 spores.
Using antisera raised against formalin-inactivated CD630 spores, we probed spore coat proteins extracted from CD630, B. subtilis, and B. clausii (Fig. 5C). C. difficile serum showed no cross-reaction against either B. subtilis or B. clausii spore coat proteins, the latter of which carries an exosporium (7, 13). Similarly, antiserum raised against formalin-inactivated B. subtilis spores showed no reaction against CD630 spores but some cross-reaction to B. clausii (Fig. 5D). These results support results from previously reported bioinformatic analyses that have shown little conservation between C. difficile and Bacillus spores (19, 30).
Effect of sonication on spore coat proteins.
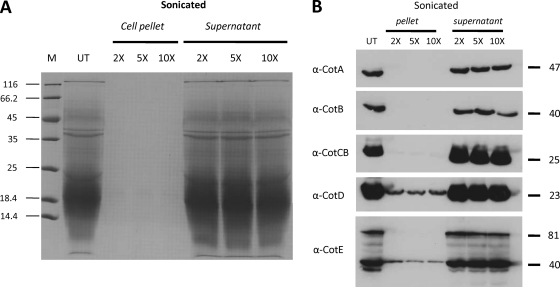
An exosporium has been documented for C. difficile spores prepared in liquid medium (19, 24). The exosporium remains the least-understood component of the bacterial endospore (13), and harsh physical methods, such as sonication and shear stress, have been reported to remove the exosporial layer (27). Although the majority of spores present in our preparations of CD630 did not appear to carry a recognizable exosporium, we subjected CD630 spores to repeated cycles of sonication. Spore pellets and supernatants were then separated. Pellets were then extracted with Na borate-SDS-DTT buffer, and extracts were run on 12.5% SDS-PAGE gels together with supernatant fractions (Fig. 6A). Our results showed that as few as two cycles of sonication were sufficient to remove almost the entire component of sodium borate-solubilized proteins, all of which were found in the supernatant fraction. Examination of the sonicated spores by phase-contrast microscopy revealed that phase-bright spores remained intact, and analyses of CFU before and after sonication demonstrated no change in viability. Analysis by TEM revealed no clear-cut differences between sonicated spores (Fig. 1C) and unsonicated spores (Fig. 1D). Although precise quantification was not possible, we observed that in sonicated samples, many spores carried angular projections on the surface layers (shown in Fig. 1D). These crystalline-like structures might indicate an underlying layer of coat resulting from sonication and the removal of one or more layers of coat material.
Fig. 6.
Removal of Cot proteins using sonication. (A) C. difficile spores were sonicated (30-s cycles) 2 times, 5 times, and 10 times, and pellets and supernatants were separated. Pellets were treated with Na borate-SDS-DTT extraction buffer, and extracted proteins were mixed with SDS-PAGE loading dye (4×) and fractionated by SDS-PAGE (12.5%). Supernatants were mixed with loading dye and run directly. (B) Spores were sonicated, and pellet and supernatant fractions (from panel A) were probed with antiserum to each of the five Cot proteins. Molecular masses of Cot proteins are shown. UT, untreated spores; M, markers.
Following sonication, the spore pellet and supernatant fractions were probed with antiserum to CotA to CotE (Fig. 6B). CotA, CotB, and CotCB were not detectable in the spore pellets and were found only in the supernatant fractions. CotD and CotE, although not visibly apparent in Coomassie-stained gels, were present in both the spore pellet and supernatant fractions by immunoanalysis. If CotD was still present in the spore coat fraction, then why was it not detected by use of anti-CotCB serum, since CotCB and CotD share related epitopes? We reason that although the recombinant proteins, at high concentrations, could be detected, this does not reflect the composition and abundance of CotCB and CotD in the spore coat but rather the different binding strengths and specificities of the respective antibodies. In the case of CotE, only the 40-kDa CotE fragment was found in the spore pellet fraction. We used confocal imaging to examine antibody-labeled sonicated spores (Fig. 4). This analysis revealed that in each case, following sonication, surface labeling was massively reduced. Labeling correlated well with the Western blotting data (Fig. 6B), with CotA, CotB, and CotCB showing almost no labeling and CotD and CotE weak labeling, suggesting that some CotD and CotE remained in the sonicated spores. These results then showed that all five Cot proteins are located on the spore surface and can be liberated either by sonication or by use of a sodium borate-SDS-DTT extraction buffer.
