Abstract
The widely conserved phage shock protein (Psp) extracytoplasmic stress response has been studied extensively in Escherichia coli and Yersinia enterocolitica. Both species have the PspF, -A, -B, and -C proteins, which have been linked to robust phenotypes, including Y. enterocolitica virulence. PspB and PspC are cytoplasmic membrane proteins required for stress-dependent induction of psp gene expression and for bacterial survival during the mislocalization of outer membrane secretin proteins. Previously, we reported that Y. enterocolitica PspB functions to positively control the amount of PspC by an uncharacterized posttranscriptional mechanism. In this study, we have discovered that the cytoplasmic membrane protease FtsH is involved in this phenomenon. FtsH destabilizes PspC in Y. enterocolitica, but coproduction of PspC with its binding partner PspB was sufficient to prevent this destabilization. In contrast, FtsH did not affect any other core component of the Psp system. These data suggested that uncomplexed PspC might be particularly deleterious to the bacterial cell and that FtsH acts as an important quality control mechanism to remove it. This was supported by the observation that toxicity caused by PspC production was reduced either by coproduction of PspB or by increased synthesis of FtsH. We also found that the phenomenon of FtsH-dependent PspC destabilization is conserved between Y. enterocolitica and E. coli.
INTRODUCTION
The phage shock protein (Psp) system is a highly conserved extracytoplasmic stress response triggered by events likely to compromise the integrity of the cytoplasmic membrane (reviewed in references 5, 20, and 35). The system has been studied extensively in the Gram-negative bacteria Escherichia coli and Yersinia enterocolitica, but homologues of some of its components are also present in many Gram-negative/positive bacteria, as well as archaea and plants (e.g., see references 3, 29, 43, and 45). The Psp system is required for the virulence of Y. enterocolitica and Salmonella enterica serovar Typhimurium (6, 23) and for biofilm formation in E. coli (2), and its production is highly induced during macrophage infection by S. enterica serovar Typhimurium and Shigella flexneri (10, 30).
The complement of Psp proteins differs between species, but E. coli and Y. enterocolitica each have PspF, -A, -B, and -C. Removing any of these causes a robust phenotype, meaning that they can perhaps be considered the core Psp system in these two species. PspF is a transcription factor that binds to the pspABC control region and activates its σ54-dependent promoter (12, 22). The PspA, -B, and -C proteins form a putative signal transduction pathway that modulates PspF activity. The integral cytoplasmic membrane proteins PspB and PspC respond to Psp-inducing stress by causing the sequestration of PspA to the cytoplasmic membrane (46). This presumably prevents PspA from forming an inhibitory complex with PspF in the cytoplasm (8, 9).
In addition to their regulatory roles, the increase in PspABC concentration after an inducing trigger is encountered reflects the fact that these proteins also have physiological roles in mediating stress tolerance. PspA has been linked to maintaining the proton motive force in E. coli (e.g., see references 26 and 27). In Y. enterocolitica, PspB and PspC (but not PspA) are essential for survival when outer membrane secretin proteins mislocalize within the cell envelope, which is a potent and specific Psp-inducing trigger (15, 32). Secretin sensitivity explains why a pspC null mutant is sensitive to native Ysc type 3 secretion system production and also avirulent in mice (6).
The critical roles of PspB and PspC have motivated us to investigate their function in Y. enterocolitica. During a genetic investigation, we reported that the steady-state concentration of PspC was higher when PspB was present, even when the genes were expressed from a non-psp promoter (15). We speculated that this phenomenon involved PspB protecting PspC from proteolysis, consistent with the fact that these two proteins interact in vivo (14, 32). In E. coli, the essential cytoplasmic membrane protein FtsH is a protease with several known targets (19). These targets include the integral cytoplasmic membrane proteins SecY and AtpB when they are produced in the absence of their normal binding partners (1, 24). Therefore, FtsH represented a promising candidate to destabilize PspC in the absence of its binding partner, PspB.
In this study, we tested the above hypothesis. Our data reveal that FtsH destabilizes PspC in Y. enterocolitica and that coproduction with PspB is sufficient to prevent this. FtsH does not affect any other core component of the Psp system. Therefore, we speculated that uncomplexed PspC is deleterious to the bacterial cell and that FtsH acts as a quality control mechanism to rapidly remove it. Consistent with this, toxicity caused by PspC production could be reduced by PspB coproduction or by increased production of FtsH. Our studies also indicate that the phenomenon of FtsH-mediated PspC destabilization is conserved between Y. enterocolitica and E. coli.
MATERIALS AND METHODS
Bacterial strains, plasmids, and routine growth.
Strains and plasmids are listed in Table 1. Primer sequences will be supplied upon request (please contact the corresponding author). PCR-generated fragments were verified by DNA sequencing. Strains were routinely grown in Luria-Bertani (LB) broth or on LB agar plates (34). Antibiotics were used as previously described (31).
Table 1.