Enzymatic properties of spores.
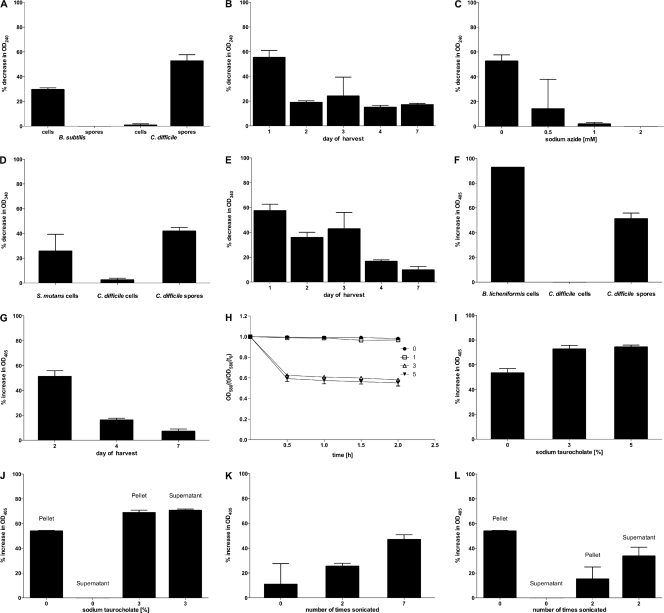
Based on the amino acid sequences of CotC, CotD, and CotE and their surface location, we predict that spores carry enzymatic activity, either latent or active. Accordingly, we conducted a number of assays to measure catalase, peroxiredoxin, and chitinase activities. In each case, we used suspensions of purified spores that had been checked microscopically to confirm greater than 99.99% free spores. Catalase activity was measured (Fig. 7A) by a photometric assay of H2O2 breakdown using suspensions of CD630 spores and vegetative cells and, as useful comparators, B. subtilis spores and vegetative cells. CD630 spores had noticeable catalase activity, while vegetative cells were completely negative. In comparison, B. subtilis spores were catalase negative, and vegetative cells were catalase positive. We next focused on CD630 spores, and we heated the spores at different temperatures for 20 min, allowed the spore suspension to return to an ambient temperature, and then conducted the catalase assay. We found that heating at 50°C had no effect on enzyme activity but that enzyme activity was reduced by 40% at 60°C and by 60% at 70°C, showing that although spores were heat stable, the enzymatic activity was not.
Fig. 7.
Enzymatic activities of C. difficile 630 spores. (A) Catalase activity in CD630 and B. subtilis spores or vegetative cells. (B) Catalase activities of CD630 spores at different stages of maturation on solid agar. (C) Inhibition of catalase activity by sodium azide. (D) Peroxiredoxin activity of CD630 spores and vegetative cells and S. mutans cells. (E) Peroxiredoxin activity of CD630 spores at different stages of maturation on solid agar. (F) chitinase activity in CD630 spores and vegetative cells of CD630 and B. licheniformis. (G) Chitinase activity of CD630 spores at different stages of maturation on solid agar. (H) Germination of CD630 spores in sodium taurocholate solutions. (I) Chitinase activity in response to spore germination using 3% or 5% sodium taurocholate solutions. (J) Chitinase activity obtained in cell pellet and supernatant fractions of CD630 spores following incubation with 0% and 3% sodium taurocholate for 30 min. (K) Chitinase activity of CD630 spores in response to sonication (30-s cycles). (L) Chitinase activity obtained in cell pellet and supernatant fractions of CD630 spores following sonication (30-s cycles).
The maturity of spores may affect spore-associated enzyme activity, since in other spore formers, notably B. subtilis, the spore coat physically changes over time, with the spore coat shrinking and forming distinctive surface corrugations (6). Spore suspensions were assessed for catalase activity at 1-day intervals postpurification, and we observed a marked decline in enzymatic activity after just 1 day of maturation (Fig. 7B). Since the substrate for catalase activity, H2O2, was the same as that used in the peroxiredoxin assay, we measured the effect of sodium azide on catalase activity, since catalase is sensitive to sodium azide, while peroxiredoxin activity is not (21). Using increasing concentrations of sodium azide, the catalase activity of CD630 spores was inhibited, demonstrating that we were measuring spore-associated catalase and not that of peroxiredoxin (Fig. 7C).
Peroxiredoxin activity was assessed by using CD630 spores and vegetative cells together with a suspension of S. mutans cells that are known to produce this enzyme (26). CD630 vegetative cells carried barely detectable levels of activity, while spores clearly were positive and had levels of activity equivalent to that of S. mutans (Fig. 7D). Peroxiredoxin activity exhibited a marked decline (60%) when spores were heated at above 60°C, and at 80°C, activity was abolished. The effects of spore maturity were also assessed, with activity gradually declining over time, with 7-day-old spores losing 48% of the activity exhibited by 1-day-old preparations (Fig. 7E). The peroxiredoxin activity of 1-day-old spores was measured in the presence of 0.5, 1, and 2 mM sodium azide, and no decline in activity was observed, indicating that the activity was that of peroxiredoxin and was not due to catalase.
The chitinase activities of CD630 spores and vegetative cells were assessed by using vegetative cells of chitinase-producing cells of B. licheniformis as a positive control (34). CD630 cells had no activity, but spores carried activity equivalent to that of B. licheniformis (Fig. 7F). As with catalase and peroxiredoxin activities, the age of the spores had a marked effect on spore-associated activity, with 7-day-old spores carrying 20% of the activity found for 2-day-old spores (Fig. 7G). Chitinase, as an enzyme involved in macromolecular degradation, would serve no obvious benefit to a dormant spore, but this would not be the case for a germinating spore. The release and subsequent activation of a latent enzyme could provide nutrients to an outgrowing cell, so we asked whether chitinase activity might be enhanced by the disruption of the spore coat and exosporium. We measured activity first following spore germination, which would rupture the spore coat, and second following the sonication of spores, which, as shown above, would remove the surface coat layers. The spore germination of 1-day-old spores was evaluated by using different solutions of sodium taurocholate as the germinant (38). By measuring the change in the OD580 attributed to the phase darkening of spores, we identified 3% and 5% sodium taurocholate as being optimal for spore germination, with a 38% reduction (at 3%) to a 50% reduction (at 5%) in the OD580 in 30 min (Fig. 7H). Next, 30 min following germination using 3% and 5% sodium taurocholate, we measured chitinase activity. We found that compared to untreated spores, the germinant produced a marked increase (21%) in chitinase activity with both 3% and 5% solutions (Fig. 7I). Using commercially obtained chitinase (catalog number C6242; Sigma) of the same type (family 18) as that predicted for CotE, we determined that sodium taurocholate had no effect on enzyme activity (data not shown). We also found that the chitinase activity was released into the medium following spore germination. Using 3% germinant, activity was clearly detectable in the supernatant fraction following centrifugation, in contrast to spore samples that had not been germinated (Fig. 7J). In support of this finding, we subjected suspensions of 7-day-old spores to increasing cycles of sonication. Seven 3-s bursts of sonication yielded more chitinase activity than two bursts, which in turn yielded more activity than untreated spores (Fig. 7K). As was the case during spore germination, sonication was sufficient to release chitinase activity into the medium (Fig. 7L). These results demonstrate first that catalase, peroxiredoxin, and chitinase activities are associated with spores and second that activity declines as spores mature, demonstrating that the enzyme either is not required or is rendered latent (dormant). Finally, for chitinase, activity is enhanced if the spore coat is disrupted.