Strains and plasmids
| Strain or plasmid | Genotype/features | Reference or source |
|---|---|---|
| Bacterial strains | ||
| E. coli K-12 | ||
| MG1655 | F−rph-1 | 16 |
| AR3289 | F− IN(rrnD-rrnE)1 sfhC21 zad220::Tn10 | 40 |
| AR3291 | F− IN(rrnD-rrnE)1 sfhC21 zad220::Tn10 ΔftsH3::kan | 40 |
| Y. enterocolitica | ||
| AJD3a | ΔyenR (r− m+) | 25 |
| AJD1171 | ΔyenR (r− m+) Δ(pspF-ycjF) ΔpspG | 32 |
| YVM859 | ΔyenR (r− m+) ΔpspBC | 32 |
| AJD4466b | ΔyenR (r− m+) ΔftsH [pAJD2105] | This study |
| AJD4490b | ΔyenR (r− m+) ΔpspBC ΔftsH [pAJD2105] | This study |
| AJD4625b | ΔyenR (r− m+) Δ(pspF-ycjF) ΔpspG ΔftsH [pAJD2105] | This study |
| Plasmids | ||
| pBAD18-Kan | Kmr ColE1 ori, araBp expression vector | 17 |
| pBAD33 | Cmr p15A ori, araBp expression vector | 17 |
| pSR47S | Kmr R6K ori, mob+ (RP4) sacB+ | 33 |
| pVLT35 | Smr Spr RSF1010 ori, tacp expression vector | 7 |
| pWSK29 | Apr pSC101 ori, lacZp expression vector | 44 |
| pAJD267 | araBp-pspF+ in pBAD18-Kan | 6 |
| pAJD268 | araBp-pspA+ in pBAD18-Kan | This study |
| pAJD1011 | araBp-pspBC+ in pBAD33 | 32 |
| pAJD1014 | araBp-pspBC+ in pBAD18-Kan | This study |
| pAJD1041 | araBp-pspB+ in pBAD33 | 32 |
| pAJD1085 | araBp-ΔpspB pspC+ in pBAD33 | This study |
| pAJD2065 | lacZp-ΔpspB pspC+ in pWSK29 | This study |
| pAJD2105 | tacp-ftsH+ in pVLT35 | This study |
| pAJD2138 | araBp-pspB+ in pBAD18-Kan | This study |
| pAJD2139 | araBp-ΔpspB pspC+ in pBAD18-Kan | This study |
| pAJD2142c | lacZp-ΔpspBEC pspCEC+ in pWSK29 | This study |
AJD3 is a virulence plasmid-cured derivative of strain JB580v (25). All other Y. enterocolitica strains listed are derivatives of AJD3.
Y. enterocolitica ΔftsH strains contained pAJD2105 and required IPTG for viability.
pAJD2142 insert is E. coli DNA. All other plasmid inserts are Y. enterocolitica DNA.
Polyclonal antisera and immunoblotting.
Lysates derived from equivalent amounts of bacterial cells (determined by optical density of cultures) were separated by SDS-PAGE on gels containing 12.5 to 15% polyacrylamide and then transferred to nitrocellulose by electroblotting. Equal loading was confirmed by total protein staining of the nitrocellulose with Ponceau S. Enhanced chemiluminescent detection followed sequential incubation with a diluted polyclonal antiserum followed by goat anti-rabbit IgG horseradish peroxidase conjugate (Bio-Rad) used at 1 in 5,000. Dilutions of previously described polyclonal antisera were 1 in 20,000 for anti-PspA and anti-PspF (46), 1 in 20,000 to 500,000 for anti-PspB (15), 1 in 5,000 to 40,000 for anti-PspC (32), and 1 in 10,000 for anti-FtsH (46). These antisera had been raised against Y. enterocolitica antigens but were also able to recognize the corresponding E. coli proteins.
Strain and plasmid constructions.
tacp-ftsH expression plasmid pAJD2105 was constructed by amplifying the ftsH gene from Y. enterocolitica genomic DNA and cloning it into plasmid pVLT35. araBp (pBAD33 and pBAD18-Kan derivatives) and lacZp (pWSK29 derivative) expression plasmids carrying Y. enterocolitica psp genes were constructed by transferring inserts from previously described plasmids (6, 15, 32) using standard cloning procedures. pWSK29 carrying E. coli ΔpspB pspC+ was made by amplifying the pspB pspC region from the chromosome of E. coli strain MG1655 using an upstream primer that introduced an in-frame deletion within pspB by loop-out mutagenesis and a second primer that annealed immediately downstream of pspC. The primers incorporated SacI (upstream) and XbaI (downstream) sites, which were used to clone the fragment into pWSK29. The design of this E. coli ΔpspB pspC+ plasmid was analogous to the previously described expression plasmids with ΔpspB pspC+ and pspBC+ inserts from Y. enterocolitica (15). Including the pspB in-frame deletion ensures that both plasmids maintain the overlapping stop and start codons between the pspB and pspC open reading frames and the putative ribosome binding sites upstream of each gene. This avoids potential translational polarity effects on pspC expression, which is important when comparing PspC production from pspC+ and pspBC+ plasmids. The pspBC genes are arranged similarly in the genomes of Y. enterocolitica and E. coli.
The Y. enterocolitica ΔftsH in-frame deletion mutation was made using the sacB+ suicide plasmid pSR47S; plasmid construction and mutagenesis procedures were identical to those described previously (43a). Mutagenesis was done in strains containing the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible ftsH+ expression plasmid pAJD2105, which were grown in the presence of 50 μM IPTG. Deletion of the chromosomal ftsH gene was confirmed by colony PCR analysis and an IPTG-dependent growth phenotype.
Determination of Y. enterocolitica FtsH-dependent growth.
Strains containing empty tacp expression plasmid pVLT35 or the derivative encoding FtsH (pAJD2105) were grown to saturation (optical density at 600 nm, >3) at 26°C in LB broth containing 1 μM IPTG. Four microliters of undiluted and serial 10-fold dilutions (10−1 to 10−7) was spotted onto LB agar containing appropriate antibiotics with or without 50 μM IPTG. Plates were incubated at 26°C for 30 h.
Measurement of PspC steady-state level and in vivo degradation in E. coli.
Saturated cultures were diluted into 6 ml of LB broth (+1 mM IPTG to induce lacZp-pspC expression) in 18-mm-diameter test tubes to an optical density (600 nm) of 0.1. The cultures were grown on a roller drum at 37°C for 3 h. Then, translation was blocked by adding 100 μg/ml chloramphenicol and incubation continued at 37°C. Samples for immunoblot analysis were harvested immediately before and 2 h after chloramphenicol addition.
FtsH depletion/overproduction and measurement of Psp protein steady-state level and in vivo degradation in Y. enterocolitica.
ΔftsH strains containing tacp-ftsH plasmid pAJD2105 were grown to saturation in LB broth plus 1 μM IPTG. These cultures were diluted into 5 ml of LB broth without IPTG in 18-mm-diameter test tubes to an optical density (600 nm) of 0.15. The cultures were grown on a roller drum at 26°C for 4 to 6 h. Overproduction of FtsH was achieved by including 50 to 100 μM IPTG in the growth medium. For experiments to measure PspB and PspC steady-state levels produced from araBp-pspB/C expression plasmids (pBAD33 derivatives), the growth medium contained 0.01% arabinose.