DISCUSSION
This study has provided an initial examination of the spore coats of C. difficile spores with the identification of five proteins that are exposed on the outermost layer of the spore coat. We have named these proteins Cot and their genes cot, since our study clearly shows that they are located in the outermost layers of the spore coat. However, it should be noted that with two distinct structures, the coat and exosporium, found on the outermost layers of the spore, the assignment of names should be approached with caution. With the exception of an unidentified 118-kDa species, these five proteins represented the major proteins extractable by using the procedures followed here. We believe, however, that the coats of C. difficile are far more complex and that these five proteins represent just a fraction of the total protein content of the spore coat. We base this assumption on existing bioinformatic analyses and the extraction studies performed here. In B. subtilis, more than 70 proteins are thought to be found in the coat layers (13), and 18 orthologues have been identified in C. difficile (13, 19). Using anti-CD630 serum, no cross-reaction was found with B. subtilis spore coat proteins, suggesting that the functional composition and organization of the coat may be very different in C. difficile. Interestingly, recent spore proteome studies of CD630 (19) identified five potential spore coat proteins, which included CotCB and CotD but not CotA or CotB (CotE, as a chitinase, was identified but not as a spore coat bifunctional protein). Regarding the extraction procedures, our method was based on the use of sodium borate, SDS, and DTT, used previously for Clostridium perfringens (36) and shown here to efficiently extract five proteins. However, the presence of a number of truncated protein species suggests that this method may be overly harsh (it should be noted that the inclusion of a proteinase K treatment step might contribute to this observed partial degradation of some proteins). It is also possible that these products arise from cleavage reactions occurring during spore maturation. SDS-PAGE analysis revealed that the sonication of spores was able to remove all five Cot proteins recovered by extraction with buffer. Interestingly, though, residual spores, which were still viable, released no additional protein when extracted with borate-SDS-DTT buffer. Sonicated spores, when examined by TEM, showed that the spore coat layers were still essentially intact, suggesting that sonication removes one component (possibly one or more layers of coat) of the spore coat and that the remaining underlying coat is impervious to further extraction or sonication. We believe that the deciphering of the inner layers of the spore coat will require the development of new extraction procedures. One additional structure of the spore is the exosporium, although note that this should not be considered part of the spore coat per se. We have shown here that C. difficile spores do carry an exosporium, but at best, this is loosely attached to the spore, and it is possible that the stability of the exosporium is linked to the conditions required to prepare (e.g., solid versus liquid medium) and/or store spores. In comparison, the exosporium of B. anthracis spores is reasonably well characterized, yet there are conflicting reports regarding how stable this structure is, with recent studies suggesting that the exosporium is not easily removed by either sonication or shear stress (35). If the C. difficile exosporium is particularly fragile, then what, if any, is the biological significance? Since our initial extraction and identification of proteins were made with spores carrying some exosporial material, we believe that one or more exosporial proteins may have been recovered and perhaps were present in low abundance in the protein extractions shown in Fig. 2. One candidate could be the collagen-rich glycoprotein BclA1 (CD0332), which has orthologues in a number of other spore formers, including B. anthracis, where it forms filaments that are attached to the exosporium and facilitates interactions with host cells, including enhancing spore uptake by macrophages (2). Interestingly, BclA is extracted from exosporium-containing Bacillus spores as a high-molecular-mass species, and its identification using conventional proteomic tools is problematic (5, 33). It will therefore be of interest to determine whether the 118-kDa species is in fact BclA.
Some of the most interesting findings of this work are the enzymatic properties of the spores and the identification of three enzymatic coat proteins (CotCB, CotD, and CotE) that most probably reside in the exosporium. Although absolute confirmation will require the inactivation of the chromosomal genes and, preferably, evidence of the enzyme activity of the purified proteins, this assumption is supported by several lines of evidence. First, vegetative cells were shown to exhibit no enzyme activity, so this is unlikely to arise from any contaminating cells. Second, a previous analysis of the spore proteome (19) revealed no additional genes that could encode these enzyme activities, although it must be emphasized that the spore proteome is incomplete.