To monitor PspC degradation over time, strains contained araBp-pspC expression plasmid pAJD2139 (pBAD18-Kan derivative). Saturated cultures were diluted into 50 ml of LB broth in 125-ml flasks to an optical density (600 nm) of 0.15. The cultures were shaken at 200 rpm and 26°C for 4 h. pspC expression was then induced by addition of 0.02% arabinose. Thirty minutes after arabinose addition, translation was blocked by adding 100 μg/ml chloramphenicol. Samples were taken at different time points for immunoblot analysis. Experiments to compare PspA, -B, -C, and -F stabilities were done similarly except that after chloramphenicol addition a single sample was taken for analysis after 1 h.
Determination of the effect of FtsH overproduction on PspC-dependent toxicity.
Saturated cultures were diluted into 5.5 ml of LB broth, containing 0.05% arabinose to induce pspB/C expression and 50 μM IPTG to induce ftsH expression, in 18-mm-diameter test tubes so that the initial optical density (600 nm) was approximately 0.2. The cultures were grown on a roller drum at 26°C for 8 h, and 0.1-ml samples were removed at hourly intervals for optical density measurement. At the 4-h time point, a 1-ml sample was removed for immunoblot analysis.
RESULTS
Y. enterocolitica and E. coli PspC proteins are stabilized in an E. coli ΔftsH mutant.
We reported previously that PspB elevated the PspC protein concentration in Y. enterocolitica via an uncharacterized posttranscriptional mechanism (15). A likely explanation is that PspB protects PspC from proteolysis. This would be analogous to the E. coli integral cytoplasmic membrane proteins SecY and AtpB, which are degraded by the FtsH protease in the absence of their normal binding partners (1, 24). FtsH has not been studied in any Yersinia species. Therefore, as a preliminary test for a link between FtsH and PspC, we took advantage of E. coli ftsH+ and ΔftsH strains. Both strains have the sfhC21 allele of fabZ, which suppresses the lethality caused by loss of FtsH (37), so that the only genetic difference between them is their ftsH+ versus ΔftsH genotype.
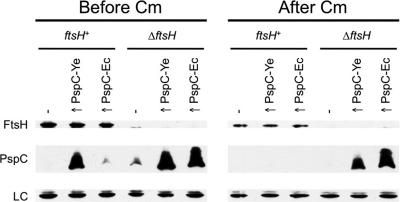
Plasmids with Y. enterocolitica or E. coli pspC genes expressed from the lacZ promoter were introduced into the E. coli ftsH+ and ΔftsH strains to overproduce PspC relative to the native E. coli Psp proteins encoded on the chromosome. Overproduced E. coli PspC was barely detectable in the ftsH+ strain but was abundant in the ΔftsH mutant (Fig. 1, left panel). In fact, even the endogenous E. coli PspC was detectable in the ΔftsH strain but not in its ftsH+ parent (compare empty vector lanes in the left panel of Fig. 1). The steady-state level of overproduced Y. enterocolitica PspC was perhaps only marginally higher in the ΔftsH mutant (Fig. 1). This might be because Y. enterocolitica PspC is an imperfect substrate for E. coli FtsH (although a trivial explanation would be different efficiencies of the two expression plasmids). However, when translation was inhibited with chloramphenicol, both the overproduced Y. enterocolitica and E. coli PspC proteins were eliminated from the ftsH+ strain but remained abundant in the ΔftsH mutant for at least 2 h (Fig. 1, right panel).
Fig. 1.
Overproduced Y. enterocolitica and E. coli PspC proteins are stabilized in an E. coli ΔftsH mutant. E. coli strain AR3289 (ftsH+) and its isogenic ΔftsH derivative AR3291 (ΔftsH) contained empty lacZp expression plasmid pWSK29 (−) or derivatives encoding Y. enterocolitica PspC (↑ PspC-Ye) or E. coli PspC (↑ PspC-Ec). Cells were grown at 37°C for 3 h (Before Cm). Chloramphenicol was then added to block translation, followed by incubation for a further 2 h at 37°C (After Cm). Cell lysates were separated by SDS-PAGE and analyzed by anti-FtsH and anti-PspC immunoblotting. An unidentified E. coli protein that cross-reacted with the anti-FtsH serum served as a convenient loading control (LC).
These data indicated that E. coli FtsH destabilized both the E. coli and Y. enterocolitica PspC proteins. Therefore, we were motivated to proceed with an investigation into the role of FtsH in our model organism, Y. enterocolitica.
FtsH is essential in Y. enterocolitica.