What, then, are the functions of these putative spore-associated enzymes? The catalase (CotCB and CotD) and peroxiredoxin (CotE) activities are potential antioxidants, and at first glance, all three would reduce the cellular toxicity of H2O2 by conversion to oxygen and water. In the case of C. difficile, which is a strict anaerobe, the presence of oxygen would in turn be harmful to the cell. Since the cell is irreversibly committed to dormancy, it is conceivable that this is not actually harmful and that C. difficile spores can be maintained in an oxic environment. Presumably, though, there is a need to remove H2O2. Previous studies of B. subtilis sporulation showed that H2O2 may play a key role in spore coat synthesis and could serve as a substrate for the oxidative cross-linking of spore coat monomers (12). Here, the enzyme superoxide dismutase (SodA) is essential to the cross-linking of tyrosine-rich spore coat proteins, and in CD630, a manganese-dependent SodA orthologue has been identified in the spore proteome (CD1631). CotE, as a 1-Cys-peroxiredoxin would be expected to have the same enzymatic activity as that of a peroxidase and could participate in the cross-linking of tyrosine-rich spore coat proteins. None of the other coat proteins identified in this work are tyrosine rich, but an examination of the C. difficile genome has revealed at least one gene (CD0597) that would encode a tyrosine-rich protein (10.34% tyrosines). This protein is homologous to CotJB of B. subtilis, and in C. difficile, its ORF lies immediately upstream of cotC, which in turn encodes an orthologue of B. subtilis CotJC. The ORFs are separated by 61 bp and probably lie within the same operon, so we propose to name CD0597 cotCA and the downstream cistron cotCB (Fig. 2B).
For chitinase activity, the presence of this enzyme in the spore coat is intriguing, since it would be expected to be involved in the breakdown of fungi and other biological matter whether in the soil or in the intestine. However, spores are dormant, so we speculate that chitinase activity may be released (or activated) during spore germination, enabling a potential source of nutrients as the C. difficile cell emerges from its coats. We have evidence to support this: the chitinase activity decreased as spores matured but increased during both spore germination and following sonication, with both of these being events that would rupture the spore coat layers. Interestingly, CotE (the putative chitinase) was detectable in the supernatant fraction as either a full-length species (81 kDa) or a single 40-kDa species following sonication, and we wonder whether the smaller species is actually the active chitinase enzyme. Another interesting aspect of CotE, based solely on its sequence prediction, is its bifunctionality and its characterization as one of a growing number of “moonlighting proteins” (15) that carry multiple functions, including a mammalian protein, 1-Cys-peroxiredoxin, that carries peroxidase and phospholipase activities (4). There is possibly a more important consequence of a chitinase and a peroxiredoxin displayed on the surface of C. difficile spores that should not be overlooked. This relates to the potential link between peroxiredoxins, chitinases, and inflammation. Peroxiredoxin 1 (a 2-Cys-peroxiredoxin), secreted from tumor cells (22), was shown previously to induce proinflammatory cytokines in macrophages via interactions with Toll-like receptor 4 and to promote chronic inflammation, which could support tumor growth (28). Regarding chitinases, it is now clear that some inflammatory conditions of the gastrointestinal (GI) tract (inflammatory bowel disease [IBD] and ulcerative colitis [UC]) lead to the induction of host cell chitinases by triggering the increased uptake of intracellular bacteria by colonic cells (16, 17) and in potentiating the development of epithelial tumorigenesis (8). Considering that some symptoms of CDAD resemble those of both IBD and UC, C. difficile chitinase may play a direct role in infection and not simply in macromolecular degradation.
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 23 September 2011.