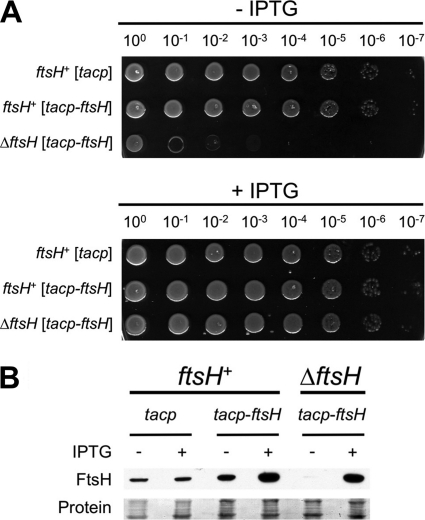
The Y. enterocolitica ftsH gene (originally annotated as YE0428) encodes a protein that is 92% identical to E. coli K-12 FtsH and is in a similar genomic context (data not shown). Predictions indicate that the E. coli and Y. enterocolitica FtsH proteins are both 647 amino acids in length, including a 26-amino-acid N-terminal Sec-dependent signal sequence, and that both ftsH genes have a TTG initiation codon (data not shown). FtsH is essential in E. coli, and we anticipated that this might be the case in Y. enterocolitica. Therefore, we used a standard sacB+ suicide vector approach to delete Y. enterocolitica ftsH in a strain with a tacp-ftsH complementation plasmid. Growth of the resulting ΔftsH mutant was dependent on tacp-ftsH expression, which confirmed that the gene is essential in Y. enterocolitica (Fig. 2A). Anti-FtsH immunoblot analysis revealed successful depletion of the FtsH protein when IPTG was omitted from the growth medium (Fig. 2B; note that a low level of FtsH protein was detectable upon prolonged exposure). These experiments were done at 26°C, which is the optimum growth temperature for Y. enterocolitica in the laboratory. At 37°C, depletion of FtsH was less efficient (data not shown), perhaps because the tacp promoter is more leaky at this temperature. Furthermore, overexpression of ftsH was toxic at 37°C, which made it impossible to find conditions under which the tacp-ftsH plasmid fully complemented the growth defect. Attempts to use an araBp-ftsH plasmid were also unsuccessful. Therefore, for our subsequent experiments we used the tacp-ftsH plasmid and a 26°C growth temperature, which allowed successful depletion or overproduction of FtsH and full restoration to a wild-type growth phenotype (Fig. 2). Importantly, Y. enterocolitica psp gene expression is regulated similarly at 26°C and 37°C and psp null strains are sensitive to secretin production at both temperatures (data not shown). Therefore, the Psp system apparently functions similarly under both temperature conditions.
Fig. 2.
FtsH depletion prevents growth of Y. enterocolitica. (A) Growth phenotypes of ftsH+ and ΔftsH strains. ftsH+ and ΔftsH strains contained empty tacp expression plasmid pVLT35 (tacp) or the derivative encoding FtsH (tacp-ftsH). Serial dilutions of saturated cultures were spotted onto LB agar with or without 50 μM IPTG and incubated at 26°C for 30 h. (B) Anti-FtsH immunoblot analysis of total cell lysates from the strains used in panel A. “Protein” indicates a Ponceau S stain of the nitrocellulose membrane used for immunoblot analysis (the region containing FtsH).
PspB prevents FtsH-dependent degradation of PspC in Y. enterocolitica.
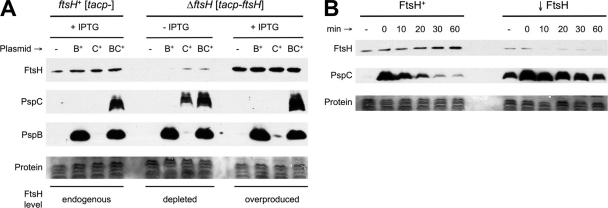
Expression of pspB and/or pspC from a plasmid promoter that it does not control (lacZp) suggested that PspB might stabilize PspC in Y. enterocolitica (15). To determine the involvement of FtsH in this phenomenon, a similar set of pBAD33 derivative araBp-pspB/C plasmids was introduced into ΔpspBC ftsH+ and ΔpspBC ΔftsH strains containing empty tacp expression plasmid pVLT35 or the tacp-ftsH derivative, respectively. These strains were grown with or without IPTG to achieve endogenous, depleted, or overproduced levels of FtsH (Fig. 3A). araBp-pspB/C expression was induced with arabinose, and immunoblot assays were used to compare steady-state levels of PspB and PspC.
Fig. 3.
FtsH-dependent degradation of PspC in Y. enterocolitica. (A) FtsH depletion increases PspC steady-state level when PspB is absent. ΔpspBC ftsH+ and ΔpspBC ΔftsH strains contained empty tacp expression plasmid pVLT35 (tacp-) or the derivative encoding FtsH (tacp-ftsH), respectively. Strains also contained empty araBp expression plasmid pBAD33 (−) or derivatives encoding PspB (B+), PspC (C+), or PspBC (BC+). Strains were grown with or without 100 μM IPTG to induce tacp-ftsH expression, and cell lysates were analyzed by anti-FtsH, anti-PspC, and anti-PspB immunoblotting. “Protein” indicates a Ponceau S stain of the nitrocellulose membrane used for immunoblot analysis (the region containing PspC). (B) PspC degradation assay. ΔpspBC ftsH+ (FtsH+) and ΔpspBC ΔftsH (↓FtsH) strains contained tacp-ftsH plasmid pAJD2105 but were grown without IPTG to repress its expression. araBp-pspC expression from pAJD2139 was induced for 30 min by addition of 0.02% arabinose, after which translation was blocked by adding chloramphenicol. Samples were taken at different times for immunoblot analysis. “−” is a sample taken before inducing araBp-pspC expression. Subsequent samples were taken at the times indicated in minutes after adding chloramphenicol.
With endogenous or overproduced FtsH, the PspC protein was detected only when it was coproduced with PspB (Fig. 3A), consistent with the previous result (15). However, when FtsH was depleted, PspC was readily detected in the absence of PspB. This suggests that PspC is subject to FtsH-dependent degradation in Y. enterocolitica and that PspB can prevent this degradation, even when FtsH is overproduced.
Next, we did a PspC degradation assay in the absence of PspB as described in Materials and Methods. For this experiment, we used a pBAD18-Kan derivative encoding only PspC. The higher copy number than that of pBAD33 allowed detection of PspC in an ftsH+ strain, and its kanamycin resistance allowed chloramphenicol to be used as a translation inhibitor. The results clearly indicated that depletion of FtsH significantly stabilized the PspC protein (Fig. 3B). Integrated density analysis of the PspC immunoblot signals with ImageJ software (http://rsb.info.nih.gov/ij) suggested that the half-life of PspC was extended from approximately 12 to 50 min upon FtsH depletion. Interestingly, in this experiment PspC was easily detected in the FtsH depletion strain even without arabinose addition (Fig. 3B). This is presumably due to a combination of leakiness of the araBp promoter and reduced proteolytic turnover of the PspC protein when FtsH was depleted.
PspC is the only core component of the Psp system affected by FtsH.
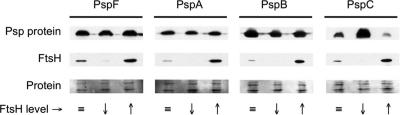
Next, we tested whether FtsH destabilized any other core component of the Psp system. This was done in ftsH+ and ΔftsH strains with all chromosomal psp genes deleted and plasmids with individual psp genes expressed from the araB promoter (pBAD18-Kan derivatives). Deletion of the chromosomal psp genes eliminated potential complications caused by their differential expression in response to the various araBp-psp expression plasmids.
After translation was blocked with chloramphenicol, immunoblot analysis revealed that the stability of the PspF, PspA, and PspB proteins was unaffected by the FtsH status of the cell (Fig. 4). This indicates that FtsH does not affect any of these Psp proteins, even when FtsH is overproduced and all other Psp proteins are absent. In contrast, and consistent with the earlier results, depletion of FtsH stabilized PspC whereas FtsH overproduction destabilized it (Fig. 4). However, as before, coproduction of PspB and PspC together in this Δ(pspF-ycjF) ΔpspG strain protected PspC from FtsH destabilization (data not shown). Therefore, among all of the Psp regulon proteins, PspB is both necessary and sufficient for this protective effect.
Fig. 4.
PspC is the only core component of the Psp system affected by FtsH in Y. enterocolitica. Strains with a Δ(pspF-ycjF) ΔpspG genotype contained araBp expression plasmids (pBAD18-Kan derivatives) encoding PspF, PspA, PspB, or PspC as indicated at the top. All strains also contained tacp-ftsH expression plasmid pAJD2105. Approximately endogenous FtsH levels ( ) were achieved using an ftsH+ strain grown without IPTG. FtsH depletion (↓) was achieved using an ΔftsH strain grown without IPTG. FtsH overproduction (↑) was achieved using an ΔftsH strain grown with 100 μM IPTG. Psp protein production was induced for 30 min by addition of 0.02% arabinose, after which translation was blocked by adding chloramphenicol. Samples were harvested 1 h after chloramphenicol addition, and cell lysates were analyzed by anti-FtsH and anti-PspF, anti-PspA, anti-PspB, or anti-PspC immunoblotting as appropriate. “Protein” indicates a Ponceau S stain of the nitrocellulose membrane used for immunoblot analysis (the region containing FtsH).
) were achieved using an ftsH+ strain grown without IPTG. FtsH depletion (↓) was achieved using an ΔftsH strain grown without IPTG. FtsH overproduction (↑) was achieved using an ΔftsH strain grown with 100 μM IPTG. Psp protein production was induced for 30 min by addition of 0.02% arabinose, after which translation was blocked by adding chloramphenicol. Samples were harvested 1 h after chloramphenicol addition, and cell lysates were analyzed by anti-FtsH and anti-PspF, anti-PspA, anti-PspB, or anti-PspC immunoblotting as appropriate. “Protein” indicates a Ponceau S stain of the nitrocellulose membrane used for immunoblot analysis (the region containing FtsH).
PspC-dependent toxicity is alleviated by increased FtsH synthesis or by coproduction of PspB.
The preceding experiments indicated that among the PspF, -A, -B, and -C proteins only PspC is subject to FtsH-dependent degradation. Furthermore, this occurs detectably only when PspC is made in the absence of its binding partner PspB. Therefore, we hypothesized that uncomplexed PspC might be particularly deleterious to the bacterial cell so that a specific FtsH-dependent mechanism has evolved to eliminate it. If this hypothesis was correct, we reasoned that conditions should be found where production of PspC is toxic and that this toxicity should be alleviated in one of two ways: first, by coproducing PspB, which would presumably allow formation of the normal and nontoxic PspBC complex, and alternatively, by increasing the synthesis of FtsH, which would more rapidly eliminate the abnormal, uncomplexed, and toxic PspC protein.
Again, these experiments were done in strains with all chromosomal psp genes deleted in order to eliminate complications caused by their differential expression. For example, in a previous study an araBp-pspBC plasmid caused overexpression of chromosomal psp genes whereas araBp-pspB and araBp-pspC plasmids did not (32). Each strain contained empty araBp expression plasmid pBAD18-Kan or pspB+ and/or pspC+ derivatives, together with either empty tacp expression plasmid pVLT35 or the ftsH+ derivative. All strains were grown with arabinose and IPTG, and optical density was monitored over time.
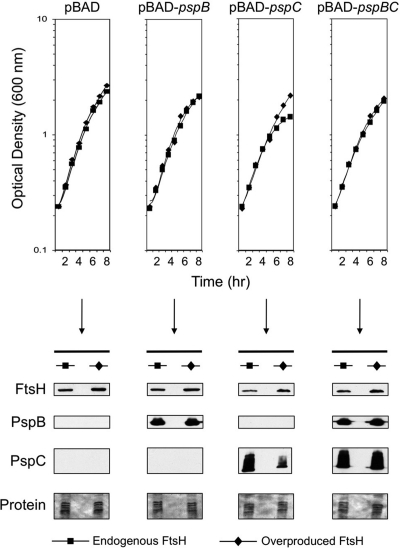
Consistent with our hypothesis, araBp-pspC expression was toxic (reduced growth yield by approximately 35%) whereas araBp-pspB was not (Fig. 5, top). However, the toxicity caused by pspC expression was abolished either by FtsH overproduction or by coexpression of pspB. Immunoblot analysis confirmed that FtsH overproduction decreased PspC concentration whereas coproduction with PspB did not (Fig. 5, bottom). Therefore, the mechanism by which each alleviates PspC-dependent toxicity is different. This is fully consistent with FtsH eliminating toxic, uncomplexed PspC and with PspB promoting the formation of a stable, nontoxic PspBC complex.
Fig. 5.