REFERENCES
- 1. Beers R. F., Jr., Sizer I. W. 1952. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J. Biol. Chem. 195:133–140 [PubMed] [Google Scholar]
- 2. Bozue J., et al. 2007. Bacillus anthracis spores of the bclA mutant exhibit increased adherence to epithelial cells, fibroblasts, and endothelial cells but not to macrophages. Infect. Immun. 75:4498–4505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carter G. P., Rood J. I., Lyras D. 2010. The role of toxin A and toxin B in Clostridium difficile-associated disease: past and present perspectives. Gut Microbes 1:58–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen J. W., Dodia C., Feinstein S. I., Jain M. K., Fisher A. B. 2000. 1-Cys peroxiredoxin, a bifunctional enzyme with glutathione peroxidase and phospholipase A2 activities. J. Biol. Chem. 275:28421–28427 [DOI] [PubMed] [Google Scholar]
- 5. Daubenspeck J. M., et al. 2004. Novel oligosaccharide side chains of the collagen-like region of BclA, the major glycoprotein of the Bacillus anthracis exosporium. J. Biol. Chem. 279:30945–30953 [DOI] [PubMed] [Google Scholar]
- 6. Driks A. 2003. The dynamic spore. Proc. Natl. Acad. Sci. U. S. A. 100:3007–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Driks A. 2007. Surface appendages of bacterial spores. Mol. Microbiol. 63:623–625 [DOI] [PubMed] [Google Scholar]
- 8. Eurich K., Segawa M., Toei-Shimizu S., Mizoguchi E. 2009. Potential role of chitinase 3-like-1 in inflammation-associated carcinogenic changes of epithelial cells. World J. Gastroenterol. 15:5249–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gerding D. N., Muto C. A., Owens R. C., Jr 2008. Measures to control and prevent Clostridium difficile infection. Clin. Infect. Dis. 46(Suppl. 1):S43–S49 [DOI] [PubMed] [Google Scholar]
- 10. Gerding D. N., Muto C. A., Owens R. C., Jr 2008. Treatment of Clostridium difficile infection. Clin. Infect. Dis. 46(Suppl. 1):S32–S42 [DOI] [PubMed] [Google Scholar]
- 11. Greetham D., Grant C. M. 2009. Antioxidant activity of the yeast mitochondrial one-Cys peroxiredoxin is dependent on thioredoxin reductase and glutathione in vivo. Mol. Cell. Biol. 29:3229–3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Henriques A. O., Melsen L. R., Moran C. P., Jr 1998. Involvement of superoxide dismutase in spore coat assembly in Bacillus subtilis. J. Bacteriol. 180:2285–2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Henriques A. O., Moran C. P., Jr 2007. Structure, assembly, and function of the spore surface layers. Annu. Rev. Microbiol. 61:555–588 [DOI] [PubMed] [Google Scholar]
- 14. Hong H. A., et al. 2009. Bacillus subtilis isolated from the human gastrointestinal tract. Res. Microbiol. 160:134–143 [DOI] [PubMed] [Google Scholar]
- 15. Jeffery C. J. 1999. Moonlighting proteins. Trends Biochem. Sci. 24:8–11 [DOI] [PubMed] [Google Scholar]
- 16. Kawada M., et al. 2008. Chitinase 3-like-1 enhances bacterial adhesion to colonic epithelial cells through the interaction with bacterial chitin-binding protein. Lab. Invest. 88:883–895 [DOI] [PubMed] [Google Scholar]
- 17. Kawada M., Hachiya Y., Arihiro A., Mizoguchi E. 2007. Role of mammalian chitinases in inflammatory conditions. Keio J. Med. 56:21–27 [DOI] [PubMed] [Google Scholar]
- 18. Lawley T. D., et al. 2009. Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect. Immun. 77:3661–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lawley T. D., et al. 2009. Proteomic and genomic characterization of highly infectious Clostridium difficile 630 spores. J. Bacteriol. 191:5377–5386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Logan C., Mayhew S. G. 2000. Cloning, overexpression, and characterization of peroxiredoxin and NADH peroxiredoxin reductase from Thermus aquaticus. J. Biol. Chem. 275:30019–30028 [DOI] [PubMed] [Google Scholar]