PspC-dependent toxicity is alleviated by increased FtsH synthesis or by coproduction of PspB in Y. enterocolitica. Δ(pspF-ycjF) ΔpspG ftsH+ strains contained the empty pVLT35 vector (Endogenous FtsH) or the derivative encoding FtsH (Overproduced FtsH). Strains also contained either the empty pBAD18-Kan vector (pBAD) or derivatives expressing pspB, pspC, or pspBC as indicated at the top. The strains were grown as described in Materials and Methods, and optical density was measured at hourly intervals. At the 4-h time point, samples were removed for the anti-FtsH, anti-PspB, and anti-PspC immunoblot analysis shown at the bottom. “Protein” indicates a Ponceau S stain of the nitrocellulose membrane used for immunoblot analysis (the region containing PspC).
DISCUSSION
This work has revealed a link between the phage shock protein system and the FtsH protease. FtsH destabilizes PspC produced in excess relative to its binding partner, PspB. The most likely explanation is that uncomplexed PspC is a degradation substrate of FtsH, although formal confirmation of that will require the establishment of an in vitro system to study PspBC. This would add PspC to the list of FtsH substrates, identified primarily in E. coli, which includes both integral cytoplasmic membrane and soluble cytoplasmic proteins (reviewed in references 19 and 36). Notably, the situation with PspC is strikingly analogous to the E. coli SecY and AtpB cytoplasmic membrane proteins, which are degraded by FtsH when produced in the absence of, or in excess over, their normal binding partners (1, 24).
SecY and AtpB function within complexes involved in moving proteins or protons, respectively, across the cytoplasmic membrane. It has been suggested that their unregulated subreactions, which might occur if they fail to assemble into their normal complexes, could be highly deleterious by compromising the permeability barrier (1). As such, FtsH-dependent degradation serves as an important quality control mechanism. Consistent with these ideas, AtpB overproduction is toxic (11, 42) and E. coli ftsH mutants are sensitized to SecY overproduction (24). Similarly, PspC overproduction was toxic, but increased FtsH production, or PspB coproduction, alleviated this (Fig. 5). Perhaps the PspBC complex might also be involved in a transmembrane conductance function such that uncontrolled PspC activity can compromise the permeability barrier. Experiments in E. coli support part of this idea. In ΔpspF E. coli cells (i.e., with an uninducible Psp regulon), PspC overproduction decreases the proton motive force, but this does not happen when PspB and PspC are coproduced (21).
To our knowledge, this is the first dedicated investigation of FtsH function in any Yersinia species, and it has revealed a lot of similarity to E. coli. The FtsH proteins are predicted to be the same length, 92% identical, and translated from a TTG initiation codon. They also share at least one probable common substrate, PspC (Fig. 1), and FtsH is essential in both species. FtsH is essential in E. coli because one of its substrates, LpxC, catalyzes a committed step in the synthesis of lipopolysaccharide (LPS) (37). In the absence of FtsH, LpxC accumulation causes a lethal imbalance between phospholipid and LPS. An L85P substitution in FabZ (the sfhC21 allele), which is involved in phospholipid biosynthesis, restores the balance and suppresses ΔftsH lethality (37). We do not know if similar phenomena occur in Y. enterocolitica. lpxC (YE0678) and fabZ (YE3273) are conserved, and the predicted FabZ protein has leucine at position 85. Recent work showed that E. coli FtsH can degrade LpxC proteins from various species, including Yersinia pseudotuberculosis (28). Removal rather than depletion of FtsH might facilitate our Y. enterocolitica studies, and so we have attempted to use sacB suicide vector technology to exchange the wild-type fabZ gene for one encoding FabZ L85P (in an ftsH+ strain). However, we could isolate wild-type segregants only from a fabZ (wild-type)/fabZ (L85P) merodiploid (S. Singh and A. J. Darwin, unpublished data). This might indicate that the L85P substitution is lethal in Y. enterocolitica, although further work will be needed to unequivocally confirm or deny that.
A link between FtsH and the Psp response fits well with several other connections between FtsH function and stress response reported in E. coli. First, ftsH expression is partially dependent on the alternate sigma factor σ32 (RpoH), which controls the cytoplasmic heat shock response, and σ32 itself is also an FtsH degradation substrate (18). Second, FtsH might be involved in shutting off the oxidative stress response by degrading the transcription factor SoxS (13). Third, compromised FtsH function leads to induction of the Cpx and RpoE extracytoplasmic responses, probably due to the proposed role of FtsH in quality control of aberrant membrane proteins (38).
We have detected FtsH-dependent PspC destabilization in Y. enterocolitica when PspC is produced in excess of PspB. Similarly, E. coli AtpB has been shown to be degraded by FtsH only when produced in excess of its binding partners (1). For some time, SecY was also thought to be degraded by FtsH only when produced in excess of its binding partner, SecE. However, it has now been found that FtsH degrades SecY when aberrant proteins block Sec translocation complexes, which might be part of a defense mechanism against such events (41). Therefore, we speculate that a situation probably arises where FtsH targeting of endogenous PspC occurs and is important. In fact, some data already support this. In an E. coli ΔftsH strain, we detected the endogenous PspC protein but not in the ftsH+ parental strain (Fig. 1). Although small, this effect is consistent with destabilization of endogenous PspC by FtsH (but cause and effect are hard to establish; see below). We did not observe this in Y. enterocolitica, but that might be because we only depleted FtsH whereas in the E. coli ΔftsH strain it is absent. FtsH-dependent degradation of σ32 and SoxS in E. coli is implicated in shutoff of the cytoplasmic heat shock and oxidative stress responses, respectively. Similarly, PspC degradation could influence shutoff of the Psp response because PspC positively controls psp gene expression. We have found that depletion of FtsH in Y. enterocolitica can slightly increase basal-level Φ(pspA-lacZ) operon fusion expression (S. Singh and A. J. Darwin, unpublished data). However, compromised FtsH function might itself cause membrane stress, at least in E. coli (38), which could explain any elevated psp gene expression. Thus, we cannot precisely establish cause and effect for this phenomenon. Similarly, extended experiments to examine effects of FtsH depletion on Psp response shutoff would be complicated by the membrane stress and eventual growth arrest caused by the FtsH depletion itself.
A 1999 publication suggested that loss of FtsH in E. coli deactivated σ54 activity by an unknown mechanism, abolishing expression of σ54-dependent promoters, including pspAp (4). Such a phenomenon would not impact our work with pspC expressed from the lacZp and araBp promoters. Furthermore, as mentioned above, FtsH depletion in Y. enterocolitica does not reduce basal Φ(pspA-lacZ) expression, nor does it prevent secretin-dependent induction (S. Singh and A. J. Darwin, unpublished data). Other work has argued against global loss of σ54 activity in an E. coli ΔftsH strain (39).
In summary, this work has uncovered a role for FtsH in destabilizing a component of the Psp stress response. This further solidifies connections between FtsH function and the bacterial cell envelope, with FtsH being linked to the biosynthesis of cell membrane components (LPS and phospholipids), quality control of integral membrane proteins, and the functioning of bacterial cell envelope stress responses.
ACKNOWLEDGMENTS
We thank Teru Ogura (Kumamoto University, Japan) for providing E. coli strains AR3289 and AR3291.
This study was supported by award number R01AI052148 from the National Institute of Allergy and Infectious Diseases (NIAID). A.J.D. holds an Investigators in Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIAID or the National Institutes of Health.
Footnotes
Published ahead of print on 30 September 2011.
REFERENCES
- 1. Akiyama Y., Kihara A., Ito K. 1996. Subunit a of proton ATPase F0 sector is a substrate of the FtsH protease in Escherichia coli. FEBS Lett. 399:26–28 [DOI] [PubMed] [Google Scholar]
- 2. Beloin C., et al. 2004. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51:659–674 [DOI] [PubMed] [Google Scholar]
- 3. Bidle K. A., Kirkland P. A., Nannen J. L., Maupin-Furlow J. A. 2008. Proteomic analysis of Haloferax volcanii reveals salinity-mediated regulation of the stress response protein PspA. Microbiology 154:1436–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carmona M., de Lorenzo V. 1999. Involvement of the FtsH (HflB) protease in the activity of σ54 promoters. Mol. Microbiol. 31:261–270 [DOI] [PubMed] [Google Scholar]
- 5. Darwin A. J. 2005. The phage-shock-protein response. Mol. Microbiol. 57:621–628 [DOI] [PubMed] [Google Scholar]
- 6. Darwin A. J., Miller V. L. 2001. The psp locus of Yersinia enterocolitica is required for virulence and for growth in vitro when the Ysc type III secretion system is produced. Mol. Microbiol. 39:429–444 [DOI] [PubMed] [Google Scholar]
- 7. de Lorenzo V., Eltis L., Kessler B., Timmis K. N. 1993. Analysis of Pseudomonas gene products using lacIqPtrp-lac plasmids and transposons that confer conditional phenotypes. Gene 123:17–24 [DOI] [PubMed] [Google Scholar]
- 8. Dworkin J., Jovanovic G., Model P. 2000. The PspA protein of Escherichia coli is a negative regulator of σ54-dependent transcription. J. Bacteriol. 182:311–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Elderkin S., Jones S., Schumacher J., Studholme D., Buck M. 2002. Mechanism of action of the Escherichia coli phage shock protein PspA in repression of the AAA family transcription factor PspF. J. Mol. Biol. 320:23–37 [DOI] [PubMed] [Google Scholar]
- 10. Eriksson S., Lucchini S., Thompson A., Rhen M., Hinton J. C. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103–118 [DOI] [PubMed] [Google Scholar]
- 11. Eya S., Maeda M., Tomochika K., Kanemasa Y., Futai M. 1989. Overproduction of truncated subunit a of H+-ATPase causes growth inhibition of Escherichia coli. J. Bacteriol. 171:6853–6858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Green R. C., Darwin A. J. 2004. PspG, a new member of the Yersinia enterocolitica phage shock protein regulon. J. Bacteriol. 186:4910–4920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Griffith K. L., Shah I. M., Wolf R. E., Jr 2004. Proteolytic degradation of Escherichia coli transcription activators SoxS and MarA as the mechanism for reversing the induction of the superoxide (SoxRS) and multiple antibiotic resistance (Mar) regulons. Mol. Microbiol. 51:1801–1816 [DOI] [PubMed] [Google Scholar]
- 14. Gueguen E., Flores-Kim J., Darwin A. J. 2011. The Yersinia enterocolitica phage shock proteins B and C can form homodimers and heterodimers in vivo with the possibility of close association between multiple domains. J. Bacteriol. 193:5747–5758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gueguen E., Savitzky D. C., Darwin A. J. 2009. Analysis of the Yersinia enterocolitica PspBC proteins defines functional domains, essential amino acids and new roles within the phage-shock-protein response. Mol. Microbiol. 74:619–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guyer M. S., Reed R. R., Steitz J. A., Low K. B. 1981. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harbor Symp. Quant. Biol. 45:135–140 [DOI] [PubMed] [Google Scholar]
- 17. Guzman L., Belin D., Carson M. J., Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Herman C., Thevenet D., D'Ari R., Bouloc P. 1995. Degradation of σ32, the heat shock regulator in Escherichia coli, is governed by HflB. Proc. Natl. Acad. Sci. U. S. A. 92:3516–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ito K., Akiyama Y. 2005. Cellular functions, mechanism of action, and regulation of FtsH protease. Annu. Rev. Microbiol. 59:211–231 [DOI] [PubMed] [Google Scholar]
- 20. Joly N., et al. 2010. Managing membrane stress: the phage shock protein (Psp) response, from molecular mechanisms to physiology. FEMS Microbiol. Rev. 34:797–827 [DOI] [PubMed] [Google Scholar]