- 21. Low F. M., Hampton M. B., Peskin A. V., Winterbourn C. C. 2007. Peroxiredoxin 2 functions as a noncatalytic scavenger of low-level hydrogen peroxide in the erythrocyte. Blood 109:2611–2617 [DOI] [PubMed] [Google Scholar]
- 22. Neumann C. A., et al. 2003. Essential role for the peroxiredoxin Prdx1 in erythrocyte antioxidant defence and tumour suppression. Nature 424:561–565 [DOI] [PubMed] [Google Scholar]
- 23. Nicholson W. L., Setlow P. 1990. Sporulation, germination and outgrowth, p. 391–450 In Harwood C. R., Cutting S. M. (ed.), Molecular biological methods for Bacillus. John Wiley & Sons Ltd., Chichester, United Kingdom [Google Scholar]
- 24. Panessa-Warren B. J., Tortora G. T., Warren J. B. 1997. Exosporial membrane plasticity of Clostridium sporogenes and Clostridium difficile. Tissue Cell 29:449–461 [DOI] [PubMed] [Google Scholar]
- 25. Paredes-Sabja D., Bond C., Carman R. J., Setlow P., Sarker M. R. 2008. Germination of spores of Clostridium difficile strains, including isolates from a hospital outbreak of Clostridium difficile-associated disease (CDAD). Microbiology 154:2241–2250 [DOI] [PubMed] [Google Scholar]
- 26. Poole L. B., Higuchi M., Shimada M., Calzi M. L., Kamio Y. 2000. Streptococcus mutans H2O2-forming NADH oxidase is an alkyl hydroperoxide reductase protein. Free Radic. Biol. Med. 28:108–120 [DOI] [PubMed] [Google Scholar]
- 27. Redmond C., Baillie L. W., Hibbs S., Moir A. J., Moir A. 2004. Identification of proteins in the exosporium of Bacillus anthracis. Microbiology 150:355–363 [DOI] [PubMed] [Google Scholar]
- 28. Riddell J. R., Wang X. Y., Minderman H., Gollnick S. O. 2010. Peroxiredoxin 1 stimulates secretion of proinflammatory cytokines by binding to TLR4. J. Immunol. 184:1022–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rupnik M., Wilcox M. H., Gerding D. N. 2009. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 7:526–536 [DOI] [PubMed] [Google Scholar]
- 30. Sebaihia M., et al. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38:779–786 [DOI] [PubMed] [Google Scholar]
- 31. Smith C. J., Markowitz S. M., Macrina F. L. 1981. Transferable tetracycline resistance in Clostridium difficile. Antimicrob. Agents Chemother. 19:997–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Songer J. G., Anderson M. A. 2006. Clostridium difficile: an important pathogen of food animals. Anaerobe 12:1–4 [DOI] [PubMed] [Google Scholar]
- 33. Sylvestre P., Couture-Tosi E., Mock M. 2002. A collagen-like surface glycoprotein is a structural component of the Bacillus anthracis exosporium. Mol. Microbiol. 45:169–178 [DOI] [PubMed] [Google Scholar]
- 34. Tharisman A., Suhartono M. T., Spindler-Barth M., Hwang J.-K., Pyun Y.-R. 2005. Purification and characterization of a thermostable chitinase form Bacillus licheniformis Mb-2. World J. Microbiol. Biotechnol. 21:733–738 [Google Scholar]
- 35. Thompson B. M., Binkley J. M., Stewart G. C. 2011. Current physical and SDS extraction methods do not efficiently remove exosporium proteins from Bacillus anthracis spores. J. Microbiol. Methods 85:143–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tsuzuki T., Ando Y. 1985. Chemical studies on the spore coat protein of Clostridium perfringens type A. Agric. Biol. Chem. 49:3221–3225 [Google Scholar]
- 37. Vonberg R. P., et al. 2008. Infection control measures to limit the spread of Clostridium difficile. Clin. Microbiol. Infect. 14(Suppl. 5):2–20 [DOI] [PubMed] [Google Scholar]
- 38. Wilson K. H., Kennedy M. J., Fekety F. R. 1982. Use of sodium taurocholate to enhance spore recovery on a medium selective for Clostridium difficile. J. Clin. Microbiol. 15:443–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.