- 21. Jovanovic G., Engl C., Mayhew A. J., Burrows P. C., Buck M. 2010. Properties of the phage shock protein (Psp) regulatory complex that govern signal transduction and induction of the Psp response in Escherichia coli. Microbiology 156:2920–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jovanovic G., Weiner L., Model P. 1996. Identification, nucleotide sequence, and characterization of PspF, the transcriptional activator of the Escherichia coli stress-induced psp operon. J. Bacteriol. 178:1936–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Karlinsey J. E., Maguire M. E., Becker L. A., Crouch M. L., Fang F. C. 2010. The phage shock protein PspA facilitates divalent metal transport and is required for virulence of Salmonella enterica sv. Typhimurium. Mol. Microbiol. 78:669–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kihara A., Akiyama Y., Ito K. 1995. FtsH is required for proteolytic elimination of uncomplexed forms of SecY, an essential protein translocase subunit. Proc. Natl. Acad. Sci. U. S. A. 92:4532–4536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kinder S. A., Badger J. L., Bryant G. O., Pepe J. C., Miller V. L. 1993. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O:8 and construction of a transformable R−M+ mutant. Gene 136:271–275 [DOI] [PubMed] [Google Scholar]
- 26. Kleerebezem M., Crielaard W., Tommassen J. 1996. Involvement of stress protein PspA (phage shock protein A) of Escherichia coli in maintenance of the protonmotive force under stress conditions. EMBO J. 15:162–171 [PMC free article] [PubMed] [Google Scholar]
- 27. Kobayashi R., Suzuki T., Yoshida M. 2007. Escherichia coli phage-shock protein A (PspA) binds to membrane phospholipids and repairs proton leakage of the damaged membranes. Mol. Microbiol. 66:100–109 [DOI] [PubMed] [Google Scholar]
- 28. Langklotz S., Schakermann M., Narberhaus F. 2011. Control of lipopolysaccharide biosynthesis by FtsH-mediated proteolysis of LpxC is conserved in enterobacteria but not in all Gram-negative bacteria. J. Bacteriol. 193:1090–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu C., et al. 2007. The chloroplast HSP70B-CDJ2-CGE1 chaperones catalyse assembly and disassembly of VIPP1 oligomers in Chlamydomonas. Plant J. 50:265–277 [DOI] [PubMed] [Google Scholar]
- 30. Lucchini S., Liu H., Jin Q., Hinton J. C., Yu J. 2005. Transcriptional adaptation of Shigella flexneri during infection of macrophages and epithelial cells: insights into the strategies of a cytosolic bacterial pathogen. Infect. Immun. 73:88–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maxson M. E., Darwin A. J. 2004. Identification of inducers of the Yersinia enterocolitica phage shock protein system and comparison to the regulation of the RpoE and Cpx extracytoplasmic stress responses. J. Bacteriol. 186:4199–4208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Maxson M. E., Darwin A. J. 2006. PspB and PspC of Yersinia enterocolitica are dual function proteins: regulators and effectors of the phage-shock-protein response. Mol. Microbiol. 59:1610–1623 [DOI] [PubMed] [Google Scholar]
- 33. Merriam J. J., Mathur R., Maxfield-Boumil R., Isberg R. R. 1997. Analysis of the Legionella pneumophila fliI gene: intracellular growth of a defined mutant defective for flagellum biosynthesis. Infect. Immun. 65:2497–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Miller J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 35. Model P., Jovanovic G., Dworkin J. 1997. The Escherichia coli phage-shock-protein (psp) operon. Mol. Microbiol. 24:255–261 [DOI] [PubMed] [Google Scholar]
- 36. Narberhaus F., Obrist M., Fuhrer F., Langklotz S. 2009. Degradation of cytoplasmic substrates by FtsH, a membrane-anchored protease with many talents. Res. Microbiol. 160:652–659 [DOI] [PubMed] [Google Scholar]
- 37. Ogura T., et al. 1999. Balanced biosynthesis of major membrane components through regulated degradation of the committed enzyme of lipid A biosynthesis by the AAA protease FtsH (HflB) in Escherichia coli. Mol. Microbiol. 31:833–844 [DOI] [PubMed] [Google Scholar]
- 38. Shimohata N., Chiba S., Saikawa N., Ito K., Akiyama Y. 2002. The Cpx stress response system of Escherichia coli senses plasma membrane proteins and controls HtpX, a membrane protease with a cytosolic active site. Genes Cells 7:653–662 [DOI] [PubMed] [Google Scholar]
- 39. Sze C. C., Bernardo L. M., Shingler V. 2002. Integration of global regulation of two aromatic-responsive σ54-dependent systems: a common phenotype by different mechanisms. J. Bacteriol. 184:760–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tatsuta T., et al. 1998. Heat shock regulation in the ftsH null mutant of Escherichia coli: dissection of stability and activity control mechanisms of σ32 in vivo. Mol. Microbiol. 30:583–593 [DOI] [PubMed] [Google Scholar]
- 41. van Stelten J., Silva F., Belin D., Silhavy T. J. 2009. Effects of antibiotics and a proto-oncogene homolog on destruction of protein translocator SecY. Science 325:753–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. von Meyenburg K., Jorgensen B. B., Michelsen O., Sorensen L., McCarthy J. E. 1985. Proton conduction by subunit a of the membrane-bound ATP synthase of Escherichia coli revealed after induced overproduction. EMBO J. 4:2357–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vrancken K., Van Mellaert L., Anne J. 2008. Characterization of the Streptomyces lividans PspA response. J. Bacteriol. 190:3475–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43a. Walker K. A., Miller V. L. 2004. Regulation of the Ysa type III secretion system of Yersinia enterocolitica by YsaE/SycB and YsrS/YsrR. J. Bacteriol. 186:4056–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang R. F., Kushner S. R. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195–199 [PubMed] [Google Scholar]
- 45. Wolf D., et al. 2010. In-depth profiling of the LiaR response of Bacillus subtilis. J. Bacteriol. 192:4680–4693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yamaguchi S., Gueguen E., Horstman N. K., Darwin A. J. 2010. Membrane association of PspA depends on activation of the phage-shock-protein response in Yersinia enterocolitica. Mol. Microbiol. 78:429–443 [DOI] [PMC free article] [PubMed] [Google Scholar]